Research Article Open Access
Molecular Diversity of Microbes with Probable Degradative Genes in Agricultural Soil Contaminated with Bonny Light Crude Oil
Ogbulie TE1* and Nwaokorie FO21Department of Biotechnology, School of Science, Federal University of Technology Owerri (FUTO), Imo State, Nigeria
2Molecular biology and Biotechnology Division, Nigerian Institute for Medical Research [NIMR], Yaba Lagos State, Nigeria
- *Corresponding Author:
- Ogbulie TE
Department of Biotechnology, School of Science
Federal University of Technology Owerri (FUTO) Imo State, Nigeria
Tel: +2348035472379
E-mail: ogbulie_toochi@yahoo.com
Received date: August 15, 2015; Accepted date: February 09, 2016; Published date: February 17, 2016
Citation: Ogbulie TE, Nwaokorie FO (2016) Molecular Diversity of Microbes with Probable Degradative Genes in Agricultural Soil Contaminated with Bonny LightCrude Oil. J Ecosys Ecograph S5:002. doi:10.4172/2157-7625.S5-002
Copyright: © 2016 Ogbulie TE, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Ecosystem & Ecography
Abstract
This study looked at the diversity of microorganisms persistent in agricultural soil sample polluted with 100 ml of 100% Nigerian Bonny light crude oil left for four years. DNA from crude oil polluted agricultural soil sample was extraction using ZYMO soil DNA extraction Kit. DNA sequencing was performed by Next Generation Sequencing Technique [NGST] using automated PCR cycle- Genome Sequencer™ FLX System from 454 Life Sciences™ and Roche Applied. Sequence analysis and alignment was performed using Vecton NTI suite 9 (InforMax, Inc.). The resulting nucleotide sequences were compared to sequences obtained from GenBank by BLASTx analysis using CLO Bio software as well as BLASTn using NCBI. Molecular Identities of microbial community was obtained by creating different dendrograms. Gene sequencing carried out read 513 different nucleotide sequences. Seven phyla with 47 corresponding culture-dependent species and 169 culture-independent bacteria clone were obtained. The resultant tree showed cladogram of proteobacteria ( b and g - proteobacteria), bacteria/enterobacteria, firmicutes, plantomycetes, acidobacteria group/ fibrobacteres, Bacteriodetes/chlorobi Actinobacteria/high G + C and chloriflexi phyla. Furher taxonomical classification was carried out with reads of sufficient Q scores (> q30) and lengths and a total of 420 read count of top kingdom classification of 100% bacteria kingdom was obtained. Proteobacteria phyla of class betaproteobacteria, order Burkholderiales and family Comamonadaceae had the highest read count with percentage diversity of 57.14%, 53.81%, 53.81 and 53.57% respectively. The nucleotide sequences with no hit (208) was sent to Genbank for asigning of ascension number. The detection of these diverse organisms from crude oil polluted agricultural soil left for four years, depict that the organism probably, have degradative genes which aided their survival.
Keywords
Microbes; Diversity; Degradative genes; Crude oil; Agricultural soil; Sequence analysis
Introduction
Incidence of environmental pollution due to high rate of petroleum related activities in Nigeria and other oil producing areas of the world has been associated with frequent oil spills, especially through oil wells blow out, tanker accidents and bunkering. Disasters arising from such incidence results in the discharge of crude oil into the environment affecting both soil, air and water bodies. This threatens human health and that of organisms that are dependent on soil. Soil contains a variety of microorganisms including bacteria that can be found in any natural ecosystem. Microbial survival in polluted soil depends on intrinsic biochemical and structural properties, physiological and genetic adaptation including morphological changes of cells as well as environmental modifications [1]. Over the years, isolation and identification of hydrocarbon-degrading microorganisms have been carried out using isolation techniques. Previous studies on population dynamics showed that bacteria genera such as Pseudomonas, Bacillus, Brevibacterium, Corynebacterium, Acinetobacter and Mycobacterium are potential organisms for hydrocarbon degradation [2-4]. Shi et al. [5] compared culture-based diversity of agricultural soil communities with diversity obtained by molecular means and found that molecular methods revealed a much higher bacterial diversity than classical isolation techniques.
A variety of molecular methods have been developed to assay the presence of micro-organisms in soil. Most recently, the method of choice to determine what micro-organisms are present in environmental sample is to amplify the conserved small subunit rRNA gene; where DNA is isolated from the soil using bead beating and Polymerase Chain Reaction (PCR) with universal or gene-specific primers used to amplify the specific gene from the sample. This study looked at the diversity of microorganisms persistent in agricultural soil sample polluted with 100 ml of 100% Nigerian Bonny light crude oil and left for four years with a view to ascertain the presence of microbes with probable degradative gene for crude oil degradation which can be harnessed for the creation of superbugs for faster clean up opertaions and to confirm similarities in microbial identities.
Material and Method
Procurement of samples
The crude oil used was bonny light Crude and was collected with sterile containers from Akiri in Oguta, Imo State, Nigeria. The agricultural soil subjected to pollution was obtained from Federal University of Technology Owerri (FUTO) farm land using surface sterilized soil auger at the depth of 15-30 cm.
Treatment of test soil sample
Surface sterilized plastic pot with no drainage holes was filled with 450 g of soil. Thereafter, 100 ml of crude oil was used to pollute the soil and 300 ml of sterile water added biweekly following modified method of Yee et al. [6]. The setup was kept in a chamber for a period of four years with a light cycle of 11 h darkness and 13 h light.
Molecular analysis
Molecular analysis was performed at the GS FLX Titanium Sequencing Service company- Inqaba, South Africa. Methodology was based on PCR and metagenomics analysis.
DNA extraction from soil sample
DNA extraction form soil sample was performed using ZYMO soil DNA extraction kit according to the manufactures. According to the method, genomic DNA was extracted by weighing out 0.25 grams of soil sample using an analytical Balance. The sample was the added into a ZR Bashing Bead™ Lysis Tube followed by the addition of 750 μ Lysis Solution to the tube. The content of the 2 ml tube disrupted by mixing in a vortex mixer at maximum speed for 5 minutes. The ZR Bashing Bead™ Lysis Tube was centrifuged in a micro centrifuge at ≤10,000 xg for 1 minutes. 400 μl of the filtrate was added to a Zymo- Spin™ IV Spin Filter in a Collection Tube and centrifuge at 7,000 rmp (˜7,000 xg) for 1 minutes. This was followed by the addition of 1,200 μl of soil DNA Binding Buffer to the filtrate in the Collection Tube. 800 μl of the mixture from above was added to a Zymo-Spin™ IIC Column in a Collection Tube and centrifuge at 10,000 xg for 1 minute. Flow through from the collection tube was discarded and this particular step was repeated with the remaining filtrate. 200 μl of DNA Pre-Wash Buffer was thereafter added to the Zymo-Spin™ IIC Column in a new collection tube and centrifuged at 10,000 xg for 1 minute. Thereafter, 500 μl of Soil DNA Wash Buffer was added to the Zymo-Spin™ IIC Column and centrifuged at 10,000 xg for 1 minute. The Zymo-Spin™ IIC Column was transferred into a clean 1.5 ml micro centrifuge tube and 100 μl of DNA Elution Buffer added directly to the column matrix. This was centrifuged at 10,000 xg for 30 seconds to elute the DNA. The eluted DNA was transferred into a filter unit of Zymo-Spin™ IV-HRC Spin Filter in a clean 1.5 ml micro centrifuge tube and centrifuged at exactly 8,000 xg for 1 minute. The filtered DNA was then used for PCR and DNA sequencing.
