Research Article Open Access
Molecular Docking: Approaches, Types, Applications and Basic Challenges
Ayaz Mahmood Dar1,2* and Shafia Mir21Department of Chemistry, Aligarh Muslim University, Aligarh, Uttar Pradesh, India
2Department of Chemistry, Government Degree College Kulgam, University of Kashmir, Jammu and Kashmir, India
- *Corresponding Author:
- Ayaz Mahmood Dar
Department of Chemistry
Aligarh Muslim University
Aligarh-202 002, Uttar Pradesh, India
Tel: +919286990247
E-mail: ayazchem09@gmail.com
Received Date: March 16, 2017 Accepted Date: March 22, 2017 Published Date: March 27, 2017
Citation: Dar AM, Mir S (2017) Molecular Docking: Approaches, Types, Applications and Basic Challenges. J Anal Bioanal Tech 8: 356. doi: 10.4172/2155-9872.1000356
Copyright: © 2017 Dar AM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Abstract
Abstract
Molecular docking is a kind of bioinformatic modelling which involves the interaction of two or more molecules to give the stable adduct. Depending upon binding properties of ligand and target, it predicts the three-dimensional structure of any complex. Molecular docking generates different possible adduct structures that are ranked and grouped together using scoring function in the software. Docking simulations predict optimized docked conformer based upon total energy of the system. In spite of all potential approaches, ligand chemistry (tautomerism and ionization), receptor flexibility (single conformation of rigid receptor) and scoring function (differentiate true binding mode) still remained the challenge. Many important aspects of molecular docking in terms of its approaches, types, applications and challenges are briefly discussed in this article.
Keywords
Docking; Conformation; Ligand; Receptor; Optimization
Introduction
Molecular docking is an attractive scaffold to understand drugbiomolecular interactions for the rational drug design and discovery, as well as in the mechanistic study by placing a molecule (ligand) into the preferred binding site of the target specific region of the DNA/protein (receptor) mainly in a non-covalent fashion to form a stable complex of potential efficacy and more specificity [1,2]. The information obtained from the docking technique can be used to suggest the binding energy, free energy and stability of complexes. At present, docking technique is utilized to predict the tentative binding parameters of ligand-receptor complex beforehand.
The main objective of molecular docking is to attain ligand-receptor complex with optimized conformation and with the intention of possessing less binding free energy. The net predicted binding free energy (ΔGbind) is revealed in terms of various parameters, hydrogen bond (ΔGhbond), electrostatic (ΔGelec), torsional free energy (ΔGtor), dispersion and repulsion (ΔGvdw), desolvation (ΔGdesolv), total internal energy (ΔGtotal) and unbound system’s energy (ΔGunb). Therefore, good understanding of the general ethics that govern predicted binding free energy (ΔGbind) provides additional clues about the nature of various kinds of interactions leading to the molecular docking [3].
Practical application of molecular docking requires data bank for the search of target with proper PDB format and a methodology to prepare ligand as a PDB file. To do this, there are various software’s (Discovery studio, etc.,) available from where the ligand can be made in PDB format. These tools provide the organization to ligands based upon their ability to interact with given target proteins/DNA. Molecular docking of small molecules to a target includes a pre-defined sampling of possible conformation of ligand in the particular groove of target in an order to establish the optimized conformation of the complex. This can be made possible using scoring function of software [4]. Since the infrared spectroscopy, X-ray crystallography and Nuclear Magnetic Resonance (NMR) spectroscopy are the techniques for the investigation and establishment of three dimensional structures of any organic molecule/ biomolecular targets. Hence homology modeling makes it possible to determine the tentative structure of proteins of unknown structure with high sequence homology to known structure. This provides a substitute approach for target structure establishment, which forms starting point for in silico drug discovery.
