Review Article Open Access
Molecular Diagnosis of Vector-Borne Parasitic Diseases
Carolina Hernández1 and Juan David Ramírez2*1Carolina Hernández, Red Chagas Colombia, Group of Parasitology, National Institute of Health, Bogota, Colombia
2Juan David Ramírez, Red Chagas Colombia, Group of Parasitology, National Institute of Health, Bogota, Colombia
- *Corresponding Author:
- Juan David Ramírez
Red Chagas Colombia, Group of Parasitology
National Institute of Health, Bogota, Colombia
E-mail: jdramirez85@gmail.com
Received Date: June 21, 2013; Accepted Date: July 24, 2013; Published Date: July 26, 2013
Citation: Hernández C, Ramírez JD (2013) Molecular Diagnosis of Vector- Borne Parasitic Diseases. Air Water Borne Diseases 2:110. doi:10.4172/2167-7719.1000110
Copyright: © 2013 Hernández C, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Air & Water Borne Diseases
Abstract
Vector-borne diseases still represent a serious problem in public health despite of the efforts of the surveillance and public health systems to mitigate these pathologies. Among these infections, those diseases caused by parasitic protozoa such as Chagas disease, malaria, leishmaniasis and African trypanosomiasis still represent serious issues in public health. These diseases are also named as neglected diseases affecting vulnerable populations around the world. One of the main goals in the basic research of these diseases is the deployment of accurate, reliable and feasible methods for the diagnosis of the etiological agents. Due to the rise of DNA technologies, several researchers have been able to develop rapid methods for the molecular diagnosis of vector-borne parasitic diseases. Herein we conducted a systematic review about the old-fashioned and current methodologies for the diagnosis of Chagas disease, leishmaniasis, sleeping sickness and malaria with special emphasis on molecular diagnosis to update the reader about their availability and feasibility. One of the purposes of this review is to encourage the researchers to deploy new methodologies that can be easily applied in endemic countries with no sophisticated methodology. This is imperative since an early diagnosis will enhance an early, rapid and appropriate treatment for populations that suffer these complex diseases.
Keywords
Chagas Disease; African Trypanosomiasis; Leishmaniasis; Malaria; PCR; Diagnosis; DNA
Introduction
Parasitic diseases represent one of the most important issues in public health. More than 20 million people are infected by diseasecausing parasites. Parasites represent a broad group of eukaryotic organisms that can cause severe disease in human populations, including malaria, leishmaniasis, Chagas disease, Schistosomiasis and Filariasis. Closely related to these parasitic diseases are geographic, social and economic factors that modulate the prevalence and incidence of these pathologies [1]. The use of technologies based on molecular biology has allowed the scientific community to unravel the disease determinants and the genetic structures of some parasites that provoke disease and cause millions of deaths each year.
The vector-borne parasitic diseases constitute a group of pathologies caused by eukaryotic pathogens transmitted by arthropods such as Anopheles, sand flies, triatomines and tsetse flies. Malaria, Chagas disease, African trypanosomiasis and leishmaniasis are the most widespread and devastating of the vector-borne parasitic diseases due to their high prevalence and the amount of deaths they cause each year. These pathologies are distributed at a wide-range geographical level affecting vulnerable and non-vulnerable human populations. Due to their relevance in terms of epidemiological trends, the diagnosis of these infections and the deployment of new diagnostic methods are needed to identify and treat the symptoms of these diseases, or to treat the diseases themselves. The rise of molecular techniques and DNA detection has played a prominent role in detecting infections, treatment follow-ups, disease installations and infection outcomes. Herein, we conducted a systematic review of the former and current techniques employed for the diagnosis of vector-borne parasitic diseases to update the scientific community with regard to potential areas of basic and applied research in Malaria, African trypanosomiasis, Chagas disease and Leishmaniasis. We are confident that the use of high-throughput whole genome sequencing technologies will facilitate the deployment of novel techniques of potential benefit in the public health system.
Malaria
Malaria is considered the most relevant parasitic infection worldwide, and its treatment is one of the biggest public health challenges for developing countries. In 2010, an estimated 219 million cases of malaria occurred worldwide, with an accompanying 660.000 deaths resulting there from. Most deaths (91%) occurred in the African Region, however, Asia and Latin America, as well as parts of the Middle East and Europe has also been affected [2]. This disease is produced by 5 species of the genus Plasmodium: Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae, Plasmodium ovale and Plasmodium knowlesi. Each species is associated with a different geographical distribution. The parasite is spread to people through the bites of infected Anopheles mosquito vectors. Transmission also depends on climatic conditions, such as rainfall patterns, temperature and humidity. In many places, transmission is seasonal, with peak rates of transmission occurring during and just after the rainy season [3]. Another important factor in the transmission of malaria is human immunity among adults who are capable of developing partial immunity in areas of moderate or intense transmission given the high rate of exposure to the disease. Consequently, those with the greatest risk of contracting malaria in locations of moderate or intense transmission (namely Africa), are young children, whereas all age groups are at risk for infection in areas with less transmission and low immunity [2].
Malaria is an acute febrile disease with an incubation period of 7 days or more after the initiating infective mosquito bite. Initial symptoms may be mild, such as fever, headache, chills and vomiting, but these symptoms are not easily recognized as malaria. Of the five parasite species that cause malaria in humans, P. falciparum, is the most severe, and thus most deadly. Therefore, a proper diagnosis of infection brought on by P. falciparum is especially important. Among the clinical manifestations of severe malaria, we have identified the following: severe anemia, cerebral malaria, renal failure, acute respiratory distress syndrome, acidosis and cardiopulmonary complications [4]. In addition, other species, like P. vivax and P. ovale, can produce clinical relapses that occur weeks to months after the first infection as a result of their dormant liver forms, known as hypnozoites [5]. The number of methods for malaria diagnosis is limited. The first step for diagnosis is the clinical diagnosis, which mainly takes into account the presence of fever in patients who have been in endemic areas. After such a finding, additional tests follow.
Manual microscopic examination of blood slide (thick and thin films) is the gold standard for detecting malaria. This method is widely used in most endemic zones because it does not require a sophisticated infrastructure [6]. In addition, it demonstrates high sensitivity (approximately 90%) [7], is the most economic of malariadetecting tests, makes species identification possible, permits definition of the parasitemia and facilitates the follow-up of therapy with antimalarial drugs [8-10]. However, the test also holds some disadvantages, being both time and labor intensive, as well as requiring skilled and experienced microscopists to ensure precise diagnoses [10,11]. In recognition of the limitations of microscopic examination of blood slides, alternative techniques for the diagnosis of malaria have been developed. For example, fluorescent microscopy [12], including: Quantitative Buffy Coat (QBC) [13,14], Kawamoto Acridine-Orange process [15,16]; and the benzothiocarboxypurine (BCP) procedure [17,18]. However, these techniques require a fluorescent microscope and their implementation is limited to a small number of laboratories.
Recently, the attention of researchers has focused mainly on the detection of antigens or enzymes by Malaria Rapid Diagnostic Tests (MRDTs). Currently, two antigens are used: Histidine-rich protein-2 (HRP-2), which is only produced by P. falciparum, and the parasite lactate-dehydrogenase (pLDH) antigen, produced by all five Plasmodium species infecting humans. Both of these antigens are secreted into the blood by all asexual stages of the parasite [19,20]. MRDTs utilizing immune chromatographic lateral-flow-strip technology were introduced in the early 1990’s [8] and remain in use today [10]. These assays involve the mixing of the clinical sample with lysing agents, buffer, and antibodies (capture antibodies and detection antibodies). This mix later migrates across the surface of a nitrocellulose membrane. The antibodies captured are sprayed by machine as a stripe onto the nitrocellulose membrane and bind to the membrane in an immobile phase. The parasite antigen is fixed from sample. The detection antibodies are conjugated to an indicator in a mobile phase and bind to the parasite antigen that has been captured by the capture antibodies on the membrane, producing a visible line if the targeted antigen is present in the clinical sample [10,19].
MRDTs have several advantages. They are performed with little skill and quickly produce results that are easily interpreted. Additionally, they do not require refrigeration, are reproducible and permit species differentiation. The last enumerated advantage is very important for providing adequate treatment. MRDT (HRP-2) detects P. falciparum at a rate of sensitivity of 95.0% and at a rate of specificity of 95.2%, whereas the sensitivity and specificity for pLDH detection is 93.2 and 98.5%, respectively [21]. However, this assay also has limitations. Mass production and widespread use has led to an inability to perform adequate quality control of its manufacturing, leading to false diagnoses [10].