Polymerase Chain Reaction [PCR]
The PCR was carried out in a 20 μl reaction mixture containing a 5X HOT FIRE Pol blend master mix (ready to use) composed of FIREPol® DNA polymerase Proof-reading enzyme, 5X reaction buffer, 7.5 mM MgCl2, 1 mM dNTPs of each have 200 μM of dATP, dCTP, dGTP, dTTP. A combination of 4 μl of master mix, 0.2 μl each of forward and reverse 16S rRNA primer and 2 μl of template gDNA constituted 6.4 μl. Hence 13.6 μl of sterile distilled water was added to make it up to the recommended PCR reaction mix of 20 μl .The entire mixture was then vortexed and loaded together with positive and negative control (dH20) into the thermal cycler (eppendorf vapor protect). The PCR reaction was carried out with an initial denaturation at 95°C for 5 min, followed by 30 consecutive cycles at 95°C for 30 sec, and annealing temperature of 55°C for 1 minute and then 72°C holding for 1 minute. This was then followed by a final extension step at 72°C for 10 minute.
DNA Sequencing
DNA sequencing was performed by Next Generation Sequencing Technique to determine the nucleotide sequence of all microorganisms present in the soil sample using automated PCR cycle-Genome Sequencer™ FLX System from 454 Life Sciences™ and Roche Applied. Sequence analysis and alignment was performed using Vector NTI suite 9 (InforMax, Inc.) and the resulting nucleotide sequences were compared to sequences obtained from GenBank1 by BLASTx. Analysis using CLO Bio software as well as BLASTn2 using NCBI. For every sample set, every read was BLASTED and the result file saved. The top 5 hits for every BLAST result (i.e., species name) was counted and a record was kept of how many times each species appeared as a hit. The number in the last column basically is the number of times a read hit/ matched to that species. The frequency (i.e., count/total number of reads) and absolute count of each species were reported and used to name the specific organism (Table 1).
| gb|GU599159.1| Uncultured soil bacterium clone HB_Ca_M_285 | 1A | 0.001592357 | 1 |
| gb|HQ120802.1| Uncultured bacterium isolate 1112865261764b 16S | 2A | 0.003184713 | 2 |
| gb|GQ918974.1| Uncultured soil bacterium clone 21_77KE06 | 3A | 0.001592357 | 1 |
| gb|JX186586.1| Uncultured bacterium clone YB61 16S | 4A | 0.011146497 | 7 |
| gb|JN168313.1| Uncultured bacterium clone WLBL550 16S | 5A | 0.001592357 | 1 |
| gb|HQ322838.1| Uncultured bacterium clone W4-84 16S | 6A | 0.001592357 | 1 |
| emb|FR716374.1| Uncultured bacterium partial 16S rRNA | 7A | 0.001592357 | 1 |
| gb|JN865443.1| Uncultured bacterium isolate DS-3 16S | 8A | 0.001592357 | 1 |
| gb|HM019522.1| Burkholderiaphymatum strain GR06 16S | 9A | 0.001592357 | 1 |
| gb|HQ119629.1| Uncultured bacterium isolate 1112842460007a 16S | 10A | 0.001592357 | 1 |
| gb|EF667534.1| Uncultured bacterium clone LaC15L18 16S | 11A | 0.003184713 | 2 |
| gb|HM069772.1| Uncultured bacterium clone Bacteria_Clone_157 16S... | 12A | 0.00477707 | 3 |
| gb|JN168399.1| Uncultured bacterium clone WLCLC424 16S | 13A | 0.001592357 | 1 |
| gb|JN168229.1| Uncultured bacterium clone WLBL429 16S | 14A | 0.001592357 | 1 |
| gb|JQ476801.1| Uncultured bacterium clone 071071_067 16S | 15A | 0.007961783 | 5 |
| gb|JQ710440.1| Nevskia sp. F2-63 16S ribosomal | 16A | 0.003184713 | 2 |
| emb|AM773969.1| uncultured bacterium partial 16S rRNA | 17A | 0.001592357 | 1 |
| gb|JX041839.1| Uncultured proteobacterium clone APC_4_G1 16S | 18A | 0.001592357 | 1 |
| emb|FR687596.1| Uncultured bacterium partial 16S rRNA | 19A | 0.003184713 | 2 |
| gb|GU598830.1| Uncultured soil bacterium clone HB_R_M_212 | 20A | 0.001592357 | 1 |
| gb|JF911130.1| Uncultured bacterium clone Bbf10-02C12 16S | 21A | 0.003184713 | 2 |
| gb|EU382007.1| Uncultured rumen bacterium clone P5_B07 | 22A | 0.001592357 | 1 |
| gb|JQ861367.1| Uncultured Acidobacteria bacterium clone XH15 | 23A | 0.001592357 | 1 |
| gb|HQ674808.1| Uncultured Acidisphaera sp. clone LWM1-70 | 24A | 0.001592357 | 1 |
| gb|GU375188.1| Uncultured soil bacterium clone Bact.wet.ACETE09 | 25A | 0.001592357 | 1 |
| gb|JN168182.1| Uncultured bacterium clone WLBL342 16S | 26A | 0.001592357 | 1 |
| gb|JQ769640.1| Uncultured bacterium clone YB-14 16S | 27A | 0.003184713 | 2 |
| gb|DQ463275.1| Uncultured bacterium clone ES3-56 16S | 28A | 0.003184713 | 2 |
| gb|GQ376581.1| Uncultured bacterium clone D1G_F09 16S | 29A | 0.001592357 | 1 |
| gb|AF018067.1| Uncultured bacterium OSW1 16S ribosomal | 30A | 0.00477707 | 3 |
