Molecular Evaluation of Class 1 Integron in Drug-Resistant Shigella Strains Isolated from Clinical Samples
Received: 28-Dec-2022 / Manuscript No. CMB-22-84958 / Editor assigned: 30-Dec-2022 / PreQC No. CMB-22-84958(PQ) / Reviewed: 13-Jan-2023 / QC No. CMB-22-84958 / Revised: 18-Jan-2023 / Manuscript No. CMB-22-84958(R) / Accepted Date: 20-Jan-2023 / Published Date: 25-Jan-2023 DOI: 10.4172/1165-158X.1000251
Abstract
Background: The aim of this study was molecular investigation of class 1 integron in drug-resistant Shigella strains isolated from clinical samples.
Materials and Methods: 118 samples were collected from the West of Iran. The antibiotic susceptibility, the pattern of antibiotic resistance and investigation the int1 gene was conducted.
Results: From 118 samples taken, 27.12% samples were confirmed as Shigella. Also, 62.5, 21.875, 9.375 and 6.25% as S. sonnei, S. flexneri, S. boydii and S. dysenteria were known, respectively. The highest sensitivity was related to carbapenem and tetracycline (97%) and Shigella has the most resistant to ampicillin (94%). Also, 14 samples (44.8% of the isolates) were found to be positive for the presence of integrase gene. This association was significant for S. flexneri and integron 1 (P = 0.0001).
Conclusion: Carbapenem and tetracycline were the most effective antibiotics. There was also a significant association between integrase 1 gene and MDR strain (P = .00001).
Keywords: Int1 gene; Shigella; Antibiotic Resistance; Carbapenem
Keywords
Int1 gene; Shigella; Antibiotic Resistance; Carbapenem
Introduction
Class 1 and 2 integrons have the highest prevalence in Gramnegative bacteria [1]. The structure of class 1 integron consists of 5' and 3' protected regions and a variable region containing gene cassettes.
Majority of the previous studies have shown that class 2 integron consists of the same arrays of 4 gene cassettes containing 3 antibiotic resistance gene cassettes (tfrA1, sat, aadA1) including trimethoprim, streptothricin and spectinomycin/streptomycin resistance as well as the orfX gene cassette with unknown function [2].
Integrons have an important proven role in disseminating resistance among pathogenic gram-negative bacteria, therefore, they are considered as an indicator of antibiotic resistance [3]. In Gramnegative bacteria, resistance to ampicillin is primarily related to beta-lactamase enzymes, which break down the beta-lactam ring and inactivate the antibiotic [1]. So far, various beta-lactamases have been described, but the predominant types exist in Gram-negative bacteria [3]. Oral mecillinam and cephalosporins are used to treat Shigella infection, but with increasing extended-spectrum B-lactamases [EBLs] and plasmid of AmpC B-lactamase treatment of Shigella infections using these antibiotics has become more difficult [3, 4]. ESBLs are enzymes responsible for the resistance to beta-lactam antibiotics such as ceftriaxone, cefotaxime [CTX], and aztreonam, and are found in Gram-negative organisms [5-8].
Although Shigella strains are resistant to various antibiotics, only a few of them produce ESBLs, such as CTX-M-14 [9], SHV-11 [10] and OXA [11, 12].
The aim of the present study was to investigate the molecular nature of class 1 integron in drug-resistant Shigella strains isolated from the clinical samples in Sanandaj City, Kurdistan.
In this cross-sectional descriptive study, 118 clinical stool samples from the patients in the West of the country were collected and reviewed while maintaining the ethical principles and patient satisfaction. The main criteria for entering the laboratory diagnosis included macroscopic examination for the blood clots, mucus and stool consistency, as well as microscopic examination by preparing a thin smear of the stool and performing Gram staining and white blood cells observation. The predominance of a bacterial morphology, the presence of yeast cells or the absence of intestinal Gram-negative bacilli in the faces were also determined. Stool sampling was performed in the early stages of the disease, before the start of antibiotic treatment.