Molecular tools such as nucleic acid amplification tests have been performed for malaria diagnosis despite some disadvantages of microscopy, including the inability to identify lower levels of parasitemia due to a detection limit that hovers being 50 to 100 parasites/μl [22] and the inability to identify MRDTs due to the lack of the hrp-2 gene in some P. falciparum parasites [23]. From the first assays of PCR in 1990 to the present, it has been used to target and amplify the multicopy small subunit 18S rRNA gene (18S rRNA) that is found in all Plasmodium species. This marker presents hundreds of identical copies in the genome and is present in 4-8 divergent copies, depending upon the species [24]. Several types of analysis have been used to differentiate species, including Southern blot and restriction length polymorphism assays. This marker has been used in a fragment polymorphism (RFLP) analysis or Nested PCR [25]. This technique was subsequently modified using a multiplex format in the second reaction [26-28]; even using blood samples collected in filter papers [29].
Despite that PCR techniques demonstrated better detection of low parasitemias (<1 parasite/μL) and mixed infections than microscopy, the method did not allow quantification of parasitemia [22]. Therefore, to improve molecular diagnosis, a multiplex real-time PCR was developed with the addition of species-specific fluorescent probes [30-32] and other assays of real-time that performed amplification of 18S rRNA with Sybr Green and species identification by high resolution melting analyses [33]. These real-time PCR tests improved the sensitivity and specificity, reduced the risk of cross contamination, developed results more quickly than conventional PCR, and allowed for the quantification of DNA and, therefore, therapeutic monitoring [34].
However, these molecular tools are limited in laboratories located in endemic zones. Therefore, the latest advances have focused on the development of Loop-mediated isothermal amplification technique (LAMP). LAMP performs amplification that causes the products to fold into looped structures, which in turn produce turbidity in the reaction mixture facilitating immediately the production of the result of the lecture reading by visual inspection of the turbidity [35] or the fluorescence by Sybr Green [24]. The target used has been 18S rRNA, evidencing 98. 5% sensitivity and 94.3% specificity compared to microscopy [36]. Therefore, LAMP is promising as a test because of its sensitivity, speed and fewer performance requirements than other molecular tests. However, the conduction of clinical trials to validate and optimize its utility for diagnosis is necessary.
Due to the genome sequencing of the P. falciparum, P. vivax and P. ovalespecies [37,38], microarrays have been designed to detect drug resistance markers and genetic virulence factors [39,40]. Also, Laser- Desorption Mass Spectrometry (LDMS) is a powerful tool that has been suggested for infection screening due to its ability to identify hemozoin (Malaria pigment) produced by malaria parasites [41,42]. This test was evaluated for malaria diagnosis in asymptomatic pregnant women in Zambia and it demonstrated a rate of sensitivity at 52% and a rate of specificity at 92% [43]. However, these techniques are limited to reference laboratories and are very expensive.
Chagas Disease
Chagas Disease, or American Trypanosomiasis, is a complex anthropozoonosis caused by Trypanosoma cruzi. It is considered a zoonotic and neglected disease that represents a pressing public health problem in the Americas, where 7.7 to 10 million people are infected [44]. In addition to its concerning presence in the Americas, Chagas disease is expanding to other continents, such as Australia, North America and Europe, becoming a serious issue in non-endemic countries [45]. The main mechanism of transmission of Chagas disease is through contact with triatomine feces, although other methods of transmission exist including blood transfusion, organ transplantation, oral and congenital transmission, and laboratory accidents [46]. Chagas disease develops in two phases. The first is an acute phase that normally is asymptomatic, or that can present symptoms around 8 to 10 days after invasion by the parasite as febrile illness or signs of portal of entry of T. cruzi (Chagom in the skin or Romaña’s sign in ocular mucosa). After some years (10-30 years), about 30-40% of the patients in acute phase develop symptoms of the chronic phase. These symptoms can be cardiac type (Cardiomyopathy), digestive (megaesophagus/megacolon) or mixed (cardiodigestive) [46].
T. cruzi displays a relevant genetic variability, different cycles of transmission and the possibility of recombination exchanges. Therefore, based on different molecular markers and biological features, the taxon has been classified into six Discrete Typing Units (DTU’s) or near-clades from TcI to TcVI [47]. The DTUs are associated with different geographical distributions and are present in diverse ecotopes. Additionally, some studies have detected a correlation with clinical manifestations and parasite DTU [47-49]. These previously mentioned aspects of the genetic variability of the parasite are very important for a proper diagnosis.
The diagnosis of Chagas disease is different with respect to the phase of the disease. During the acute phase, the parasitemia is elevated, therefore the aim of the diagnosis is detection of trypomastigotes in blood, mainly by microscopic tests such as direct parasitological fresh blood test and smear and thick drop blood tests, and concentration tests such as microhematocrite or Strout that demonstrate sensitivity rates of 80 to 90% in patients with Chagas disease [50-52]. In addition, other methods can be used, including Hemoculture and Xenodiagnosis, but despite that these methods allow isolation and detection of parasites, they fail to provide immediate results.
In the chronic phase, the parasitaemia is scarce, therefore methods like hemoculture and Xenodiagnosis have low sensitivity (20-50%) in the chronic phase [53,54]. Instead, the diagnosis of Chagas disease in the chronic stage is perfomed with serological tests due to the necessity of detecting IgG antibodies against T. cruzi antigens. The most common serological methods used are the conventional serological tests that include, primarily, Immunofluorescence Assays (IFA), Enzyme-Linked Immunosorbent Assays (ELISA) and indirect hemagglutination assays (IHA), these assays can employ recombinant and/or crude antigenic T. cruzi preparations [55-57]. Some authors have reported that the conventional tests prepared with crude antigenic T. cruzi preparation scan produce a certain number of false positives and false negatives [58,59], whereby the use of recombinant antigens is promising. However, recent systematic reviews report conventional- ELISA sensitivity and specificity of 97.7% and 96.3%, respectively and for ELISA-Recombinant 99.3% sensitivity and 97.5% specificity [60,61].
The use of recombinant antigens has allowed the development of diagnosis by Rapid Diagnosis Tests (RDTs) for antibodies detection. RDTs offer multiple advantages: they are relatively easy to perform, require minimal amounts of sample, impose only a short time delay, and require only minimal amount of trained staff to execute them smoothly in soil lacking laboratory facilities given their easy handling. These tests have been evaluated in several studies in different countries finding high sensitivity and specificity [57,62-68]. Immunoblot assays have been also conducted; among these the most important is TESAblot that consists of detection of antibodies against the antigens TESA (trypomastigote excreted-secreted antigen). This assay is currently commercialized [55,69-71] and has been widely used because of its high sensitivity and specificity and ability to resolve doubtful serology and cross-antigenicity issues with related protozoan parasites in regions where the disease is endemic. The TESA antigen assay also has been used for other tests like ELISA [72].
Molecular tools have been applied to the diagnosis of Chagas disease; PCR is the main molecular tool and allows the diagnosis and discrimination of the DTU’s in acute and chronic phases. However, its usefulness is primarily focused in the chronic phase, congenital Chagas diagnosis and post therapeutic follow-up due to its ability to detect low parasitemias. Several targets for the detection of T. cruzi by PCR have been widely used, including: Mini-exon gene, kDNA minicircle constant regions, 195 bp and 1025 bp Nuclear repetition, Flagellum protein gene and Variable domain LSU RNA [73]. However, the variable region of the minicircle kinetoplast DNA (kDNA) and a repeat tandem sequence of nuclear DNA (stDNA) of the parasite have been the most used regions as target sequences for diagnosis via PCR because of their high number of copies [55,73-75].
Some studies implementing the k-PCR reaction (polymerase chain of kinetoplast) technique demonstrated diagnosed chronic phase sensitivity of 100% and 83. 5% over serological methods [76,77]. Other studies showed that the sensitivity improved by 50% to 92.2%, with a specificity of 97.7% when guanidine hydrochloride is used for storage of samples [61]. Sensitivities of 70% were observed for kPCR and 75% for the Sat-PCR reaction (Polymerase Chain satellite) as compared with TESA-blot serological test [55]. Other studies that used k-PCR for monitoring patients post-treatment and for monitoring animal models showed that this technique is much more sensitive than xenodiagnosis and Hemoculture [53,55,78,]. Additionally, the PCR and qPCR have been widely used in studies to follow-up post therapeutic efficacy [79-85]. These studies showed that the PCR is a useful tool that can be used as an early marker of chemotherapy failure before serological reversal [55,79-86]. In fact, currently a clinical trial is being conducted using PCR as a technique for monitoring treatment efficacy with benznidazole [87].