There are various databases available, which offer information on small ligand molecules such as CSD (Cambridge Structural Database), ACD (Available Chemical Directory), MDDR (MDL Drug Data Report) and NCI (National Cancer Institute Database). While performing docking, different interacted conformers are generated and compared with each other. In the condition of rejection, new conformers are obtained and again search procedure continues to its endpoint after acceptance of one conformation. The docked conformers according to their experimental binding affinities and binding free energies seem to be more difficult than their binding orientation. To overcome this problem, different scoring functions are employed such as consensus scoring; appliance of number of score functions to the same docked pose in order to eliminate false positives [5]. A huge number of attempts has been made for the development of efficient docking protocols. No doubt, significant progress has been made in the computational prediction of docking modes. This mini review article is dedicated to recent computational approaches, types, applications and its challenges.
Approaches of Molecular Docking
For performing molecular docking, primarily two types of approaches are used.
Simulation approach
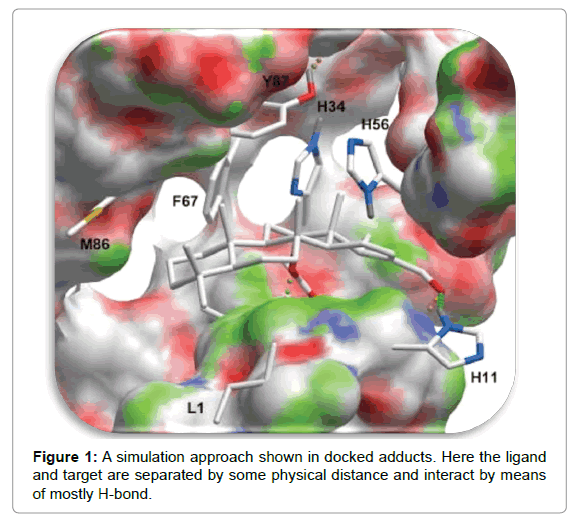
Here the ligand and target is being separated by physical distance and then ligand is allowed to bind into groove of target after “definite times of moves” in its conformational space (Figure 1). The moves involve variations to the structure of ligand either internally (torsional angle rotations) or externally (rotations and translations). The ligand in every move in the conformational limit releases energy, as “Total Energy”. This approach is more advantageous in the sense that it is more compatible to accept ligand flexibility. Additionally, it is more real to assess the molecular recognition between ligand and target. However, this approach takes longer duration to estimate optimal docked conformer due to the large energy dissipating for each conformation. Recently, fast optimization method and grid-based tools have dominantly revolutionized this drawback to make simulation approach more userfriendly [6].
\
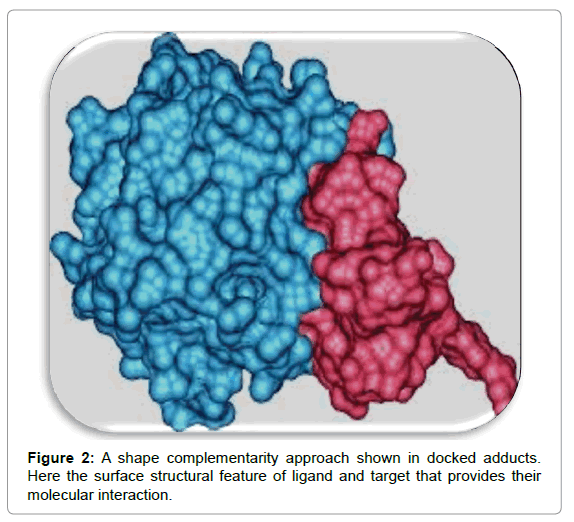
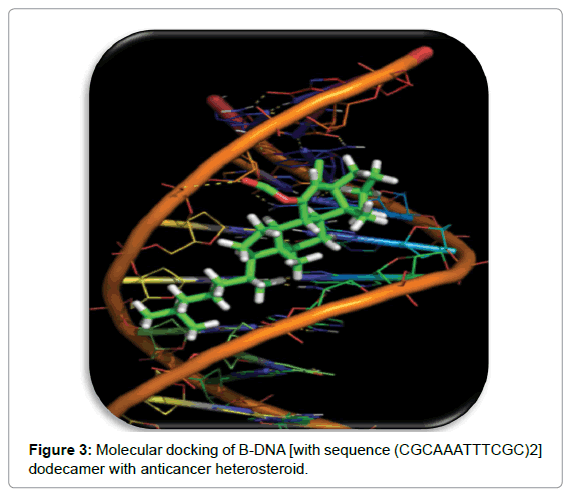
Shape complementarity approach
This approach employs ligand and target as surface structural feature that provides their molecular interaction (Figures 2 and 3). Here the surface of target is shown with respect to its solvent-accessible surface area and ligand’s molecular surface is showed in terms of matching surface illustration. The complementarity between two surfaces based on shape matching illustration helps in searching the complementary groove for ligand on target surface. For example, in protein target molecules, hydrophobicity is estimated by employing number of turns in the main-chain atoms. This approach is rather quick and involves the rapid scanning of numerous thousands of ligands in a few seconds to find out the possible binding properties of ligand on target molecular surface [5,6].