| gb|HM439297.1| Uncultured Acidobacteria bacterium clone BG25-1 | 31A | 0.062101911 | 39 |
| gb|HQ674837.1| Uncultured Rhodanobacter sp. clone LWM1-59 | 32A | 0.001592357 | 1 |
| gb|JX174218.1| Dyella sp. 2341 16S ribosomal | 33A | 0.00477707 | 3 |
| gb|JN168198.1| Uncultured bacterium clone WLBL366 16S | 34A | 0.001592357 | 1 |
| gb|JF910554.1| Uncultured bacterium clone Bfb08-H4 16S | 35A | 0.001592357 | 1 |
| gb|EF020266.1| Uncultured Acidobacteriaceae bacterium clone Elev... | 36A | 0.00477707 | 3 |
| gb|HQ445747.1| Uncultured bacterium clone Luq_GS470_003 16S | 37A | 0.001592357 | 1 |
| gb|JX172839.1| Uncultured bacterium clone PB17026-1A_G11 16S | 38A | 0.015923567 | 10 |
| emb|HE660678.1| Uncultured bacterium partial 16S rRNA | 39A | 0.001592357 | 1 |
| gb|JX171869.1| Uncultured bacterium clone PB17007-2_E01 16S | 40A | 0.001592357 | 1 |
| gb|HQ264667.1| Uncultured bacterium clone SCP117 16S | 41A | 0.003184713 | 2 |
| gb|JF440522.1| Uncultured bacterium clone CG364 16S | 42A | 0.001592357 | 1 |
| gb|HQ023258.1| Burkholderiaunamae strain CACua-11 16S | 43A | 0.003184713 | 2 |
| gb|JQ968935.1| Uncultured bacterium clone Gra-Bac073 16S | 44A | 0.001592357 | 1 |
| gb|JN911353.1| Uncultured microorganism clone GF13U7304JZZZD 16S... | 45A | 0.001592357 | 1 |
| gb|FJ648701.2| Burkholderia sp. SWF66247 16S ribosomal | 46A | 0.003184713 | 2 |
| gb|GQ140333.1| Comamonastestosteroni strain SJ89 16S | 47A | 0.001592357 | 1 |
| gb|JQ665348.1| Pseudomonas aeruginosa strain CSMCRI-1069 16S | 48A | 0.001592357 | 1 |
| emb|HE856926.1| Uncultured bacterium partial 16S rRNA | 49A | 0.001592357 | 1 |
| gb|HQ730653.1| Uncultured Acidobacterium sp. clone JL123_4 | 50A | 0.001592357 | 1 |
| gb|FJ625119.1| Uncultured bacterium clone H_C_122 16S | 51A | 0.001592357 | 1 |
| gb|HQ010155.1| Uncultured Acidobacteria bacterium clone An45_C4 | 52A | 0.001592357 | 1 |
| gb|FJ451723.1| Uncultured bacterium clone ORFRC-FW102-670d-2.8 1... | 53A | 0.001592357 | 1 |
| gb|JN172802.1| Uncultured soil bacterium clone em_emp435 | 54A | 0.001592357 | 1 |
| gb|JF797204.1| Pseudomonas aeruginosa strain ITCC B0030 | 55A | 0.001592357 | 1 |
| gb|EU881261.1| Uncultured bacterium clone KGB200711-007 16S | 56A | 0.001592357 | 1 |
| gb|EF600579.1| Uncultured bacterium clone E5-47 16S | 57A | 0.007961783 | 5 |
| gb|JX083379.1| Burkholderiakururiensis strain PR1 16S | 58A | 0.003184713 | 2 |
| gb|EU680360.1| Uncultured bacterium clone S7-68 16S | 59A | 0.003184713 | 2 |
| gb|FJ166807.1| Uncultured bacterium clone R_LQ3_C01 16S | 60A | 0.001592357 | 1 |
| gb|JN817761.1| Firmicutes bacterium enrichment culture clone | 61A | 0.001592357 | 1 |
| gb|DQ264622.1| Uncultured bacterium clone BANW684 16S | 62A | 0.001592357 | 1 |
| gb|GQ376582.1| Uncultured bacterium clone D1G_F10 16S | 63A | 0.001592357 | 1 |
| gb|EF516204.1| Uncultured bacterium clone FCPP711 16S | 64A | 0.001592357 | 1 |
| gb|JF440427.1| Uncultured bacterium clone CG208 16S | 65A | 0.001592357 | 1 |
| gb|JX047141.1| Uncultured bacterium clone KWB121 16S | 66A | 0.006369427 | 4 |
| gb|JN391993.1| Uncultured bacterium clone Q7591-HYSO 16S | 67A | 0.001592357 | 1 |
| gb|EU265982.1| Uncultured bacterium clone Nit2A0626_56 16S | 68A | 0.00477707 | 3 |
| gb|JN168228.1| Uncultured bacterium clone WLBL427 16S | 69A | 0.003184713 | 2 |
| gb|JN082688.1| Uncultured Schlegelella sp. clone 262 | 70A | 0.001592357 | 1 |
| gb|EU755081.1| Uncultured bacterium clone HM-51 16S | 72A | 0.001592357 | 1 |
| gb|JQ864383.1| Dyella sp. LB15 16S ribosomal | 73A | 0.020700637 | 13 |
| gb|JF833857.1| Uncultured bacterium clone E30 16S | 74A | 0.001592357 | 1 |
| emb|AM159259.1| Uncultured Chloroflexi bacterium 16S rRNA | 75A | 0.001592357 | 1 |
| gb|EF471223.1| Dyella sp. CHNCT13 16S ribosomal | 76A | 0.00477707 | 3 |
| gb|JN873119.1| Uncultured bacterium isolate DGGE gel | 77A | 0.017515924 | 11 |
| gb|JF361451.1| Uncultured soil bacterium clone GO0VNXF07H1XC7 | 78A | 0.001592357 | 1 |
| gb|HM545452.1| Uncultured bacterium clone ZM9-198 16S | 79A | 0.003184713 | 2 |
| gb|GQ376832.1| Uncultured bacterium clone D10H_G08 16S | 80A | 0.001592357 | 1 |
| gb|HQ433554.1| Uncultured bacterium clone GOP_C 16S | 81A | 0.020700637 | 13 |
| gb|HM663734.1| Uncultured bacterium clone GB7N87003GA6J8 small | 82A | 0.02388535 | 15 |
| gb|HM488701.1| Uncultured Myxococcales bacterium clone BOM_f02 | 83A | 0.001592357 | 1 |
| gb|DQ450730.1| Uncultured Chloroflexi bacterium clone B12_WMSP1 | 84A | 0.001592357 | 1 |