Recently, in order to perform a validation of PCR procedures for detection of T. cruzi DNA in human blood samples, a multicentric international study was performed in which PCR techniques used by different PCR laboratories from 16 countries were compared. This study evidenced high variability among laboratories and methods confirming that the lack of standardization led to poor reproducibility, precluding the possibility of comparing findings among different laboratories. However, four methods demonstrated the best rates of sensitivity between 83.3-94.4%, specificity of 85-95%, accuracy of 86.8-89.5% and kappa index of 0.7-0.8 compared to consensus PCR reports of the 16 good performing tests which demonstrated respective rates at 63-69%, 100%, 71.4-76.2% and 0.4-0.5, as compared to serodiagnosis. Furthermore, some methods showed an important reduction of the analytical sensitivity when spiked blood samples were tested in comparison to purified parasite DNA, suggesting that the DNA purification step is crucial for the PCR yield and the necessity of using an internal amplification control [88]. These results generated the standardized and validated multiplex Real-Time PCR using TaqMan probes targeting the Satellite DNA [89,90]. In addition, there have been designed Real-Time PCR methods using TaqMan probes for other targets such as kDNA and 18S rRNA [91]. Also, it was observed that the genetic variability could affect the molecular and serological diagnosis. Different algorithms have been deployed for the molecular characterization of DTU’s by performing RAPD, PCR-RFLP, qPCR, MLST, MLMT and DNA sequencing analyses [49,92-96]. However, new algorithms have been developed for an accurate and reliable discrimination of T. cruzi DTU’s [49,97] even though such identification has been achieved through the implementation of a High-Resolution Melting (HRM) genotyping assay that offers the highest rates of specificity and sensitivity, high performance, and low cost as compared with previously described characterization methods (Figure 1) [98].
African Trypanosomiasis
African trypanosomiasis (sleeping sickness) is caused by the infection of two subspecies of the complex Trypanosoma brucei (T. brucei gambiense and T. brucei rhodesiense) and transmitted by the bite of the tsetse fly [99]. The disease affects mostly poor populations living in remote rural areas of Sub-Saharan Africa due to tsetse fly infestation, which covers about 10 million km2 of the landmass of Africa [100]. Clinical manifestations are described as occurring in early or late stages. In the early (hemolymphatic) stage, onset 1–3 weeks after sustaining the tsetse fly bite, the parasites multiply in the blood, lymph, and peripheral tissues. At this stage, symptoms tend to be non-specific, and include malaise, headache, arthralgia, generalized weakness, and weight loss. During late (encephalitic) stage, the parasites can cross the blood– brain barrier and invade the CNS. In this late stage, symptoms can be grouped into general categories such as psychiatric, motor, and sensory abnormalities, but primarily take the form of the sleep disturbances that give the disease its name [101]. Such sleep disturbances occur in 74% of infected patients [102].
Untreated or ineffectively treated African trypanosomiasis is usually fatal. Usually, T.b. gambiense infection lasts several months or years. This infection is responsible for most reported cases, and human beings are believed to be the main host. In contrast, T.b. rhodesiense infection causes an acute disease lasting several weeks [101,103]. According to WHO, between 1999 to 2011, the reported number of new cases of the chronic form of human African trypanosomiasis (T. b. gambiense) fell by 77%, from 27.862 to 6.631. During the same period, the number of newly reported cases of the acute form of human African trypanosomiasis (T.b. rhodesiense) fell by 82% [104].
The diagnosis varies according to the subspecies. In cases produced by T.b. rhodesiense, for which screening tests is not available, diagnosis is usually conducted by direct microscopic examination of thick and thin smears of blood and lymph aspirates in patients with symptoms. Also, there are concentration methods that demonstrate increased sensitivity rates, such as the microhematocrit centrifuge technique (mHCT), quantitative buff and coat analysis, and the minianion-exchange centrifugation technique (mAECT) [105]. In cases of possible infection by T.b. gambiense, direct methods are used to search for parasites in the blood or lymphatic aspirates. However, they are ineffective since this infection is usually chronic and the parasitemia is cyclical. For that reason, the Card Agglutination Test for Trypanosomiasis (CATT), which is simple and quick to undertake, is usually useful for screening [106]. However, this test has limitations such as low sensitivity [107]. Therefore, the cases must be confirmed by lumbar puncture, which permits detection of the parasite or elevated white-blood- cell count in CSF [103].
Due to the numerous limitations in its diagnosis, a wide range of molecular amplification techniques has been developed to detect African trypanosomiasis. Techniques implemented range from FISH to gene sequencing. However, the most implemented technique is PCR. The sensitivity and specificity of a diagnostic PCR largely depends on the DNA sequence targeted by the primers. For that reason, the targets used for detection correspond to the most abundant and conserved sequences across the parasite genome. Among these we can find: fragment 177 bp of satellite DNA; expression-site-associated genes 6/7 (ESAG6/7); glycosylphosphatidylinositol phospholipase C gene (GPI-PLC);first internal transcribed spacer (ITS1); kinetoplast DNA (KDNA); paraflagellar rod protein A gene (PFRA); 18S ribosomal DNA (18S rDNA);18S ribosomal RNA (18S rRNA) and DNA Maxicircles [108-112].
In addition, other targets are used for differentiation of subspecies, such as the internal transcribed spacer (ITS1), which is amplified by conventional and nested PCR [113,114]; Trypanosoma brucei gambien-sespecific glycoprotein (TgsGP) gene, whose amplification is specific for T.b. gambiense [111,115] and serum-resistance-associated gene (SRA), whose amplification by conventional PCR [114,116] or Multiplex PCR [117] facilitate the specific identification of T.b. rhodesiense. In fact, a recent systematic review determined that the sensitivity for PCR on blood was 99.0% and the specificity 97.7% [118]. These results are due to development of modifications of the conventional PCR, such as the Real-time PCR with primers targeting the 177 bp repeat satellite DNA in T. brucei and detection by Sybr Green [119], or even better assays that use specific probes as molecular beacons probes and primers targeting 18s rRNA, facilitating specific detection of the two subspecies [120,121].
Unfortunately, the PCR does not generally exist in field conditions. Consequently, the patient populations studied were not always representative [118]. For that reason, other assays have been employed, such as a rapid test for detection of PCR products that are subsequently visualized on a dipstick in only 5 minutes through hybridization with a gold-conjugated probe (oligochromatography) [103,121] and Loopmediated isothermal amplification of DNA (LAMP), that allows rapidly amplified target DNA under isothermal conditions [122], the latter becoming a potential tool for the diagnosis in the field, especially in endemic regions [123,124]. The LAMP assay has undergone several changes, including the visualization with Sybr Green [125], and the adding of detergent to the samples prior to LAMP assay [126], considerably increasing sensitivity. Finally, molecular diagnosis in African trypanosomiasis has been successful with sensitive molecular tests that allow the detection and differentiation of the parasites. However, there are many limitations, including the facts that these tests are restricted to reference laboratories and are unavailable for the use in the field. Therefore, it is necessary to design fast, simple, economical, and reliable diagnosis and screening tests.
Leishmaniasis
Leishmaniasis is a parasitic disease caused by a complex of species of the genus Leishmania. These parasites are spread by the bite of infected sand flies occurring in two independent geographical foci; in the new world the main genus is Lutzomyia and in the old world the main genus is Phlebotomus [127]. There are several different clinical manifestations of leishmaniasis. The most common are cutaneous and visceral manifestations. The cutaneous type causes skin sores. The visceral type affects internal organs, such as the spleen, liver, and bone marrow. People with this form usually exhibit fever, weight loss, and an enlarged spleen and liver [128].
Leishmaniasis is found in regions of approximately90 countries in the tropics, subtropics, and southern Europe. This tropical disease is generally more common in rural than in urban areas. Climate and other environmental changes have the potential to expand the geographic range of the sand fly vectors and the regions of the world where leishmaniasis is found [129]. The number of new cases per year is not known with certainty. For cutaneous leishmaniasis, estimates of the number of cases range from approximately 0.7 million (700.000) to 1.2 million (1.200.000). For visceral leishmaniasis, estimates of the number of cases range from approximately 0.2 million (200,000) to 0.4 million (400,000) [127]. Additionally, infection in people is caused by more than 20 species of Leishmania parasites (L. donovani, L. infantum, L. panamensis, L. guyanensis, L. tropica, L. major, L. mexicana, L. braziliensis, L. aethiopica among others), which are spread by about 30 species of phlebotomine sand flies (Lutzomyia and Phlebotomus species) [130,131]. This epidemiological scenario demonstrates the need to pursue overall research in order to unravel the disease drivers.
Regarding the diagnosis, the techniques employed for the detection of the parasite vary from the microscopic observation of the parasite to the detection of Leishmania DNA [132]. Despite of the rise of genome sequencing and Next Generation Sequencing, nowadays there is not a standard diagnostic test for Leishmaniasis. The problem starts in the disease itself. The pathology demonstrates a tailored number of distinct phenotypes that may be caused by different species and also attributed to mixed infections of independent species [133,134]. Therefore, oldfashioned techniques and molecular tools interpose for the diagnosis of this tropical disease. As a matter of fact, microscopy (examination of giemsa-stained lesion biopsy smears) and culture (biopsy triturates and aspirates) are still considered the gold standard for leishmaniasis diagnosis due to its high specificity [135]. The big problem with this technique is that the sample can only be diagnosed until genus Leishmania and in most of the cases the species is not discriminated [136]. Additionally, the sensitivity of these techniques is low and depends on the number and dispersion of parasites in biopsy samples, the sampling procedure and the operator [137,138].