Materials and Method
Cultivation of the samples
As faces cause Shigella death due to its acidic nature, the sample should be cultured immediately. The swab samples were first placed in Gram-negative (GN) broth medium and discarded after transferring faces to the culture medium. After 8 hr incubation at 37°C, a loop was taken from the surface of the broth without mixing and cultured on XLD [xylose lysine deoxycholate] and McConkie agar. The plates were then incubated at 37°C for 24 hr. The incubation was continued for up to 48 hr if no growth or poor growth occurred.
Phenotypic evaluation and identification
After incubation, the growth rate, shape and color of the colonies were examined and recorded in each culture media. Shigella colonies on the McConkey medium were seen as lactose-negative [colourless] or the same color as the medium with a diameter of 2-3 mm and on the XLD medium were seen as red or pink without a black center with a diameter of 1-2 mm.
Pure culture preparation
Grown and confirmed colonies as well as suspected ones on McConkey and XLD media were re-cultured and purified on blood agar medium and used to continue the research. This bacterium forms Gray colonies on the surface of the blood agar medium.
A completely isolated colony suspected of Shigella was carefully selected from the 24-hr blood agar plate. The wire loop was contacted to the center of the colony, and then a suspension was prepared in 1 ml of the sterile physiology serum and used for all the biochemical tests.
Differential and biochemical tests to confirm Shigella samples TSI Agar, SIM, urea test, LIA, Simon Citrate, MR-VP sugar fermentation tests were performed for phenotypic investigations on all samples.
Preparation of the glycerol containing culture medium for long-term storage
In order to perform additional tests on bacterial samples, the bacteria must be kept in proper condition. Brain Heart Broth [BHB]- based culture medium was used for this purpose. Therefore, 7.4 gr BHB powder was dissolved in 200 ml distilled water. It was then boiled and 15% glycerol was added to the medium and shaken well to obtain a uniform solution. Then, it was aliquated into microtubes in small amounts of 1-2 ml. Then, it was autoclaved at 121ºC, 15 atm pressure for 15 min. After the autoclave, the Shigella isolates were inoculated into the medium and stored at -70°C.
Evaluation of bacterial susceptibility to antibiotics [Antibiogram]
Agar disk diffusion method was used to determine the antibiotic resistance pattern. Discs were placed on Müller Hinton agar medium [Merck, Germany] with half McFarland concentration at 2.5 cm distances. Then, the plate was incubated at 37°C for 18-24 hr. Next, the plates were examined under the lamp and the non-growth halo diameter was measured with ruler. The antibiogram test report for each antibiotic was determined as sensitive, resistant and intermediate.
Molecular experiments
DNA extraction and PCR: Bacterial culture [fresh culture] was used for the extraction. The bacterial DNA extraction was performed by conventional boiling method. Then, the presence of DNA was confirmed, measuring the OD at 260 nm wavelength and running the gel electrophoresis. Sinagene PCR kit was used according to the instructions.
The studied primers were obtained from previous studies and confirmed doing BLAST (Table 1). PCR products were electrophoresed on 2% agarose gel along with the control sample. The size and quality of the amplified fragments were confirmed with a 100 bp marker size.
| Name | Secence Sence | Target | Amplicon Size |
|---|---|---|---|
| intI F intI R | 5'-CAGTGGACATAAGCCTGTTC-3' 5'-CCCGAGGGCATAGACTGTA -3' |
5- intI gene 3-intI gene |
160-bp |
Table 1: Sequence of primers.
Statistical analysis
Statistical analysis was performed using SPSS v.19.0 statistical software. Chi-square test and Fisher's exact test (in cases where the frequencies were less than or equal to 5) were used to examine the differences in allele frequency and genotype distribution between healthy controls and patients. Logistic regression analysis was performed to calculate the odds ratio (OR) with confidence interval (95% CI) with age and sex adjustment.