| emb|AJ233524.1| uncultured eubacterium 16S ribosomal RNA, | 85A | 0.001592357 | 1 |
| gb|EF018933.1| Uncultured bacterium clone Amb_16S_1442 16S | 86A | 0.001592357 | 1 |
| gb|EF019209.1| Uncultured Caulobacteraceae bacterium clone Amb_1... | 87A | 0.001592357 | 1 |
| gb|DQ297980.1| Uncultured soil bacterium clone UC11 | 88A | 0.006369427 | 4 |
| gb|GQ918879.1| Uncultured soil bacterium clone 12-77KA07 | 89A | 0.001592357 | 1 |
| emb|HE604298.1| Uncultured beta proteobacterium partial 16S | 90A | 0.001592357 | 1 |
| gb|GQ356931.1| Uncultured bacterium clone Fe_B_114 16S | 91A | 0.027070064 | 17 |
| gb|FJ370943.1| Uncultured bacterium clone TS5_a03b01 16S | 92A | 0.00477707 | 3 |
| gb|HQ864217.1| Uncultured bacterium clone TP-SL-B-279 16S | 93A | 0.001592357 | 1 |
| gb|HM582700.1| Uncultured bacterium clone LCH_B101 16S | 94A | 0.00955414 | 6 |
| gb|JQ796741.1| Burkholderia sp. 10-18 16S ribosomal | 95A | 0.001592357 | 1 |
| gb|DQ264442.1| Uncultured bacterium clone BANW446 16S | 96A | 0.007961783 | 5 |
| gb|JQ926999.1| Uncultured beta proteobacterium clone 1-10e | 97A | 0.022292994 | 14 |
| gb|JN172798.1| Uncultured soil bacterium clone em_emp426 | 98A | 0.001592357 | 1 |
| emb|FQ684062.1| 16S rRNAamplicon fragment from | 99A | 0.001592357 | 1 |
| gb|GU548354.1| Uncultured bacterium clone F1Q32TO06G2XSI 16S | 100A | 0.003184713 | 2 |
| gb|EU465058.1| Uncultured bacterium clone AFYEL_aaj67d08 16S | 101A | 0.003184713 | 2 |
| gb|FJ004759.1| Uncultured bacterium clone M1R20 16S | 102A | 0.001592357 | 1 |
| gb|GU366823.1| Uncultured bacterium clone C2 A25 | 103A | 0.001592357 | 1 |
| gb|CP003782.1| Burkholderiapseudomallei BPC006 chromosome II, | 104A | 0.01433121 | 9 |
| gb|JQ726640.1| Frateuriaaurantia 16S ribosomal RNA | 105A | 0.001592357 | 1 |
| gb|HQ322850.1| Uncultured bacterium clone W5-12 16S | 106A | 0.001592357 | 1 |
| gb|EF018783.1| Uncultured bacterium clone Amb_16S_1246 16S | 107A | 0.001592357 | 1 |
| gb|HM990012.1| Uncultured bacterium clone U12 16S | 108A | 0.001592357 | 1 |
| gb|JQ692176.1| Burkholderia sp. RR11 16S ribosomal | 109A | 0.001592357 | 1 |
| emb|FR687637.1| Uncultured bacterium partial 16S rRNA | 110A | 0.00477707 | 3 |
| gb|HQ684418.1| Uncultured bacterium clone OI2132 16S | 111A | 0.001592357 | 1 |
| emb|CU234118.1| Bradyrhizobium sp. ORS278,complete sequence | 112A | 0.001592357 | 1 |
| gb|HQ264456.1| Uncultured bacterium clone SCD330 16S | 113A | 0.003184713 | 2 |
| gb|EU662545.1| Uncultured bacterium clone MC1B_16S_181p 16S | 114A | 0.001592357 | 1 |
| gb|HQ706109.1| Burkholderiasilvatlantica strain AB284 16S | 115A | 0.001592357 | 1 |
| gb|JN412269.1| Uncultured Oxalobacteraceae bacterium clone CM67 | 116A | 0.001592357 | 1 |
| gb|JN172799.1| Uncultured soil bacterium clone em_emp427 | 117A | 0.001592357 | 1 |
| gb|HM580555.1| Uncultured bacterium clone cs1H11 16S | 118A | 0.001592357 | 1 |
| emb|AJ292885.1| uncultured eubacterium WR828 partial 16S | 119A | 0.001592357 | 1 |
| gb|JX091743.1| Uncultured bacterium clone BAC27A5 16S | 120A | 0.003184713 | 2 |
| gb|GQ376973.1| Uncultured bacterium clone PI_C12 16S | 121A | 0.001592357 | 1 |
| ref|XM_001977801.1| Drosophila erecta GG19261 (Dere\GG19261), mRNA | 122A | 0.001592357 | 1 |
| gb|GU599104.1| Uncultured soil bacterium clone HB_Ca_M_182 | 123A | 0.001592357 | 1 |
| gb|GU172206.1| Uncultured bacterium clone DSM-R34 16S | 124A | 0.001592357 | 1 |
| gb|JX391481.1| Uncultured bacterium clone N0045 16S | 125A | 0.027070064 | 17 |
| gb|JF402916.1| Uncultured soil bacterium clone GO0VNXF07IGU40 | 126A | 0.001592357 | 1 |
| gb|HQ904139.1| Uncultured bacterium clone sa0.62 16S | 127A | 0.001592357 | 1 |
| gb|JF809205.1| Uncultured bacterium clone CPf2-B2 16S | 128A | 0.001592357 | 1 |
| gb|EU800550.1| Uncultured bacterium clone 2C228685 16S | 129A | 0.001592357 | 1 |
| emb|AJ292905.1| uncultured eubacterium WR8101 partial 16S | 130A | 0.001592357 | 1 |
| gb|JF829562.1| Uncultured bacterium clone M2_284 16S | 131A | 0.001592357 | 1 |
| emb|FR687715.1| Uncultured bacterium partial 16S ribosomal | 132A | 0.001592357 | 1 |
| gb|FJ178166.1| Uncultured bacterium clone TY-F-II-OTU3 16S | 133A | 0.003184713 | 2 |
| gb|JN172666.1| Uncultured soil bacterium clone eb_ebp111 | 134A | 0.001592357 | 1 |
| gb|CP003169.1| Mycobacterium rhodesiae NBB3, complete genome | 135A | 0.001592357 | 1 |
| gb|EU046591.1| Klebsiellapneumoniae strain ECU-21 genomic | 136A | 0.001592357 | 1 |
| gb|HQ397045.1| Uncultured Bacillus sp. clone HAHS13.81 | 137A | 0.001592357 | 1 |