Types of Docking
Comprehensively utilized docking tools employ search algorithms such as genetic algorithm, fragment-based algorithms, Monte Carlo algorithms and molecular dynamics algorithms. Besides this, there are some tools such as DOCK, GOLD, FlexX and ICM which are mainly used for high throughput docking simulations. There are various kinds of molecular docking procedures involving either ligand/target flexible or rigid based upon the objectives of docking simulations [6,7] like flexible ligand docking (target as rigid molecule), rigid body docking (both the target and ligand as rigid molecules) and flexible docking (both interacting molecules as flexible).
Applications of Molecular Docking
Molecular docking can demonstrate the feasibility of any biochemical reaction as it is carried out before experimental part of any investigation. There are some areas, where molecular docking has revolutionized the findings. In particular, interaction between small molecules (ligand) and protein target (may be an enzyme) may predict the activation or inhibition of enzyme. Such type of information may provide a raw material for the rational drug designing. Some of the major applications of molecular docking are described below: -
Lead optimization
Molecular docking can predict an optimized orientation of ligand on its target. It can predict different binding modes of ligand in the groove of target molecule. This can be used to develop more potent, selective and efficient drug candidates [5,7].
Hit identifications
Docking in combination with scoring function can be used to evaluate large databases for finding out potent drug candidate in silico, which can target the molecule of interest [8].
Drug-DNA interaction
Molecular docking plays a prominent role in the initial prediction of drug’s binding properties to nucleic acid. This information establishes the correlation between drug’s molecular structure and its cytotoxicity. Keeping this in view, medicinal chemists are constantly putting their efforts to elucidate the underlying anticancer mechanism of drugs at molecular level by investigating the interaction mode between nucleic acid and drugs [9,10] in presence of copper. Medicinal chemists are doing in silico observations where their main finding is to predict whether the compound/drug is interacting with the protein/DNA. If the docking programme is predicting the said interaction, then the experimental procedures are made available to find out the real binding mode of the complex. This leads to the development of new anticancer drug. Furthermore, this knowledge would be instrumental in the detection of those structural modifications in a drug that could result in sequence/structure specific binding to their target [11].
Basic Challenges in Molecular Docking
Certain basic challenges in docking and scoring are discussed under the following headings.
Ligand chemistry
The ligand preparation has prominent effect on the docking results because the ligand recognition by any biomolecule depends on 3-dimensional orientation and electrostatic interaction. This confirms that the conformation of both the ligand as well as ligand preparation is important. Earlier, keeping approximate pKa values, the structure being most likely optimized by removing or adding hydrogens but the tautomeric and protomeric states of the molecules which are to be docked, still remained a major discrepancy. Since almost all databases keep molecules in their neutral forms but under physiological conditions they are actually ionized. Hence it is compulsory to ionize them prior to docking. But in different programs, the standard ionization is easy to achieve. Regarding the issue of tautomers, the problem still remains there, which tautomer one should use or should one use all possible tautomers [12].
Receptor flexibility
This is a major challenge in docking i.e., handling of flexible protein. A biomolecule/protein adopts different conformations depending upon the ligand to which it binds. This confirms that docking done with a rigid receptor will give a single conformation of receptor. However, when the docking is done with flexible receptor, the ligands may require many receptor conformations to bind. In molecular docking studies, usually the most neglected aspect is different conformational states of proteins. Since the protein flexibility is important as it accounts for better affinity to be achieved between a given a drug and target. Another aspect of target flexibility is active site water molecules. Water molecules must be rectified to avoid using artifact waters in the docking process [13].