Results
Phenotypic evaluation results
In this study, from 118 samples taken, 32 samples (27.12%) were confirmed as Shigella. Most of the patients were in the age range of 7 to 14 years old. Also, among them 17 (53.13%) were male and 15 (46.87%) were female and the ratio of male to female was 1.13. Therefore, no significant difference was observed in terms of gender. Also, based on the biochemical and serological tests the isolates were known as follow: 20 isolates (62.5%) as S. sonnei, 7 (21.875%) as S. flexneri, 3 (9.375%) as S. boydii and 2 (6.25%) as S. dysenteria.
Antibiogram test results
The study of antibiotic susceptibility and the pattern of antibiotic resistance were performed on the approved samples. The results are shown in Table 2.
| [Sensitive] | [Intermediate] | [Resistant] | Antibiotic | ||||
|---|---|---|---|---|---|---|---|
| % | N | % | N | % | N | Groups | |
| 70 | 22 | 6 | 2 | 24 | 8 | [CTX 30mg] | Cephalosporins |
| 79 | 25 | 6 | 2 | 15 | 5 | [CAZ 30mg] | Cephalosporins |
| 94 | 30 | 0 | 0 | 6 | 2 | [AK30mg ] | Aminoglycoside |
| 79 | 25 | 3 | 1 | 18 | 6 | [GM 10mg ] | Aminoglycoside |
| 97 | 31 | 0 | 0 | 3 | 1 | [IMI 30mg] | Carbapenem |
| 97 | 31 | 0 | 0 | 3 | 1 | [TE 30mg] | Tetracycline |
| 94 | 30 | 3 | 1 | 3 | 1 | [CIP 5mg] | Quinolones |
| 76 | 24 | 6 | 2 | 18 | 6 | [NA 30mg] | - |
| 88 | 28 | 3 | 1 | 9 | 3 | [C 30mg] | - |
| 6 | 2 | 3 | 1 | 91 | 29 | [TS 25mg] | - |
| 3 | 1 | 3 | 1 | 94 | 30 | [AMP 10mg] | - |
Table 2: Antibiogram results in 32 Shigella samples.
According to the results, the highest sensitivity was related to carbapenem (97%) and tetracycline (97%), followed by amikacin and ciprofloxacin with 94% sensitivity, as the most effective antibiotic against Shigella. Also, chloramphenicol (88%), gentamicin (79%) and ceftazidime (79%) were the next treatments of choice. Shigella was also most resistant to ampicillin (94%) primarily and cotrimoxazole (91%) secondarily. Only one sample of 32 isolates showed resistance to imipenem, tetracycline and ciprofloxacin. One sample showed intermediate resistance to ciprofloxacin.
Also, 17 samples out of 32 isolates equivalent to 53.125% of the isolates, were resistant to at least three families of antibiotics, which were in the Multi Drug Resistance (MDR) group. From these 17 isolates, 55% belonged to S. Sonnei, 35% to S. Flexneri and 10% to S. boydii. The statistical analysis showed a significant association between S. Sonnei group and MDR (P=0.0001). In other words, antibiotic resistance was more prevalent in S. sonnei strains than other tested species.