| gb|EU335380.1| Uncultured bacterium clone BacC-u_034 16S | 138A | 0.001592357 | 1 |
| gb|JQ514083.1| Bradyrhizobium sp. R34_Vidisha 16S ribosomal | 139A | 0.001592357 | 1 |
| gb|JN168234.1| Uncultured bacterium clone WLBL437 16S | 140A | 0.001592357 | 1 |
| emb|AM773608.1| Uncultured Nitrosovibrio sp. partial 16S | 141A | 0.001592357 | 1 |
| gb|HQ433572.1| Uncultured bacterium clone GOP_V 16S | 142A | 0.003184713 | 2 |
| gb|HQ684238.1| Uncultured bacterium clone OI1112 16S | 143A | 0.001592357 | 1 |
| gb|EF073963.1| Uncultured Acidobacteria bacterium clone GASP-WB2... | 144A | 0.001592357 | 1 |
| gb|AY571493.1| Uncultured bacterium clone RsaHw485 16S | 145A | 0.031847134 | 20 |
| gb|JN178272.1| Uncultured bacterium clone TX2_4M08 16S | 146A | 0.001592357 | 1 |
| gb|HM688413.1| Uncultured bacterium clone GB7N87001BH8QO small | 147A | 0.001592357 | 1 |
| gb|JX255150.1| Uncultured bacterium clone abscm03.0.46 16S | 148A | 0.001592357 | 1 |
| gb|JQ178187.1| Uncultured Thermoanaerobacterales bacterium clone... | 149A | 0.001592357 | 1 |
| gb|JQ655798.1| Uncultured bacterium clone N24 16S | 150A | 0.00477707 | 3 |
| gb|JQ820144.1| Uncultured bacterium clone TP16S-64 16S | 151A | 0.003184713 | 2 |
| gb|DQ202202.1| Uncultured bacterium clone CJRC180 16S | 152A | 0.001592357 | 1 |
| gb|HQ598830.1| Uncultured Acidobacteria bacterium clone SEW_08_1... | 153A | 0.003184713 | 2 |
| gb|JN177845.1| Uncultured bacterium clone TX2_1F13 16S | 154A | 0.001592357 | 1 |
| gb|DQ415833.1| Uncultured bacterium clone zEL40 16S | 155A | 0.001592357 | 1 |
| gb|EU637673.1| Uncultured bacterium clone 2-58 16S | 156A | 0.001592357 | 1 |
| gb|FJ231156.1| Uncultured bacterium clone Simba-s-1 16S | 157A | 0.007961783 | 5 |
| gb|JX286412.1| Uncultured bacterium clone SW-5B_D09 16S | 158A | 0.003184713 | 2 |
| gb|FJ475454.1| Uncultured Acidobacteriaceae bacterium clone Ahed... | 159A | 0.001592357 | 1 |
| gb|GQ302576.1| Uncultured Acidobacterium sp. clone sw-xj126 | 160A | 0.001592357 | 1 |
| gb|DQ814516.1| Uncultured bacterium clone aaa30c07 16S | 161A | 0.00477707 | 3 |
| gb|JQ649209.1| Uncultured Ralstonia sp. clone PRS1-61 | 162A | 0.003184713 | 2 |
| gb|GU458296.2| Streptomyces sp. 145(2010) 16S ribosomal | 163A | 0.001592357 | 1 |
| gb|JX133661.1| Uncultured bacterium clone WB123 16S | 164A | 0.003184713 | 2 |
| gb|FJ178119.1| Uncultured bacterium clone TY-D-I-OTU5 16S | 165A | 0.001592357 | 1 |
| gb|JX255255.1| Uncultured bacterium clone abscm03.0.575 16S | 166A | 0.001592357 | 1 |
| gb|FJ405890.1| Planctomycetacia bacterium WSF3-27 16S ribosomal | 167A | 0.001592357 | 1 |
| gb|JN911190.1| Uncultured microorganism clone GF13U7304I5R3Y 16S... | 168A | 0.001592357 | 1 |
| gb|HQ322962.1| Uncultured bacterium clone W9-11 16S | 169A | 0.001592357 | 1 |
| gb|JQ684492.1| Uncultured Rhodoferax sp. clone deep95 | 170A | 0.001592357 | 1 |
| gb|EF588371.1| Uncultured Acidobacteria bacterium clone WSD-045 | 171A | 0.00477707 | 3 |
| gb|FJ193705.1| Uncultured Ralstonia sp. clone GI1-Mcs-G07 | 172A | 0.078025478 | 49 |
| gb|JQ690672.1| Uncultured bacterium clone MIG-B19 16S | 173A | 0.001592357 | 1 |
| gb|JX174263.1| Burkholderia sp. 2386 16S ribosomal | 174A | 0.003184713 | 2 |
| gb|HM108406.1| Uncultured Clostridia bacterium clone SHAI049 | 175A | 0.001592357 | 1 |
| gb|JX133575.1| Uncultured bacterium clone FB31 16S | 176A | 0.001592357 | 1 |
| gb|EF018259.1| Uncultured Verrucomicrobia bacterium clone Amb_16... | 177A | 0.001592357 | 1 |
| dbj|AB188624.1| Uncultured bacterium gene for 16S | 178A | 0.001592357 | 1 |
| gb|EU381918.1| Uncultured rumen bacterium clone P5_C17 | 179A | 0.001592357 | 1 |
| gb|HM138688.1| uncultured bacterium clone GQ25 genomic | 180A | 0.001592357 | 1 |
| gb|FJ712828.1| Uncultured bacterium clone Cvi1 16S | 181A | 0.003184713 | 2 |
| gb|JX172662.1| Uncultured bacterium clone PB17024-1_H03 16S | 182A | 0.001592357 | 1 |
| gb|HQ121355.1| Uncultured bacterium clone G40 16S | 183A | 0.001592357 | 1 |
| gb|JN172776.1| Uncultured soil bacterium clone em_ems414 | 184A | 0.052547771 | 33 |
| gb|JN868802.1| Uncultured bacterium clone MW47 16S | 185A | 0.001592357 | 1 |
| gb|GU931381.1| Lysobacter sp. RB-31 16S ribosomal | 186A | 0.001592357 | 1 |
| gb|HQ674949.1| Uncultured Acidobacteria bacterium clone MWM2-75 | 187A | 0.001592357 | 1 |
| gb|HQ433573.1| Uncultured bacterium clone GOP_H 16S | 188A | 0.027070064 | 17 |
| gb|JN168386.1| Uncultured bacterium clone WLCLC404 16S | 189A | 0.001592357 | 1 |