Scoring function
Another challenge in docking is imperfection in scoring function. Just like search algorithm is having potential to give optimum conformation, scoring function should also be able to differentiate true binding modes from all the other parallel modes. A potential scoring function would be computationally much economical, unfavourable for analyzing several binding modes. When there is accuracy, scoring functions make number of suggestions to evaluate ligand affinity. The physical phenomenon i.e., entropy and electrostatic interactions are disregarded in scoring schemes. Hence the lack of suitable scoring function, both in terms of accuracy and speed, is the main congestion in molecular docking programming [14].
Conclusion
The potential docking technique is done after thoroughly screening the target, ligands and docking method performance. The ligand flexibility however is almost resolved and does not create much problem however protein flexibility needs to be improved. Water molecules should be included to consider the hydrogen bonding with non-aqueous residues. It is evident from docking literature that it has attained a good amount of maturity and in this short review, we have focused on types, approaches, applications and challenges of molecular docking in brief but accounting for flexibility and successful scoring remain significant challenges.
Acknowledgements
Authors thank Head of Department of Chemistry, GDC Kulgam for granting the permission to publish the work.
References
- Rohs R, Bloch I, Sklenar H, Shakked Z (2005) Molecular flexibility in ab-initio drug docking to DNA: binding-site and binding-mode transitions in all-atom Monte Carlo simulations. Nucl Acids Res 33: 7048-7057.
- Guedes IA, de Magalhães CS, Dardenne LE (2014) Receptor-ligand molecular docking. Biophysical Reviews 6: 75-87.
- Agarwal S, Chadha D, Mehrotra R (2015) Molecular modeling and spectroscopic studies of semustine binding with DNA and its comparison with lomustine–DNA adduct formation. J Biomol Struct Dyn 33: 1653-1668.
- Seeliger D, de Groot BL (2010) Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des 24: 417-422.
- Shoichet BK, McGovern SL, Wei B, Irwin JJ (2002) Lead discovery using molecular docking. Curr Opin Chem Biol 6: 439-446.
- Lamb ML, Jorgensen WL (1997) Computational approaches to molecular recognition. Curr Opin Chem Biol 1: 449-457.
- Gschwend DA, Good AC, Kuntz ID (1996) Molecular docking towards drug discovery. J Mol Recognit 9: 175-186.
- Ferreira LG, dos Santos RN, Oliva G, Andricopulo AD (2015) Molecular docking and structure-based drug design strategies. Molecules 20: 13384-13421.
- Agarwal S, Jangir DK, Mehrotra R, Lohani N, Rajeswari M (2014) A Structural Insight into Major Groove Directed Binding of Nitrosourea Derivative Nimustine with DNA: A Spectroscopic Study. PLoS ONE 9: 104-115.
- Mehrotra R, Jangir DK, Agarwal S, Ray B, Singh P, et al. (2013) Interaction studies of anticancer drug lomustine with calf thymus DNA using surface enhanced Raman spectroscopy. MAPAN 28: 273-277.
- Holt PA, Chaires JB, Trent JO (2008) Molecular docking of intercalators and groove-binders to nucleic acids using Autodock and Surflex. J Chem Inf Model 48: 1602-1615.
- Elokely KM, Doerksen RJ (2013) Docking Challenge: Protein Sampling and Molecular Docking Performance. J Chem Inf Model 53: 1934-1945.
- Totrov M, Abagyan R (2008) Flexible ligand docking to multiple receptor conformations: A practical alternative. Curr Opin Struct Biol 18: 178-184.
- Cho AE, Rinaldo D (2009) Extension of QM/MM docking and its applications to metalloproteins. J Comput Chem 30: 2609-2616.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 115998
- [From(publication date):
April-2017 - Mar 31, 2025] - Breakdown by view type
- HTML page views : 107054
- PDF downloads : 8944