Molecular studies results
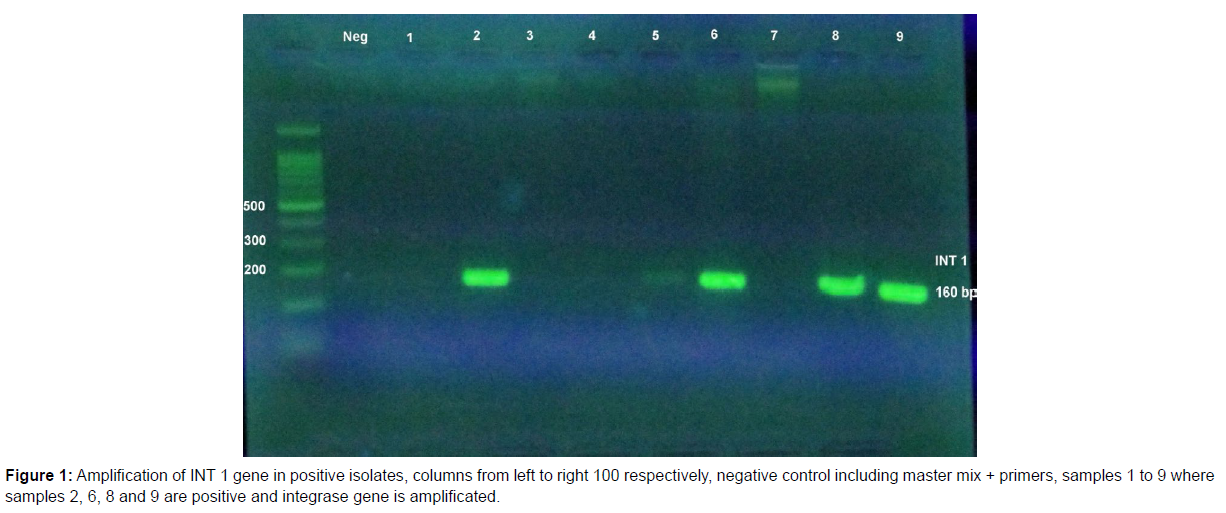
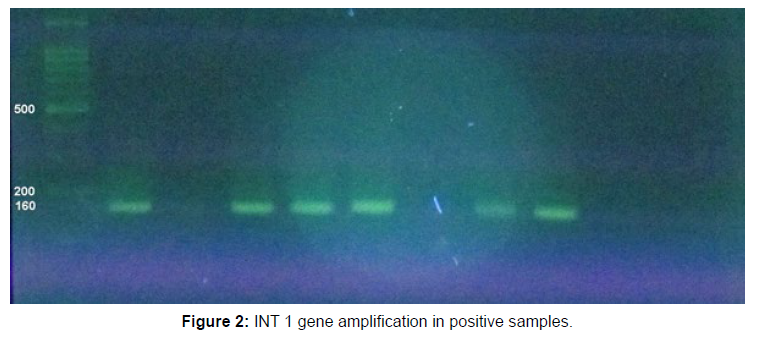
PCR on integrase gene 1 (Int1) showed that 14 out of 32 isolates of Shigella equivalent to 44.8% were positive for the presence of this gene.
Of these, 11 strains (78.6%) were S. flexneri, 2 isolates (14.3%) were S. boydii and 1 strain (7.1%) was S. sonnei. The statistical analysis showed association between Shigella strains and integrase 1 gene and this association was significant for S. flexneri and integron 1 (P=0.0001). There was also a significant association between integrase 1 gene and MDR strain (P=0.00001). Also, a significant association was observed between integron class and resistance to antibiotics chloramphenicol, cotrimoxazole, gentamicin and ceftazidime (Figures 1 and 2).
Discussion
In this study, 32 out of 118 samples (27.12%) were confirmed as Shigella based on the phenotypic tests. Most of them were in the age group of 7 to 14 years old. Also, 17 (53.13%) were male and 15 (46.87%) were female and the ratio of male to female was 1.13 with no significant difference in terms of gender. Also, based on the biochemical and serological tests, 20 (62.5%) were S. sonnei, 7 (21.875%) were S. flexneri, 3 (9.375%) were S. boydii and 2 (6.25%) were S. dysenteria isolates.
Our results differed from the studies in India, which reported frequencies close to 2% [13]. This difference in Shigella abundance reports may be related to the age, the level of economic development, geography, climate, as well as many other environmental conditions.
Facilities such as public water supply and sewerage systems with close association with personal hygiene, food contamination, and medical hygiene may not be available or might have poor quality. S. sonnei is generally found in industrialized countries, while S. flexneri is highly prevalent in developing countries [14]. Our results showed that S. sonnei was the main species in this study with 62.5% of all Shigella species and then S. flexneri with 21.875% was in the second degree. This finding was similar to data reported from Iran (Shiraz) as well as other countries, including Thailand [15]. This pattern was different from other studies in Iran (Abadan), southern part of Iran and countries such as India with S. flexneri as the most common species [16-17].