| gb|CP000494.1| Bradyrhizobium sp. BTAi1, complete genome | 190A | 0.001592357 | 1 |
| gb|JQ770095.1| Uncultured bacterium clone YT-47 16S | 191A | 0.001592357 | 1 |
| gb|CP002299.1| Frankia sp. EuI1c, complete genome | 192A | 0.00477707 | 3 |
| gb|JF829579.1| Uncultured bacterium clone M2_247 16S | 193A | 0.085987261 | 54 |
| gb|JQ389743.1| Streptomyces tanashiensis strain BAB1573 16S | 194A | 0.001592357 | 1 |
| gb|HQ684430.1| Uncultured bacterium clone OI2144 16S | 195A | 0.001592357 | 1 |
| gb|AY773105.1| Uncultured bacterium clone 133 16S | 196A | 0.00955414 | 6 |
| gb|JX133612.1| Uncultured bacterium clone WB19 16S | 197A | 0.001592357 | 1 |
| dbj|AB473920.1| Uncultured endolithic bacterium gene for | 198A | 0.001592357 | 1 |
| gb|JX172085.1| Uncultured bacterium clone PB17012-2_G11 16S | 199A | 0.003184713 | 2 |
| gb|FJ936857.1| Uncultured bacterium clone kab140 16S | 200A | 0.001592357 | 1 |
| gb|JQ349505.1| Bradyrhizobium sp. DW6.4 16S ribosomal | 201A | 0.001592357 | 1 |
| gb|GU598779.1| Uncultured soil bacterium clone HB_R_M_105 | 202A | 0.001592357 | 1 |
| gb|JF800677.1| Uncultured bacterium clone BT41 16S | 203A | 0.00477707 | 3 |
| gb|HQ852986.1| Uncultured bacterium clone C8 16S | 204A | 0.003184713 | 2 |
| gb|JF412274.1| Lysobacter sp. ATCC 53042 lysobactin | 205A | 0.001592357 | 1 |
| gb|EF588337.1| Uncultured Acidobacteria bacterium clone WSD-011 | 206A | 0.001592357 | 1 |
| gb|JX240938.1| Uncultured Planococcus sp. clone ONGS77 | 207A | 0.001592357 | 1 |
| gb|EU471806.1| Uncultured bacterium clone AE2_aaa02a11 16S | 208A | 0.003184713 | 2 |
| gb|GU482873.1| Uncultured bacterium clone F1Q32TO05GFAJR 16S | 209A | 0.001592357 | 1 |
| gb|JQ183105.1| Uncultured bacterium clone 10 16S | 210A | 0.001592357 | 1 |
| gb|EF018834.1| Uncultured bacterium clone Amb_16S_1315 16S | 211A | 0.001592357 | 1 |
| gb|JN428405.1| Uncultured organism clone SBXY_2668 16S | 212A | 0.003184713 | 2 |
| gb|HM581210.1| Uncultured bacterium clone mg6H04 16S | 213A | 0.001592357 | 1 |
| gb|CP002521.1| Acidovoraxavenae subsp. avenae ATCC | 214A | 0.001592357 | 1 |
| gb|HQ684408.1| Uncultured bacterium clone OI2121 16S | 215A | 0.001592357 | 1 |
| gb|JQ684400.1| Uncultured bacterium clone HWGB-142 16S | 216A | 0.003184713 | 2 |
| gb|HQ397486.1| Uncultured bacterium clone BSS101 16S | 217A | 0.001592357 | 1 |
Table 1: Report on the frequency ( counts/total number of reads of each species).
Results and Discussion
The study of identification of bacteria for the biodegrading capabilities is important in microbial ecology, especially with molecular techniques. In particular, analysis of the microbial communities that take part during in-situ hydrocarbon biodegradation activities has been a challenge to microbiologist. Interest in this area has been catalyzed by the rapid advancement of molecular ecological methodologies. Thus, the ability of an organism to degrade a specific substrate and persist within such environment is clear evidence that its genome harbored the relevant degrading gene [7]. The previous studies by Bindu and Satish [8] and Jyothi et al. [9] on hydrocarbon degradation by bacteria reveal that catechol 2, 3 dioxygenase is one of the enzyme involved in hydrocarbon degradation.
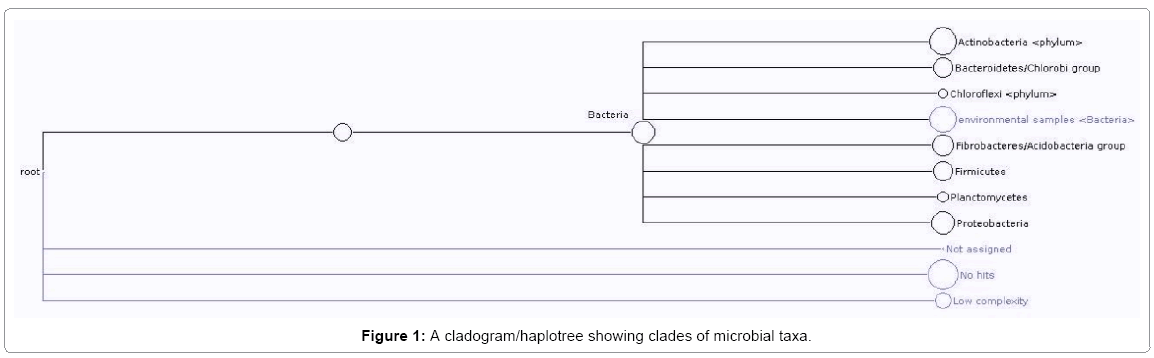
Molecular confirmation of similarities in microbial Identities was obtained by creating different dendrograms/distance trees. Gene sequencing carried out read 513 different nucleotide sequences. Every read was BLASTED and the result file saved. The report on the frequency of reads of each species is as shown in Table 1. Seven phylum with 47 corresponding culture-dependent species and 169 culture-independent bacteria clone was obtained. The resultant haplotree/cladogram however, showed clades of proteobacteria (b-and g-proteobacteria), bacteria/enterobacteria, firmicutes, plantomycetes, acidobacteria group/ fibrobacteres, Bacteriodetes/chlorobi, Actinobacteria/high G+C and chloriflexi phyla (Figure 1). The nucleotide sequences with no hit was sent to Genbank for asigning of ascension number. The isolation of the aforementioned organisms from crude oil polluted agricultural soil left for four years, depict that the organism probably, have degradative genes coding for enzymes for hydrocarbon catabolism which aided their survival. These have been confirmed by the presence of plasmid DNA in culture - dependent isolates obtained and published elsewhere [10].