Immunological diagnosis has also been attempted in leishmaniasis, especially in visceral leishmaniasis, which is foreseen in the presence of currently available direct agglutination tests and immunochromatographic dipsticks [139]. Serological tests do not show a good performance in cutaneous leishmaniasis because sensitivity can be variable and because the number of circulating antibodies tends to be low. The specificity is the most critical point in leishmaniasis diagnosis because in most of the endemic areas of leishmaniasis co-exists Trypanosoma cruzi and Trypanosoma brucei, where cross-reaction may be observed, producing a high amount of false-positive results [136,140,141]. Lastly, but not less important, is the Montenegro skin test (MST) that is occasionally used in cutaneous leishmaniasis diagnosis (epidemiological surveys and vaccine studies). This test has been widely used because of its sensitivity and specificity. Unfortunately, the main disadvantage of this method is the requirement of antigen preparations and the impossibility of establishing the etiological agent of the infection [142]. Due to all the disadvantages that the parasitological and immunological techniques hold, it is necessary to deploy molecular tools for a rapid, feasible and accurate diagnosis of cutaneous and visceral leishmaniasis.
Molecular tools have also been deployed to detect Leishmania infections and to discriminate the species involved. Different molecular approaches have been developed that range from pulse-field gel electrophoresis to genome sequencing [137,143]. Most of these efforts have been focused on discriminating Leishmania species, but recently competitive and accurate PCR methods have been developed as molecular diagnosis tools of leishmaniasis [144]. In the last decade, PCR analysis has been successfully introduced and has been proven to be the most sensitive molecular tool for direct detection and parasite characterization of Leishmania species in clinical samples [145,146]. PCR allows a rapid detection of the parasite without the need of culturing. The PCR sensitivities in skin aspiration, and even in blood in the cases of cutaneous and visceral leishmaniasis, range from 60% to 100% [137]. This is clearly dependent on different factors, such as the DNA extraction method employed, the quality of the samples and of most relevance, the target selected for the detection of Leishmania DNA. Currently a broad range of markers have been employed to detect and discriminate Leishmania; among these, we can find the Internal Transcribed Spacer 1 (ITS1), Kinetoplast DNA (kDNA), The Small Unit or rRNA (SSU rDNA), The Heat Shock Protein 70 (HSP70) and the 7SL rRNA (7SL) [146-149].
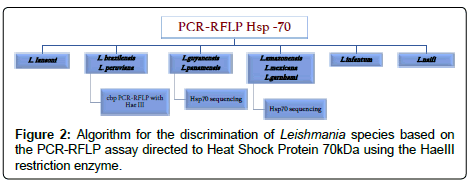
An important issue with regard to the selection of the method for the diagnosis of Leishmania is the number of copies of each target, which indeed is different for most of the species [150]. Therefore, when applied to biological samples, the absence of amplification is not always related to the absence of parasite, just for the fact of the low parasite burden that the biological sample persists. This reason suggests that the most used markers for this detection are the most abundant on parasite genome (kDNA and ribosomal genes). In fact PCR-RFLPs algorithms have been deployed with the purpose of detecting the parasite and also to discriminate the Leishmania species with HaeIII digestion (Figure 2) [151,152]. Lately, the quantitative real-time PCR (qPCR) has emerged as an appropriate alternative for the diagnosis of Leishmaniasis [153-155]. This method permits an absolute quantification of the parasitic load in blood and tissues from infected patients. Also, due to the properties of the assay, it is possible to discriminate the different species using High-Resolution Melting analyses or even conducting analysis of denaturation with FRET probes [156,157]. Therefore, molecular diagnosis is highly efficient for the screening of patients infected with Leishmania species.
Despite efforts in the deployment of molecular tools for the diagnosis of Leishmania, there are six clinical/biological issues, as mentioned by Reithinger and Dujardin in 2006 [137]. i) The first is the cure criteria in treated patients. PCR has produced percentages of sensitivity and specificity close to 100%. In patients with visceral leishmaniasis it has been observed that in the lack of symptomatology the PCR remains negative but in the cases of cutaneous leishmaniasis up to 80% of patients with no symptomatology present a positive PCR, even 8 years after clinical cure [158,159]. ii) Host tissue quantification of parasites should be assessed by PCR. Although, there are currently qPCR methods with analytical sensitivity around 0.0125 parasites per mL and excellent linearity, there has not been developed a multicentric comparison in order to assess the potential of this technique in biopsies [160]. iii) In some context, with the purpose of evaluating the viability of parasites submitted to chemotherapy of vaccines, assays detecting RNA are necessary. In this sense, isothermal amplifications such as reverse transcription Loop isothermal assays have been reported, but never compared across geographical regions [161]. iv) The deployment of a method that allows a rapid and accurate diagnosis of Leishmania species. Despite the current availability of PCR-RFLP methods for this purpose, they are not sensitive enough which requires the need to pursue studies on this area. v) Molecular diagnosis might allow defining parasite-specific features, such as virulence or drug resistance. This is still under study and forthcoming techniques are necessary. vi) Parasite tracking is needed for the purpose of detecting parasite strains that are susceptible or resistant to chemotherapy.
In conclusion, regarding the molecular diagnosis of leishmaniasis, there are significant advances toward the establishment of a method that can be widely used. Unfortunately there are not multicentric evaluations, such as those conducted on Chagas disease. The only one available has recently been published by Cruz et al. 2013 [162] demonstrating the high reproducibility of the current methods for PCR. The analyses of the new sequenced Leishmania strains will provide further information about candidate markers for the diagnosis of this neglected tropical disease.
Conclusions
Herein we engaged in a schematic and systematic review about the current methods available for the molecular diagnosis of vector-borne parasitic diseases. The advances in this field have been important, but novel research is needed. The most notable absence in the research thus conducted is the lack of international multicentric studies comparing the sensitivity, concordance and specificity of the methods employed by different laboratories. Such research has only been conducted for Chagas disease, and to a lesser extent on leishmaniasis. We encourage the lead researchers in every pathology to conduct international evaluations in order to propose reliable, accurate and promising technologies for the molecular diagnosis of these pathologies that still represent serious problems in public health.
References
- World Health Organization (2010) Millennium Development Goals: progress towards the health-related Millennium Development Goals.
- World Health Organization (2012) World Malaria Report 2012.
- Butler D (2008) Malaria map brings good new. Disease transmission is low throughout large areas of malarial risk. Nature news.
- Gay F, Zougbédé S, N'dilimabaka N, Rebollo A, Mazier D, et al. (2012) Cerebral malaria: what is known and what is on research. Rev Neurol (Paris) 168: 239-256.
- Shanks GD (2012) Control and elimination of Plasmodium vivax. Adv Parasitol 80: 301-341.
- Abdul-Nasir AS, Mashor MY, Mohamed Z (2012) Modified global and modified linear contrast stretching algorithms: new colour contrast enhancement techniques for microscopic analysis of malaria slide images. Comput Math Methods Med 2012: 16.
- Ross NE, Pritchard CJ, Rubin DM, Dusé AG (2006) Automated image processing method for the diagnosis and classification of malaria on thin blood smears. Med Biol Eng Comput 44: 427-436.
- Makler MT, Palmer CJ, Ager AL (1998) A review of practical techniques for the diagnosis of malaria. Ann Trop Med Parasitol 92: 419-433.
- Mitiku K, Mengistu G, Gelaw B (2003) The reliability of blood film examination for malaria at the peripheral health unit. Ethiop J Health Dev17: 197–204.
- Wilson ML (2013) Laboratory diagnosis of malaria: conventional and rapid diagnostic methods. Arch Pathol Lab Med 137: 805-811.
- Hänscheid T (1999) Diagnosis of malaria: a review of alternatives to conventional microscopy. Clin Lab Haematol 21: 235-245.
- Avila SL, Ferreira AW (1996) Malaria diagnosis: a review. Braz J Med Biol Res 29: 431-443.
- Baird JK, Purnomo, Jones TR (1992) Diagnosis of malaria in the field by fluorescence microscopy of QBC capillary tubes. Trans R Soc Trop Med Hyg 86: 3-5.
- Benito A, Roche J, Molina R, Amela C, Alvar J (1994) Application and evaluation of QBC malaria diagnosis in a holoendemic area. Appl Parasitol 35: 266-272.
- Kawamoto F (1991) Rapid diagnosis of malaria by fluorescence microscopy with light microscope and interference filter. Lancet 337: 200-202.