In recent decades, most cases of Shigella in Iran have been caused by S. flexneri, although recent studies have shown an increasing number of S. sonnei infections [15-18]. Economic and public health conditions affect the frequency of Shigella groups [19]. In the current study, the highest age of infection is under 15 years old. One of the most important reasons for this outcome is not following the health instructions that are less considered by children at this age. Differences in the severity and clinical signs of pathogenicity of Shigella species is related to the differences in Shigella species in terms of the presence of pathogenic genes, the amount of bacteria in the patient, and the physiological characteristics of the patient such as the ability of the immune system to cope with the infection, age and the lack of autoimmune diseases such as Acquired Immune Deficiency Syndrome (AIDS).
Due to the importance of knowing the antibiotic resistance pattern, in the present study, antibiotic susceptibility testing was performed on 32 isolates taken from the patients. The highest sensitivity was related to carbapenem (97%) and tetracycline (97%), followed by amikacin and ciprofloxacin with 94% sensitivity as the most effective antibiotics against Shigella. Chloramphenicol (88%), gentamicin (79%) and ceftazidime (79%) can be the next treatments of choice. Shigella is also resistant to ampicillin (94%) primarily and cotrimoxazole (91%) secondarily.
Cotrimoxazole is a drug often used for the experimental treatment of diarrheal diseases [20]. Widespread use of this drug has led to the emergence of resistant strains of Shigella. In this study, Shigella showed high resistance to cotrimoxazole (91%). Previous reports in Iran have mentioned the resistance level of 92.2 to 94% against cotrimoxazole [21-22]. High resistance to cotrimoxazole has also been reported from Turkey (95%) [23]. According to our regional reports from Iran, ampicillin resistance was 94% and the previous study reported 57% [24]. These results strongly indicated that the use of cotrimoxazole and ampicillin is not appropriate for the treatment of severe diarrhea and dysentery in the West of Iran. S. flexneri resistant to ciprofloxacin was also identified from parts of India (46.25%) [23]. In the present study, 6 patients (18%) with Shigella isolates were resistant to nalidixic acid. In two reports from Tehran and Tabriz, 17.4% and 31% of the isolated Shigella strains were resistant to nalidixic acid, respectively [25].
Previous studies in Iran have also shown Shigella resistance to cephalosporins in the range of 7.57-3.7% between 2008 and 2018 [26]. This shows increasing resistance to cephalosporins in Iran compared to the other countries [27]. This finding is worrying, as previous studies in China, the Middle East and Southeast Asia have reported less resistance to cephalosporins (15.1-20%). In the present study, the resistance to cephalosporins was in the range of 15 to 24% among the isolates. In this study, class 1 integrons were observed in 14 isolates (44.8%).
The statistical analysis also showed that there was an association between Shigella strains and integrase 1 gene and this association was significant for S. flexneri and integron 1(P=0.0001). There was also a significant association between integrase 1 gene and MDR strain (P=0.00001). On the other hand, a significant association was observed between integron class and resistance to chloramphenicol, cotrimoxazole, gentamicin and ceftazidime antibiotics.
In Brazil, class 1 integrons were found in two strains, while class 2 integrons were obtained in 56 (90.3%) strains [28]. From 3 classes of integrons associated with antimicrobial resistance, class 1 has the highest load in Gram-negative bacteria [29]. Class 2 integrons are the most dominant integrons in S. sonnei [30].
One of the limitations of the present study was the lack of isolated shigella bacteria typing. The recent emergence of S. sonnei in developing countries increases the need for the effective epidemiological surveillance systems. The study showed that the frequency of Shigella isolates production in ESBL is higher than other countries. Our results raise the concerns about the spread of ESBL among endemic S. sonnei strains nationwide because the most isolated Shigella species occur in Iran. Therefore, to prevent the spread of these resistant isolates, the antimicrobial resistance of the isolated shigella bacteria should be monitored continuously and experimental antibiotic therapy should be adapted accordingly.