Taxonomical classification and percentage diversity
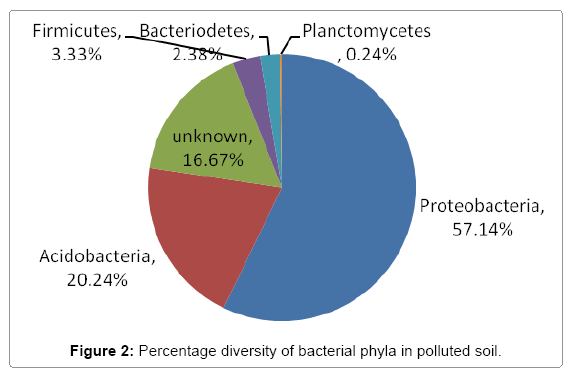
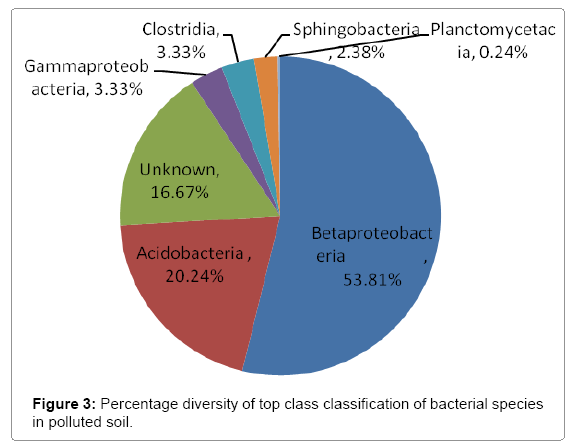
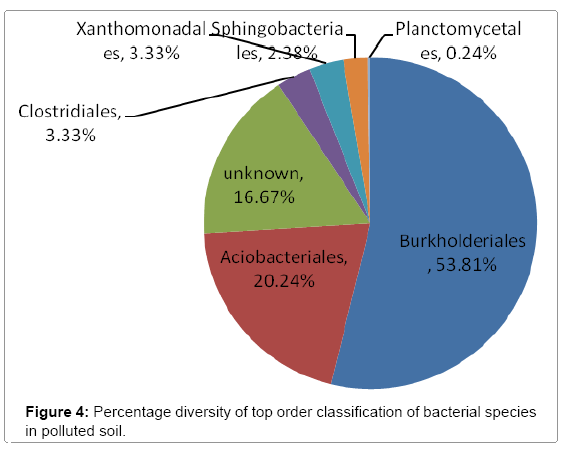
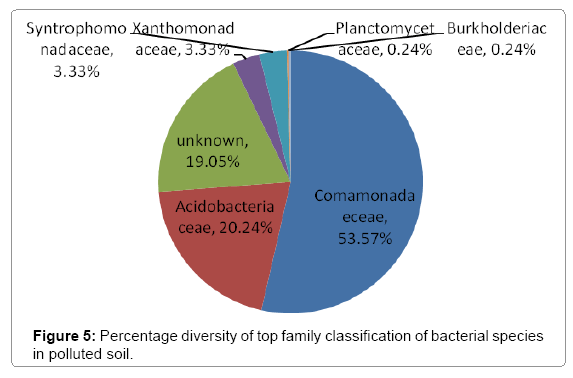
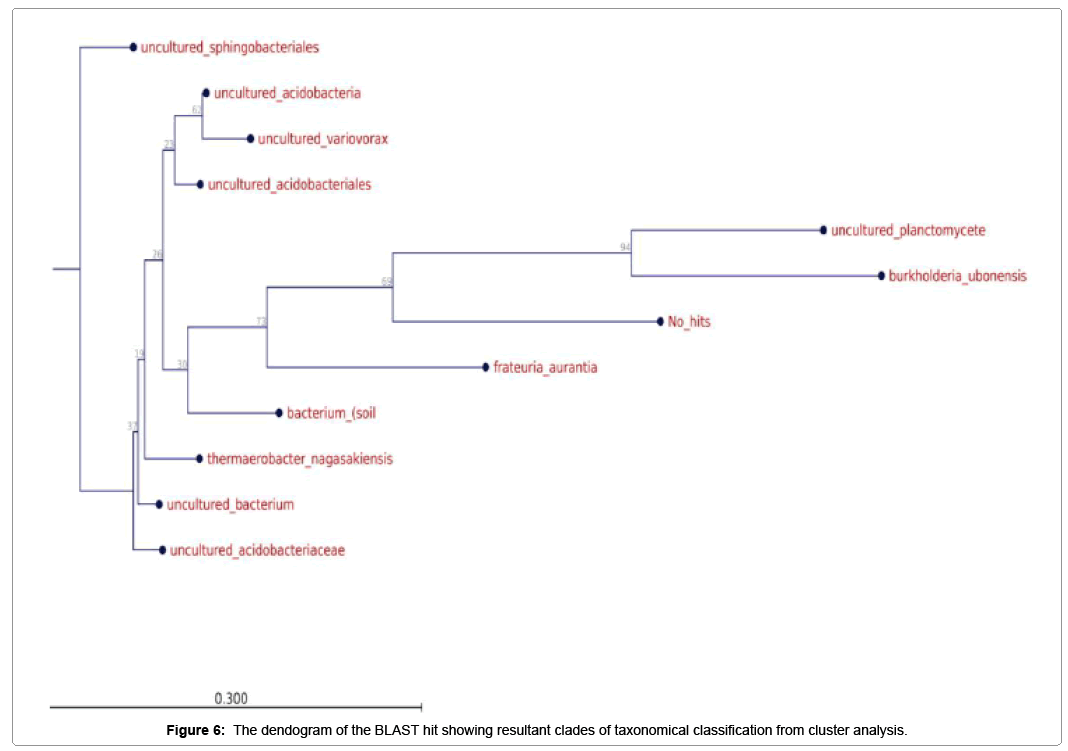
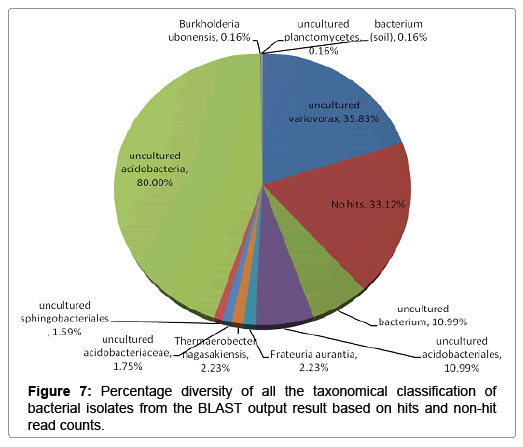
Further taxonomical analysis was carried out with reads of sufficient Q scores (>q30) and lengths and a total of 420 read count of top kingdom classification of 100% bacteria kingdom was obtained. Top phyla classification depict that that Proteobacteria had the highest diversity of 57.14% followed by Acidobacteria (20.24%), Unknown (16.67%), Firmicutes (3.33%), Bacteroidetes (2.38%), and Planctomycetes (0.24%) in that order (Figure 2). Top class and order classification of phylum proteobacteria, class beta proteobacteria and order Burkholderiales also had similar highest values of 53.81% (Figure 3 and Figure 4) whereas in top family classification, Burkholderiaceae recorded the lowest diversity of 0.24% (Figure 4). Furthermore, the family of unknown increased by 2.38% while diversity of Acidobacteria phyla, class, order and family remained constant (20.24%). Generally, the dendogram of the BLAST hit showing resultant clades with their leaves and height of the branch points indicating the similarity and differences of isolates from each other (the greater the height, the greater the difference) as well as the percentage diversity of all the taxonomical classification of bacterial isolates from the BLAST output result based on hits and non-hit read counts are shown in Figure 6 and Figure 7 respectively. The result (Figure 5) depict that about 33.12% had no hit.