- Lowe BS, Jeffa NK, New L, Pedersen C, Engbaek K, et al. (1996) Acridine orange fluorescence techniques as alternatives to traditional Giemsa staining for the diagnosis of malaria in developing countries. Trans R Soc Trop Med Hyg 90: 34-36.
- Makler MT, Ries LK, Ries J, Horton RJ, Hinrichs DJ (1991) Detection of Plasmodium falciparum infection with the fluorescent dye, benzothiocarboxypurine. Am J Trop Med Hyg 44: 11-16.
- Cooke AH, Moody AH, Lemon K, Chiodini PL, Horton J (1992) Use of the fluorochrome benzothiocarboxypurine in malaria diagnosis. Trans R Soc Trop Med Hyg 86: 378.
- Murray CK, Gasser RA Jr, Magill AJ, Miller RS (2008) Update on rapid diagnostic testing for malaria. Clin Microbiol Rev 21: 97-110.
- Gillet P, Maltha J, Hermans V, Ravinetto R, Bruggeman C, et al. (2011) Malaria rapid diagnostic kits: quality of packaging, design and labelling of boxes and components and readability and accuracy of information inserts. Malar J 10: 39.
- Abba K, Deeks JJ, Olliaro P, Naing CM, Jackson SM, et al. (2011) Rapid diagnostic tests for diagnosing uncomplicated P. falciparum malaria in endemic countries. Cochrane Database Syst Rev 7: CD008122.
- Erdman LK, Kain KC (2008) Molecular diagnostic and surveillance tools for global malaria control. Travel Med Infect Dis 6: 82-99.
- Koita OA, Doumbo OK, Ouattara A, Tall LK, Konaré A, et al. (2012) False-negative rapid diagnostic tests for malaria and deletion of the histidine-rich repeat region of the hrp2 gene. Am J Trop Med Hyg 86: 194-198.
- Lucchi NW, Demas A, Narayanan J, Sumari D, Kabanywanyi A, et al. (2010) Real-time fluorescence loop mediated isothermal amplification for the diagnosis of malaria. PLoS One 5: e13733.
- Snounou G, Viriyakosol S, Jarra W, Thaithong S, Brown KN (1993) Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol Biochem Parasitol 58: 283-292.
- Rubio JM, Benito A, Berzosa PJ, Roche J, Puente S, et al. (1999) Usefulness of seminested multiplex PCR in surveillance of imported malaria in Spain. J Clin Microbiol 37: 3260-3264.
- Humar A, Harrington MA, Kain KC (1997) Evaluation of a non-isotopic polymerase chain reaction-based assay to detect and predict treatment failure of Plasmodium vivax malaria in travellers. Trans R Soc Trop Med Hyg 91: 406-409.
- Calderaro A, Piccolo G, Zuelli C, Galati L, Ricci L, et al. (2004) Evaluation of a new plate hybridization assay for the laboratory diagnosis of imported malaria in Italy. New Microbiol 27: 163-171.
- Singh B, Cox-Singh J, Miller AO, Abdullah MS, Snounou G, et al. (1996) Detection of malaria in Malaysia by nested polymerase chain reaction amplification of dried blood spots on filter papers. Trans R Soc Trop Med Hyg 90: 519-521.
- Perandin F, Manca N, Calderaro A, Piccolo G, Galati L, et al. (2004) Development of a real-time PCR assay for detection of Plasmodium falciparum, Plasmodium vivax, and Plasmodium ovale for routine clinical diagnosis. J Clin Microbiol 42: 1214-1219.
- Swan H, Sloan L, Muyombwe A, Chavalitshewinkoon-Petmitr P, Krudsood S, et al. (2005) Evaluation of a real-time polymerase chain reaction assay for the diagnosis of malaria in patients from Thailand. Am J Trop Med Hyg 73: 850-854.
- Mangold KA, Manson RU, Koay ES, Stephens L, Regner M, et al. (2005) Real-time PCR for detection and identification of Plasmodium spp. J Clin Microbiol 43: 2435-2440.
- Fabre R, Berry A, Morassin B, Magnaval JF (2004) Comparative assessment of conventional PCR with multiplex real-time PCR using SYBR Green I detection for the molecular diagnosis of imported malaria. Parasitology 128: 15-21.
- Tjitra E, Anstey NM (2001) Will the high rates of post-treatment sexual stage parasitaemia seen in malaria-endemic areas make the optiMAL antigen test unreliable in predicting malaria treatment outcome? Br J Haematol 113: 255-257.
- Poon LL, Wong BW, Ma EH, Chan KH, Chow LM, et al. (2006) Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin Chem 52: 303-306.
- Han ET, Watanabe R, Sattabongkot J, Khuntirat B, Sirichaisinthop J, et al. (2007) Detection of four Plasmodium species by genus- and species-specific loop-mediated isothermal amplification for clinical diagnosis. J Clin Microbiol 45: 2521-2528.
- Gardner MJ, Hall N, Fung E, White O, Berriman M, et al. (2002) Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419: 498-511.
- Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, et al. (2008) Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455: 757-763.
- Holland CA, Kiechle FL (2005) Point-of-care molecular diagnostic systems--past, present and future. Curr Opin Microbiol 8: 504-509.
- Gilbert GL (2002) Molecular diagnostics in infectious diseases and public health microbiology: cottage industry to postgenomics. Trends Mol Med 8: 280-287.
- Demirev PA, Feldman AB, Kongkasuriyachai D, Scholl P, Sullivan D Jr, et al. (2002) Detection of malaria parasites in blood by laser desorption mass spectrometry. Anal Chem 74: 3262-3266.
- Scholl PF, Kongkasuriyachai D, Demirev PA, Feldman AB, Lin JS, et al. (2004) Rapid detection of malaria infection in vivo by laser desorption mass spectrometry. Am J Trop Med Hyg 71: 546-551.
- Nyunt M, Pisciotta J, Feldman AB, Thuma P, Scholl PF, et al. (2005) Detection of Plasmodium falciparum in pregnancy by laser desorption mass spectrometry. Am J Trop Med Hyg 73: 485-490.
- Rassi A Jr, Rassi A, Marcondes de Rezende J (2012) American trypanosomiasis (Chagas disease). Infect Dis Clin North Am 26: 275-291.
- Lee BY, Bacon KM, Bottazzi ME, Hotez PJ (2013) Global economic burden of Chagas disease: a computational simulation model. Lancet Infect Dis 13: 342-348.
- Rassi A Jr, Rassi A, Marin-Neto JA (2010) Chagas disease. Lancet 375: 1388-1402.
- Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, et al. (2012) The revised Trypanosoma cruzi subspecific nomenclature: rationale, epidemiological relevance and research applications. Infect Genet Evol 12: 240-253.
- Ramírez JD, Guhl F, Rendón LM, Rosas F, Marin-Neto JA, et al. (2010) Chagas cardiomyopathy manifestations and Trypanosoma cruzi genotypes circulating in chronic Chagasic patients. PLoS Negl Trop Dis 4: e899.
- Burgos JM, Diez M, Vigliano C, Bisio M, Risso M, et al. (2010) Molecular identification of Trypanosoma cruzi discrete typing units in end-stage chronic Chagas heart disease and reactivation after heart transplantation. Clin Infect Dis 51: 485-495.
- STROUT RG (1962) A method for concentrating hemoflagellates. J Parasitol 48: 100.
- Flores MA, Trejos A, Paredes AR, Ramos AY (1966) Strout's concentration method in the diagnosis of acute Chagas disease. Bol Chil Parasitol 21: 38-39.
- Feilij H, Muller L, Gonzalez Cappa SM (1983) Direct micromethod for diagnosis of acute and congenital Chagas' disease. J Clin Microbiol 18: 327-330.
- Britto C, Cardoso MA, Vanni CM, Hasslocher-Moreno A, Xavier SS, et al. (1995) Polymerase chain reaction detection of Trypanosoma cruzi in human blood samples as a tool for diagnosis and treatment evaluation. Parasitology 110 : 241-247.
- Fernandes CD, Tiecher FM, Balbinot MM, Liarte DB, Scholl D, et al. (2009) Efficacy of benznidazol treatment for asymptomatic chagasic patients from state of Rio Grande do Sul evaluated during a three years follow-up. Mem Inst Oswaldo Cruz 104: 27-32.
- Ramírez JD, Guhl F, Umezawa ES, Morillo CA, Rosas F, et al. (2009) Evaluation of adult chronic Chagas' heart disease diagnosis by molecular and serological methods. J Clin Microbiol 47: 3945-3951.
- da Silveira JF, Umezawa ES, Luquetti AO (2001) Chagas disease: recombinant Trypanosoma cruzi antigens for serological diagnosis. Trends Parasitol 17: 286-291.