Conclusion
Based on the results, carbapenem (97%) and tetracycline (97%), followed by amikacin and ciprofloxacin with 94% sensitivity, were the most effective antibiotics against Shigella. There was also a significant association between integrase 1 gene and MDR strain (P = 0.00001).
References
- Wei J, Goldberg MB, Burland V, Venkatesan MM, Deng W, et al. (2003) Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect Immun 71: 2775-2786.
- Kuo CY, Su LH, Perera J, Carlos C, Tan BH, et al. (2008) Antimicrobial susceptibility of Shigella isolates in eight Asian countries, 2001-2004. J Microbiol Immunol Infect; 41: 107-11.
- Gupta A, Polyak CS, Bishop RD, Sobel J, Mintz ED (2004) Laboratory-confirmed shigellosis in the United States, 1989- 2002: Epidemiologic trends and patterns. Clin Infect Dis 38: 1372-1377.
- Murugesan P, Revathi K, Elayaraja S, Vijayalakshmi S, Balasubramanian T (2012) Distribution of enteric bacteria in the sediments of Parangipettai and Cuddalore coast of India. J Environ Biol 33: 705-11.
- Torres AG (2004) Current aspects of Shigella pathogenesis. Rev Latinoam Microbiol 46: 89-97.
- Bhattacharya D, Bhattacharya H, Thamizhmani R, Sayi DS, Reesu R, et al. (2014) Shigellosis in Bay of Bengal Islands, India: Clinical and seasonal patterns, surveillance of antibiotic susceptibility patterns, and molecular characterization of multidrug-resistant Shigella strains isolated during a 6-year period from 2006 to 2011. Eur J Clin Microbiol Infect Dis; 33: 157-170.
- Bachand N, Ravel A, Onanga R, Arsenault J, Gonzalez JP (2012) Public health significance of zoonotic bacterial pathogens from bushmeat sold in urban markets of Gabon, Central Africa. J Wildl Dis 48: 785-789.
- Saeed A, Abd H, Edvinsson B, Sandström G (2009) Acanthamoeba castellanii an environmental host for Shigella dysenteriae and Shigella sonnei. Arch Microbiol 191: 83-88.
- Iwamoto M, Ayers T, Mahon BE, Swerdlow DL (2010) Epidemiology of seafood-associated infections in the United States. Clin Microbiol Rev 23: 399-411.
- Von-Seidlein L, Kim DR, Ali M, Lee HH, Wang X, Thiem VD, et al. (2006) A multicentre study of Shigella diarrhoea in six Asian countries: Disease burden, clinical manifestations, and microbiology. PLoS Med 3: e353.
- Germani Y, Sansonetti PJ (2006) The genus Shigella. The prokaryotes In: Proteobacteria: Gamma Subclass Berlin: Springer 6: 99-122.
- Aggarwal P, Uppal B, Ghosh R, Krishna Prakash S, Chakravarti A, et al. (2016) Multi drug resistance and extended spectrum beta lactamases in clinical isolates of Shigella: a study from New Delhi, India. Travel Med Infect Dis 14: 407–413.
- Taneja N, Mewara A (2016) Shigellosis: epidemiology in India. Indian J Med Res 143: 565-576.
- Farshad S, Sheikhi R, Japoni A, Basiri E, Alborzi A (2006) Characterizationof Shigella strains in Iran by plasmid profile analysis and PCR amplification of ipa genes. J Clin Microbiol 44: 2879–2883.
- Jomezadeh N, Babamoradi S, Kalantar E, Javaherizadeh H (2014) Isolation and antibiotic susceptibility of Shigella species from stool samplesamong hospitalized children in Abadan, Iran. Gastroenterol Hepatol Bed Bench 7: 218.
- Sangeetha A, Parija SC, Mandal J, Krishnamurthy S (2014) Clinical and microbiological profiles of shigellosis in children. J Health Popul Nutr 32: 580.
- Ranjbar R, Dallal MMS, Talebi M, Pourshafie MR (2008) Increased isolation and characterization of Shigella sonnei obtained from hospitalized children in Tehran, Iran. J Health Popul Nutr 26: 426.