Indeed, microbial degradation is the major and ultimate natural mechanism by which one can clean up the petroleum hydrocarbon pollutants from the environment [10,11]. The recognition of biodegraded petroleum hydrocarbons in the environment as observed in previous studies [2,3,9,12,13], which was evident through detectable biodegradation of n-alkane profile of the crude oil by microorganisms supports the findings of this study. The microbial genera, namely, Arthrobacter, Alcaligenes, Burkholderia, Mycobacterium, Micrococcus, Pseudomonas, Acinetobacter, Bacillus, Sphingomonas, Corynebacterium and Rhodococcus have been incriminated to be involved in hydrocarbon degradation as observed in the percentage diversity of the taxonomical classification in this study; as these organisms fall within similar identified phyla, class, order and family of bacterial isolate during this metagenomic analysis. From the findings of previous studies and in line with this study, bacteria are the most active agents in petroleum degradation, and they work as primary degraders of spilled oil in the environment having in them enzymes for hydrocarbon degradation. This corroborates the report made by Rahman et al. [14] and Brooijmans et al. [15] who studied on hydrocarbon degrading bacterial in petroleum sludge. The persistence of the identified bacterial isolates in this study could also be due to the ability of the isolates to produce bio surfactants which aids in the formation of micelles to enhance uptake of hydrocarbons. Studies have also shown that total bacteria population in polluted soil are more than that in unpolluted soil [16-18] which implies that those organisms are the active degraders of that oil.
Metagenomic analysis carried out in this study have actually helped in detection of Acidobacteria phyla which are under- represented in culture even though they are physiologically diverse and ubiquitous as well as so many uncultured genera. The low diversity of Planctomycetes is not surprising since they are aquatic bacteria phyla and are found in samples of brackish, marine and fresh water. Further molecular studies are therefore needed to detect specific catabolic genes resident in these hydrocarbon degrading isolates. This can help to produce superbugs required for faster remediation, cost effective and efficient bioremediation protocol for Nigerian oil polluted soil.
Conclusion
The isolation of the aforementioned organisms from crude oil polluted agricultural soil left for four years, depict that the organism probably, have degradative genes which aided their survival.
1http://www.lahey.org/studies/webt.html
2http://blast.ncbi.nlm.nih.gov/Blast.cgi
References
- Chan Y (2006) Utilization of simple phenolics for dinitrogen fixation by soil diazotrophic bacteria.Plant and soil 90: 141-150.
- Kuiper I, Lagendijk EL, Bloemberg GO, Lugtenberg BJJ (2004) Rhizoremediation. A beneficial plant microbe interaction.Mol Plant Microbe In 17: 6-15.
- Ogbulie TE, Nwigwe HC, Iwuala MOE, Okpokwasili GC (2010) Study on the use of Monoculture and Multispecies in Bioaugumentation of crude oil contaminated Agricultural soil. Nigerian Journal of Microbiology 24: 2160-2167.
- Ogbulie TE, Nwigwe HC, Okpokwasili GC, Iwuala MOE (2011) Comparative study on the effect of symbiotic interaction between plants and non-indigenous isolates on crude oil remediation. Analele Universitãtii din Oradea- Fascicula Biologie Tom XVIII: 15-22.
- Shi L, Muller S, Harms H, Wicks L (2008) Effect of electrokinetic transport on the vulnerability of PAH-degrading bacteria in a model aquifer. Environ Geochem Health 30: 177-182.
- Yee DC, Maynard JA, Wood TK (1998) Rhizoremediation of trichloroethylene by a recombinant root-colonizing Pseudomonas fluorescens strain expressing toluene ortho-monoxygenase constitutively. Appl Environ Microbiol 64: 112-118.
- Cowan DA, Strafford W (2007) Metagenomic methods for the identification of active microorganisms and genes in biodegradation processes. In Environmental Microbiology 3rd edition. ASM Press, USA.
- Bindu J, Satish W (1996) PCR amplification of catechol 2,3-dioxygenase gene sequences from naturally occurring hydrocarbon degrading bacteria isolated from petroleum hydrocarbon contaminated groundwater FEMS Microbiology Ecology 19: 5-15.
- Jyothi. K, Surendra Babu K, Nancy Clara K, Amita K (2012) Identification and Isolation of Hydrocarbon Degrading Bacteria by Molecular Characterization. Helix 2: 105-111.
- Atlas RM (1992) Petroleum microbiology, In Encyclopedia of Microbiology, Academic Press, Baltimore, Md, USA, pp: 363-369.
- Lal B, Khanna S (1996) Degradation of crude oil by Acinetobacter calcoaceticus and Alcaligenes Odorans. J Appl Bacteriol 81: 355-362.
- Jones DM, Douglas AG, Parkes RJ, Taylor J, Giger W, et al. (1983) The recognition of biodegraded petroleum-derived aromatic hydrocarbons in recent marine sediments. Marine Poll Bull 14: 103-108.
- Adebusoye SA, Ilori MO, Amund OO, Teniola OD, Olatope OS (2007) Microbial degradation of petroleum hydrocarbons in a polluted tropical stream. World J Microb Biot 23: 1149-1159.
- Rahman KSM, Rahman TJ, Kourkoutas Y, Petsas I, Marchant R, et al. (2003) Enhanced bioremediation of n-alkane in petroleum sludge using bacterial consortium amended with rhamnolipid and micronutrients. Bioresource Technology 90: 159-168.
- Brooijmans RJW, Pastink MI, Siezen RJ (2009) Hydrocarbon-degrading bacteria: the oil- spill clean-up crew. Microbial Biotechnology 2: 587-594.
- Okerentugba P, Ezeronye O (2003) Petroleum degrading potentials of single and mixed microbial cultures isolated from rivers and refinery effluent in Nigeria. Afr J Biotechnol 2: 288-292.
- Onifade A, Abubakar F (2007) Characterization of hydrocarbon-degrading microorganisms isolated from crude oil contaminated soil and remediation of the soil by enhanced natural attenuation. Res J Microbiol 2: 149-155.
- Boboye B, Olukunle OF, Adetuyi FC (2010) Degradative activity of bacteria isolated from hydrocarbon- polluted site in Ilaje, Ondo State, Nigeria. Afr J Microbiol Res 4: 2484-2491.
Relevant Topics
- Aquatic Ecosystems
- Biodiversity
- Conservation Biology
- Coral Reef Ecology
- Distribution Aggregation
- Ecology and Migration of Animal
- Ecosystem Service
- Ecosystem-Level Measuring
- Endangered Species
- Environmental Tourism
- Forest Biome
- Lake Circulation
- Leaf Morphology
- Marine Conservation
- Marine Ecosystems
- Phytoplankton Abundance
- Population Dyanamics
- Semiarid Ecosystem Soil Properties
- Spatial Distribution
- Species Composition
- Species Rarity
- Sustainability Dynamics
- Sustainable Forest Management
- Tropical Aquaculture
- Tropical Ecosystems
Recommended Journals
Article Tools
Article Usage
- Total views: 11471
- [From(publication date):
specialissue-2016 - Nov 21, 2024] - Breakdown by view type
- HTML page views : 10734
- PDF downloads : 737