- Umezawa ES, Bastos SF, Coura JR, Levin MJ, Gonzalez A, et al. (2003) An improved serodiagnostic test for Chagas' disease employing a mixture of Trypanosoma cruzi recombinant antigens. Transfusion 43: 91-97.
- Malchiodi EL, Chiaramonte MG, Taranto NJ, Zwirner NW, Margni RA (1994) Cross-reactivity studies and differential serodiagnosis of human infections caused by Trypanosoma cruzi and Leishmania spp; use of immunoblotting and ELISA with a purified antigen (Ag163B6). Clin Exp Immunol 97: 417-423.
- Caballero ZC, Sousa OE, Marques WP, Saez-Alquezar A, Umezawa ES (2007) Evaluation of serological tests to identify Trypanosoma cruzi infection in humans and determine cross-reactivity with Trypanosoma rangeli and Leishmania spp. Clin Vaccine Immunol 14: 1045-1049.
- Afonso AM, Ebell MH, Tarleton RL (2012) A systematic review of high quality diagnostic tests for Chagas disease. PLoS Negl Trop Dis 6: e1881.
- Brasil PE, De Castro L, Hasslocher-Moreno AM, Sangenis LH, Braga JU (2010) ELISA versus PCR for diagnosis of chronic Chagas disease: systematic review and meta-analysis. BMC Infect Dis 10: 337.
- Ponce C, Ponce E, Vinelli E, Montoya A, de Aguilar V, et al. (2005) Validation of a rapid and reliable test for diagnosis of chagas' disease by detection of Trypanosoma cruzi-specific antibodies in blood of donors and patients in Central America. J Clin Microbiol 43: 5065-5068.
- Verani JR, Seitz A, Gilman RH, LaFuente C, Galdos-Cardenas G, et al. (2009) Geographic variation in the sensitivity of recombinant antigen-based rapid tests for chronic Trypanosoma cruzi infection. Am J Trop Med Hyg 80: 410-415.
- Luquetti AO, Ponce C, Ponce E, Esfandiari J, Schijman A, et al. (2003) Chagas' disease diagnosis: a multicentric evaluation of Chagas Stat-Pak, a rapid immunochromatographic assay with recombinant proteins of Trypanosoma cruzi. Diagn Microbiol Infect Dis 46: 265-271.
- Reithinger R, Grijalva MJ, Chiriboga RF, de Noya BA, Torres JR, et al. (2010) Rapid detection of Trypanosoma cruzi in human serum by use of an immunochromatographic dipstick test. J Clin Microbiol 48: 3003-3007.
- Roddy P, Goiri J, Flevaud L, Palma PP, Morote S, et al. (2008) Field evaluation of a rapid immunochromatographic assay for detection of Trypanosoma cruzi infection by use of whole blood. J Clin Microbiol 46: 2022-2027.
- Chappuis F, Mauris A, Holst M, Albajar-Vinas P, Jannin J, et al. (2010) Validation of a rapid immunochromatographic assay for diagnosis of Trypanosoma cruzi infection among Latin-American Migrants in Geneva, Switzerland. J Clin Microbiol 48: 2948-2952.
- Barfield CA, Barney RS, Crudder CH, Wilmoth JL, Stevens DS, et al. (2011) A highly sensitive rapid diagnostic test for Chagas disease that utilizes a recombinant Trypanosoma cruzi antigen. IEEE Trans Biomed Eng 58: 814-817.
- Umezawa ES, Nascimento MS, Kesper N Jr, Coura JR, Borges-Pereira J, et al. (1996) Immunoblot assay using excreted-secreted antigens of Trypanosoma cruzi in serodiagnosis of congenital, acute, and chronic Chagas' disease. J Clin Microbiol 34: 2143-2147.
- Umezawa ES, Bastos SF, Camargo ME, Yamauchi LM, Santos MR, et al. (1999) Evaluation of recombinant antigens for serodiagnosis of Chagas' disease in South and Central America. J Clin Microbiol 37: 1554-1560.
- Umezawa ES, Souza AI, Pinedo-Cancino V, Marcondes M, Marcili A, et al. (2009) TESA-blot for the diagnosis of Chagas disease in dogs from co-endemic regions for Trypanosoma cruzi, Trypanosoma evansi and Leishmania chagasi. Acta Trop 111: 15-20.
- Matsumoto TK, Cotrim PC, da Silveira JF, Stolf AM, Umezawa ES (2002) Trypanosoma cruzi: isolation of an immunodominant peptide of TESA (Trypomastigote Excreted-Secreted Antigens) by gene cloning. Diagn Microbiol Infect Dis 42: 187-192.
- Guhl F, Jaramillo C, Carranza JC, Vallejo GA (2002) Molecular characterization and diagnosis of trypanosoma cruzi and T. rangeli. Arch Med Res 33: 362-370.
- Moser DR, Kirchhoff LV, Donelson JE (1989) Detection of Trypanosoma cruzi by DNA amplification using the polymerase chain reaction. J Clin Microbiol 27: 1477-1482.
- Vallejo GA, Guhl F, Chiari E, Macedo AM (1999) Species specific detection of Trypanosoma cruzi and Trypanosoma rangeli in vector and mammalian hosts by polymerase chain reaction amplification of kinetoplast minicircle DNA. Acta Trop 72: 203-212.
- Avila HA, Pereira JB, Thiemann O, De Paiva E, DeGrave W, et al. (1993) Detection of Trypanosoma cruzi in blood specimens of chronic chagasic patients by polymerase chain reaction amplification of kinetoplast minicircle DNA: comparison with serology and xenodiagnosis. J Clin Microbiol 31: 2421-2426.
- Gomes ML, Galvao LM, Macedo AM, Pena SD, Chiari E (1999) Chagas' disease diagnosis: comparative analysis of parasitologic, molecular, and serologic methods. Am J Trop Med Hyg 60: 205-210.
- Carvalho CM, Andrade MC, Xavier SS, Mangia RH, Britto CC, et al. (2003). Chronic chagas' disease in rhesus monkeys (macaca mulatta): evaluation of parasitemia, serology, electrocardiography, echocardiography, and radiology. Am.J. Trop.Med. Hyg 68: 683-691.
- Meira WS, de Castro AM, Gontijo ED, Rassi A, Luquetti AO, et al. (2006) Evaluation of blood tests, complement-mediated lysis and polymerase chain reaction in the verification of therapeutic efficacy in Chagas disease. Rev Soc Bras Med Trop 39 Suppl 3: 107-109.
- Aguiar C, Batista AM, Pavan TB, Almeida EA, Guariento ME, et al. (2012) Serological profiles and evaluation of parasitaemia by PCR and blood culture in individuals chronically infected by Trypanosoma cruzi treated with benzonidazole. Trop Med Int Health 17: 368-373.
- Murcia L, Carrilero B, Muñoz MJ, Iborra MA, Segovia M (2010) Usefulness of PCR for monitoring benznidazole response in patients with chronic Chagas' disease: a prospective study in a non-disease-endemic country. J Antimicrob Chemother 65: 1759-1764.
- Flores-Chavez M, Bosseno MF, Bastrenta B, Dalenz JL, Hontebeyrie M, et al. (2006) Polymerase chain reaction detection and serologic follow-up after treatment with benznidazole in Bolivian children infected with a natural mixture of Trypanosoma cruzi I and II. Am J Trop Med Hyg 75: 497-501.
- Britto C, Silveira C, Cardoso MA, Marques P, Luquetti A, et al. (2001) Parasite persistence in treated chagasic patients revealed by xenodiagnosis and polymerase chain reaction. Mem Inst Oswaldo Cruz 96: 823-826.
- Lauria-Pires L, Braga MS, Vexenat AC, Nitz N, Simões-Barbosa A, et al. (2000) Progressive chronic Chagas heart disease ten years after treatment with anti-Trypanosoma cruzi nitroderivatives. Am J Trop Med Hyg 63: 111-118.
- Braga MS, Lauria-Pires L, Argañaraz ER, Nascimento RJ, Teixeira AR (2000) Persistent infections in chronic Chagas' disease patients treated with anti-Trypanosoma cruzi nitroderivatives. Rev Inst Med Trop Sao Paulo 42: 157-161.
- Russomando G, de Tomassone MM, de Guillen I, Acosta N, Vera N, et al. (1998) Treatment of congenital Chagas' disease diagnosed and followed up by the polymerase chain reaction. Am J Trop Med Hyg 59: 487-491.
- Britto CC (2009) Usefulness of PCR-based assays to assess drug efficacy in Chagas disease chemotherapy: value and limitations. Mem Inst Oswaldo Cruz 104 Suppl 1: 122-135.
- Marin-Neto JA, Rassi A Jr, Morillo CA, Avezum A, Connolly SJ, et al. (2008) Rationale and design of a randomized placebo-controlled trial assessing the effects of etiologic treatment in Chagas' cardiomyopathy: the BENznidazole Evaluation For Interrupting Trypanosomiasis (BENEFIT). Am. Heart J 156: 37-43.