- Zhang J, Jin H, Hu J, Yuan Z, Shi W, Yang X, et al. (2014) Antimicrobial resistance of Shigella spp. from humans in Shanghai, China, 2004–2011. Diagn Microbiol Infect Dis 78: 282–286.
- Pourakbari B, Mamishi S, Mashoori N, Mahboobi N, Ashtiani MH, Afsharpaiman S, et al. (2010) Frequency and antimicrobial susceptibility of Shigella species isolated in children medical center hospital, Tehran, Iran, 2001–2006. Braz J Infect Dis 14: 153–157.
- Nikfar R, Shamsizadeh A, Darbor M, Khaghani S, Moghaddam M. (2017) A Study of prevalence of Shigella species and antimicrobial resistance patterns in paediatric medical center, Ahvaz, Iran. Iran J Microbiol 9: 277.
- Kacmaz B, Unaldi O, Sultan N, Durmaz R (2014) Drug resistance profiles and clonality of sporadic Shigella sonnei isolates in Ankara, Turkey. Braz J Microbiol 45: 845–849.
- Akcali A, Levent B, Akbaş E, Esen B (2008) Typing of Shigella sonnei strains isolated in some provinces of Turkey using antimicrobial resistance and pulsed field gel electrophoresis methods. Mikrobiyol Bul 42: 563–572.
- Jafari F, Hamidian M, Rezadehbashi M, Doyle M, Salmanzadeh-Ahrabi S, et al. (2009) Prevalence and antimicrobial resistance of diarrheagenic Escherichia coli and Shigella species associated with acute diarrhea in Tehran, Iran. Can J Infect Dis Med Microbiol 20: 56–62.
- Ranjbar R, Behnood V, Memariani H, Najafi A, Moghbeli M, et al. (2016) Molecular characterisation of quinolone-resistant Shigella strains isolated in Tehran, Iran. J Glob Antimicrob Resist 5: 26–30.
- Zamanlou S, Ahangarzadeh Rezaee M, Aghazadeh M, Ghotaslou R (2018) Characterization of integrons, extended-spectrum β-lactamases, AmpC cephalosporinase, quinolone resistance, and molecular typing of Shigella spp. Infect Dis 50: 616–624.
- Varghese S, Aggarwal A (2011) Extended spectrum beta-lactamase production in Shigella isolates-A matter of concern. Indian J Med Microbiol 29: 76.
- Peirano G, Agersø Y, Aarestrup FM, Dos Prazeres Rodrigues D (2005) Occurrence of integrons and resistance genes among sulphonamide-resistant Shigella spp. from Brazil. J Antimicrob Chemother 55: 301–305.
- Kang HY, Jeong YS, Oh JY, Tae SH, Choi CH, et al. (2005) Characterization of antimicrobial resistance and class 1 integrons found in Escherichia coli isolates from humans and animals in Korea. J Antimicrob Chemother 55: 639-644.
- Pan J-C, Ye R, Meng D-M, Zhang W, Wang H-Q, et al. (2006) Molecular characteristics of class 1 and class 2 integrons and their relationships to antibiotic resistance in clinical isolates of Shigella sonnei and Shigella flexneri. J Antimicrob Chemother 58: 288–296.
- The HC, Thanh DP, Holt KE, Thomson NR, Baker S (2016) The genomic signatures of Shigella evolution, adaptation and geographical spread. Nat Rev Microbiol 14: 235.
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Google Scholar, Crossref, Indexed at
Citation: Abdulkareem AA, Lahourpour F, Keshavarzi F(2023) MolecularEvaluation of Class 1 Integron in Drug-Resistant Shigella Strains Isolated fromClinical Samples. Cell Mol Biol, 69: 251. DOI: 10.4172/1165-158X.1000251
Copyright: © 2023 Abdulkareem AA, et al. This is an open-access articledistributed under the terms of the Creative Commons Attribution License, whichpermits unrestricted use, distribution, and reproduction in any medium, providedthe original author and source are credited.