- Schijman AG, Bisio M, Orellana L, Sued M, Duffy T, et al. (2011) International study to evaluate PCR methods for detection of Trypanosoma cruzi DNA in blood samples from Chagas disease patients. PLoS Negl Trop Dis 5: e931.
- Duffy T, Cura CI, Ramirez JC, Abate T, Cayo NM, et al. (2013) Analytical performance of a multiplex Real-Time PCR assay using TaqMan probes for quantification of Trypanosoma cruzi satellite DNA in blood samples. PLoS Negl Trop Dis 7: e2000.
- Moreira OC, Ramírez JD, Velázquez E, Melo MF, Lima-Ferreira C, et al. (2013) Towards the establishment of a consensus real-time qPCR to monitor Trypanosoma cruzi parasitemia in patients with chronic Chagas disease cardiomyopathy: a substudy from the BENEFIT trial. Acta Trop 125: 23-31.
- Qvarnstrom Y, Schijman AG, Veron V, Aznar C, Steurer F, et al. (2012) Sensitive and specific detection of Trypanosoma cruzi DNA in clinical specimens using a multi-target real-time PCR approach. PLoS Negl Trop Dis 6: e1689.
- Lewis MD, Ma J, Yeo M, Carrasco HJ, Llewellyn MS, et al. (2009) Genotyping of Trypanosoma cruzi: systematic selection of assays allowing rapid and accurate discrimination of all known lineages. Am J Trop Med Hyg 81: 1041-1049.
- Rozas M, De Doncker S, Adaui V, Coronado X, Barnabé C, et al. (2007) Multilocus polymerase chain reaction restriction fragment--length polymorphism genotyping of Trypanosoma cruzi (Chagas disease): taxonomic and clinical applications. J Infect Dis 195:1381-1388.
- Duffy T, Bisio M, Altcheh J, Burgos JM, Diez M, et al. (2009) Accurate real-time PCR strategy for monitoring bloodstream parasitic loads in chagas disease patients. PLoS Negl Trop Dis 3: e419.
- Yeo M, Mauricio IL, Messenger LA, Lewis MD, Llewellyn MS, et al. (2011) Multilocus sequence typing (MLST) for lineage assignment and high resolution diversity studies in Trypanosoma cruzi. PLoS Negl Trop Dis 5: e1049.
- Llewellyn MS, Miles MA, Carrasco HJ, Lewis MD, Yeo M, et al. (2009) Genome-scale multilocus microsatellite typing of Trypanosoma cruzi discrete typing unit I reveals phylogeographic structure and specific genotypes linked to human infection. PLoS Pathog 5: e1000410.
- Ramírez JD, Turriago B, Tapia-Calle G, Guhl F (2013) Understanding the role of dogs (Canis lupus familiaris) in the transmission dynamics of Trypanosoma cruzi genotypes in Colombia. Vet Parasitol .
- Higuera SL, Guhl F, Ramírez JD (2013) Identification of Trypanosoma cruzi Discrete Typing Units (DTUs) through the implementation of a High-Resolution Melting (HRM) genotyping assay. Parasit Vectors 6: 112.
- Brun R, Blum J, Chappuis F, Burri C (2010) Human African trypanosomiasis. Lancet 375: 148-159.
- Kennedy PG (2004) Human African trypanosomiasis of the CNS: current issues and challenges. J Clin Invest 113: 496-504.
- Blum J, Schmid C, Burri C (2006) Clinical aspects of 2541 patients with second stage human African trypanosomiasis. Acta Trop 97: 55-64.
- Deborggraeve S, Büscher P (2010) Molecular diagnostics for sleeping sickness: what is the benefit for the patient? Lancet Infect Dis 10: 433-439.
- World Health Organization (2013). Human African trypanosomiasis. Global Health Observatory (GHO).
- Truc P, Lejon V, Magnus E, Jamonneau V, Nangouma A, et al. (2002) Evaluation of the micro-CATT, CATT/Trypanosoma brucei gambiense, and LATEX/T b gambiense methods for serodiagnosis and surveillance of human African trypanosomiasis in West and Central Africa. Bull World Health Organ 80: 882-886.
- Chappuis F, Loutan L, Simarro P, Lejon V, Büscher P (2005) Options for field diagnosis of human african trypanosomiasis. Clin Microbiol Rev 18: 133-146.
- Mathieu-Daude F, Bicart-See A, Bosseno MF, Breniere SF, Tibayrenc M (1994) Identification of Trypanosoma brucei gambiense group I by a specific kinetoplast DNA probe. Am J Trop Med Hyg 50: 13-19.
- Kabiri M, Franco JR, Simarro PP, Ruiz JA, Sarsa M, et al. (1999) Detection of Trypanosoma brucei gambiense in sleeping sickness suspects by PCR amplification of expression-site-associated genes 6 and 7. Trop Med Int Health 4: 658-661.
- Kyambadde JW, Enyaru JC, Matovu E, Odiit M, Carasco JF (2000) Detection of trypanosomes in suspected sleeping sickness patients in Uganda using the polymerase chain reaction. Bull World Health Organ 78: 119-124.
- Penchenier L, Simo G, Grébaut P, Nkinin S, Laveissière C, et al. (2000) Diagnosis of human trypanosomiasis, due to Trypanosoma brucei gambiense in central Africa, by the polymerase chain reaction. Trans R Soc Trop Med Hyg 94: 392-394.
- Solano P, Jamonneau V, N'Guessan P, N'Dri L, Dje NN, et al. (2002) Comparison of different DNA preparation protocols for PCR diagnosis of Human African Trypanosomosis in Côte d'Ivoire. Acta Trop 82: 349-356.
- Deborggraeve S, Lejon V, Ekangu RA, Mumba Ngoyi D, Pati Pyana P, et al. (2011) Diagnostic accuracy of PCR in gambiense sleeping sickness diagnosis, staging and post-treatment follow-up: a 2-year longitudinal study. PLoS Negl Trop Dis 5: e972.
- Cox A, Tilley A, McOdimba F, Fyfe J, Eisler M, et al. (2005) A PCR based assay for detection and differentiation of African trypanosome species in blood. Exp Parasitol 111: 24-29.
- Adams ER, Malele II, Msangi AR, Gibson WC (2006) Trypanosome identification in wild tsetse populations in Tanzania using generic primers to amplify the ribosomal RNA ITS-1 region. Acta Trop 100: 103-109.
- Radwanska M, Claes F, Magez S, Magnus E, Perez-Morga D, et al. (2002) Novel primer sequences for polymerase chain reaction-based detection of Trypanosoma brucei gambiense. Am J Trop Med Hyg 67: 289-295.
- Welburn SC, Picozzi K, Fèvre EM, Coleman PG, Odiit M, et al. (2001) Identification of human-infective trypanosomes in animal reservoir of sleeping sickness in Uganda by means of serum-resistance-associated (SRA) gene. Lancet 358: 2017-2019.
- Picozzi K, Carrington M, Welburn SC (2008) A multiplex PCR that discriminates between Trypanosoma brucei brucei and zoonotic T. b. rhodesiense. Exp Parasitol 118: 41-46.
- Mugasa CM, Adams ER, Boer KR, Dyserinck HC, Büscher P, et al. (2012) Diagnostic accuracy of molecular amplification tests for human African trypanosomiasis--systematic review. PLoS Negl Trop Dis 6: e1438.
- Becker S, Franco JR, Simarro PP, Stich A, Abel PM, et al. (2004) Real-time PCR for detection of Trypanosoma brucei in human blood samples. Diagn Microbiol Infect Dis 50: 193-199.
- Mugasa CM, Schoone GJ, Ekangu RA, Lubega GW, Kager PA, et al. (2008) Detection of Trypanosoma brucei parasites in blood samples using real-time nucleic acid sequence-based amplification. Diagn Microbiol Infect Dis 61: 440-445.
- Mugasa CM, Laurent T, Schoone GJ, Kager PA, Lubega GW, et al. (2009) Nucleic acid sequence-based amplification with oligochromatography for detection of Trypanosoma brucei in clinical samples. J Clin Microbiol 47: 630-635.
- Kuboki N, Inoue N, Sakurai T, Di Cello F, Grab DJ, et al. (2003) Loop-mediated isothermal amplification for detection of African trypanosomes. J Clin Microbiol 41: 5517-5524.
- Kennedy PG (2013) Clinical features, diagnosis, and treatment of human African trypanosomiasis (sleeping sickness). Lancet Neurol 12: 186-194.
- Njiru ZK, Mikosza AS, Armstrong T, Enyaru JC, Ndung'u JM, et al. (2008) Loop-mediated isothermal amplification (LAMP) method for rapid detection of Trypanosoma brucei rhodesiense. PLoS Negl Trop Dis 2: e147.
- Njiru ZK, Mikosza AS, Matovu E, Enyaru JC, Ouma JO, et al. (2008) African trypanosomiasis: sensitive and rapid detection of the sub-genus Trypanozoon by loop-mediated isothermal amplification (LAMP) of parasite DNA. Int J Parasitol 38: 589-599.
- Grab DJ, Nikolskaia OV, Inoue N, Thekisoe OM, Morrison LJ, et al. (2011) Using detergent to enhance detection sensitivity of African trypanosomes in human CSF and blood by loop-mediated isothermal amplification (LAMP). PLoS Negl Trop Dis 5: e1249.
- Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, et al. (2012) Leishmaniasis worldwide and global estimates of its incidence. PLoS One 7: e35671.
- Desjeux P (2004) Leishmaniasis: current situation and new perspectives. Comp Immunol Microbiol Infect Dis 27: 305-318.
- Alvar J, Yactayo S, Bern C (2006) Leishmaniasis and poverty. Trends Parasitol 22: 552-557.
- Pratlong F, Lami P, Ravel C, Balard Y, Dereure J, et al. (2013) Geographical distribution and epidemiological features of Old World Leishmania infantum and Leishmania donovani foci, based on the isoenzyme analysis of 2277 strains. Parasitology 140: 423-434.
- Ready PD (2013) Biology of phlebotomine sand flies as vectors of disease agents. Annu Rev Entomol 58: 227-250.
- Montalvo AM, Fraga J, Monzote L, García M, Fonseca L (2012) Leishmaniasis diagnosis: going from microscopic observation of parasite to DNA detection. Rev Cubana Med Trop 64: 108-131.
- Bastrenta B, Mita N, Buitrago R, Vargas F, Flores M, et al. (2003) Human mixed infections of Leishmania spp. and Leishmania-Trypanosoma cruzi in a sub Andean Bolivian area: identification by polymerase chain reaction/hybridization and isoenzyme. Mem Inst Oswaldo Cruz 98: 255-264.
- Madeira MF, Schubach A, Schubach TM, Pacheco RS, Oliveira FS, et al. (2006) Mixed infection with Leishmania (Viannia) braziliensis and Leishmania (Leishmania) chagasi in a naturally infected dog from Rio de Janeiro, Brazil. Trans R Soc Trop Med Hyg 100: 442-445.
- Bensoussan E, Nasereddin A, Jonas F, Schnur LF, Jaffe CL (2006) Comparison of PCR assays for diagnosis of cutaneous leishmaniasis. J Clin Microbiol 44: 1435-1439.
- Chappuis F, Rijal S, Jha UK, Desjeux P, Karki BM, et al. (2006) Field validity, reproducibility and feasibility of diagnostic tests for visceral leishmaniasis in rural Nepal. Trop Med Int Health 11: 31-40.
- Reithinger R, Dujardin JC (2007) Molecular diagnosis of leishmaniasis: current status and future applications. J Clin Microbiol 45: 21-25.
- Herwaldt BL (1999) Leishmaniasis. Lancet 354: 1191-1199.
- Lockwood DN, Sundar S (2006) Serological tests for visceral leishmaniasis. BMJ 333: 711-712.
- De Doncker S, Hutse V, Abdellati S, Rijal S, Singh Karki BM, et al. (2005) A new PCR-ELISA for diagnosis of visceral leishmaniasis in blood of HIV-negative subjects. Trans R Soc Trop Med Hyg 99: 25-31.
- Cota GF, de Sousa MR, Demarqui FN, Rabello A (2012) The diagnostic accuracy of serologic and molecular methods for detecting visceral leishmaniasis in HIV infected patients: meta-analysis. PLoS Negl Trop Dis 6: e1665.
- Ramírez JR, Agudelo S, Muskus C, Alzate JF, Berberich C, et al. (2000) Diagnosis of cutaneous leishmaniasis in Colombia: the sampling site within lesions influences the sensitivity of parasitologic diagnosis. J Clin Microbiol 38: 3768-3773.
- Singh S, Dey A, Sivakumar R (2005) Applications of molecular methods for Leishmania control. Expert Rev Mol Diagn 5: 251-265.
- Yehia L, Adib-Houreih M, Raslan WF, Kibbi AG, Loya A, et al. (2012) Molecular diagnosis of cutaneous leishmaniasis and species identification: analysis of 122 biopsies with varied parasite index. J Cutan Pathol 39: 347-355.
- Schönian G, Nasereddin A, Dinse N, Schweynoch C, Schallig HD, et al. (2003) PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn Microbiol Infect Dis 47: 349-358.
- Schönian G, Mauricio I, Gramiccia M, Cañavate C, Boelaert M, et al. (2008) Leishmaniases in the Mediterranean in the era of molecular epidemiology. Trends Parasitol 24: 135-142.
- Garcia L, Kindt A, Bermudez H, Llanos-Cuentas A, De Doncker S, et al. (2004) Culture-independent species typing of neotropical Leishmania for clinical validation of a PCR-based assay targeting heat shock protein 70 genes. J Clin Microbiol. 42: 2294-2297.
- Rotureau B, Ravel C, Nacher M, Couppié P, Curtet I, et al. (2006) Molecular epidemiology of Leishmania (Viannia) guyanensis in French Guiana. J Clin Microbiol 44: 468-473.
- Zelazny AM, Fedorko DP, Li L, Neva FA, Fischer SH (2005) Evaluation of 7SL RNA gene sequences for the identification of Leishmania spp. Am J Trop Med Hyg 72: 415-420.
- Downing T, Imamura H, Decuypere S, Clark TG, Coombs GH, et al. (2011) Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res 21: 2143-2156.
- Garcia AL, Parrado R, De Doncker S, Bermudez H, Dujardin JC (2007) American tegumentary leishmaniasis: direct species identification of Leishmania in non-invasive clinical samples. Trans R Soc Trop Med Hyg 101: 368-371.
- Alvarenga JS, Ligeiro CM, Gontijo CM, Cortes S, Campino L, et al. (2012) KDNA genetic signatures obtained by LSSP-PCR analysis of Leishmania (Leishmania) infantum isolated from the new and the old world. PLoS One 7: e43363.
- Bretagne S, Durand R, Olivi M, Garin JF, Sulahian A, et al. (2001) Real-time PCR as a new tool for quantifying Leishmania infantum in liver in infected mice. Clin Diagn Lab Immunol 8: 828-831.
- Wortmann G, Hochberg L, Houng HH, Sweeney C, Zapor M, et al. (2005) Rapid identification of Leishmania complexes by a real-time PCR assay. Am J Trop Med Hyg 73: 999-1004.
- Ranasinghe S, Rogers ME, Hamilton JG, Bates PA, Maingon RD (2008) A real-time PCR assay to estimate Leishmania chagasi load in its natural sand fly vector Lutzomyia longipalpis. Trans R Soc Trop Med Hyg 102: 875-882.
- Tsukayama P, Núñez JH, De Los Santos M, Soberón V, Lucas CM, et al. (2013) A FRET-based real-time PCR assay to identify the main causal agents of New World tegumentary leishmaniasis. PLoS Negl Trop Dis 7: e1956.
- de Almeida Ferreira S, Leite RS, Ituassu LT, Almeida GG, Souza DM, et al. (2012) Canine skin and conjunctival swab samples for the detection and quantification of Leishmania infantum DNA in an endemic urban area in Brazil. PLoS Negl Trop Dis 6: e1596.
- Maurya R, Singh RK, Kumar B, Salotra P, Rai M, et al. (2005) Evaluation of PCR for diagnosis of Indian kala-azar and assessment of cure. J Clin Microbiol 43: 3038-3041.
- Schubach A, Cuzzi-Maya T, Gonçalves-Costa SC, Pirmez C, Oliveira-Neto MP (1998) Leishmaniasis of glans penis. J Eur Acad Dermatol Venereol 10: 226-228.
- Mary C, Faraut F, Lascombe L, Dumon H (2004) Quantification of Leishmania infantum DNA by a real-time PCR assay with high sensitivity. J Clin Microbiol 42: 5249-5255.
- Khan MG, Bhaskar KR, Salam MA, Akther T, Pluschke G, et al. (2012) Diagnostic accuracy of loop-mediated isothermal amplification (LAMP) for detection of Leishmania DNA in buffy coat from visceral leishmaniasis patients. Parasit Vectors 5: 280.
- Cruz CF, Cruz MF, Galati EA (2013) Sandflies (Diptera: Psychodidae) in rural and urban environments in an endemic area of cutaneous leishmaniasis in southern Brazil. Mem Inst Oswaldo Cruz 108.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 16806
- [From(publication date):
June-2013 - Nov 21, 2024] - Breakdown by view type
- HTML page views : 12110
- PDF downloads : 4696