Research Article Open Access
Molecular Cloning, Characterization, and Expression Analysis of Flavanone 3-Hydroxylase (F3H) Gene during Muscadine Grape Berry Development
Jasmine Hall1, Anthony Ananga1*, Vasil Georgiev1, Joel Ochieng2, Ernst Cebert3 and Violetka Tsolova11Center for Viticulture and Small Fruit Research, College of Agriculture and Food Science, Florida A&M University, 6505 Mahan Drive, Tallahassee FL 32317, USA
2Faculties of Agriculture and Veterinary Medicine, University of Nairobi, P. O. Box 29053 Nairobi, 00625 Kenya
3Department of Biological and Environmental Sciences, Alabama A&M University, 4900 Meridian Street, Normal AL 35762, USA
- Corresponding Author:
- Anthony Ananga
Center for Viticulture and Small Fruit Research
College of Agriculture and Food Science, Florida A&M University
6505 Mahan Drive, Tallahassee FL 32317, USA
Tel: +1-850-412-7393
Fax: +1-850-561-2617
E-mail: anthony.ananga@gmail.com, Anthony.ananga@famu.edu
Received date:: April 25, 2015; Accepted date:: May 18, 2015; Published date:: May 25, 2015
Citation: Hall J, Ananga A, Georgiev V, Ochieng J, Cebert E, et al. (2015) Molecular Cloning, Characterization, and Expression Analysis of Flavanone 3-Hydroxylase (F3H) Gene during Muscadine Grape Berry Development. J Biotechnol Biomater 5:180. doi:10.4172/2155-952X.1000180
Copyright: © 2015 Hall J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Biotechnology & Biomaterials
Abstract
Flavonoids are natural antioxidants that include the groups of notable pigments such as anthocyanins and proanthocyanidins. Flavanone 3-Hydroxylase (F3H) is a key enzyme needed for the biosynthesis of flavonoids, the main ingredients of muscadine grape extracts. This study reports the first successful isolation, cloning and characterization of F3H gene from Vitis rotundifolia Michx. The full length cDNA of V. rotundifolia F3H gene (designated as VrF3H) had an open reading frame (ORF) of 1081 bp encoding 364 amino acids with a calculated molecular mass of 40.8kDa as well as an isoelectric point of 5.60. Comparative and in silico analyses revealed that the cloned VrF3H from muscadine grapes has high identity with F3H from other plant species. The deduced VrF3H protein showed similarities with other available plant F3H proteins, and the conserved amino acids ligating ferrous iron and residues participating in 2-oxo-glutarate binding were found in similar positions comparable to other F3Hs. Furthermore, three-dimensional structure modeling showed that F3H protein had the enzyme core consisting of β-sheet, a typical structure shared by all 2-oxoglutarate-dependent dioxygenases including F3Hs. Phylogenetic tree analysis indicated that VrF3H belongs to the Vitis F3H cluster. VrF3H transcripts were found to be abundantly expressed in the in-vitro red cells, véraison and physiologically mature red berries, but not expressed in the skins of the green berries. The isolation and characterization of VrF3H gene will enable further study in the role of VrF3H gene in the biosynthesis of flavonoids in V. rotundifolia.
Keywords
Flavanone 3-hydroxylase; MVrF3H; Muscadine; Muscadinia rotundifolia (Michx.) Small
Introduction
Flavonoids are secondary metabolites present throughout the plant kingdom. They have important functions in the plant’s adaptations to specific ecological niches or its responses to biotic and abiotic stresses. However, some of the secondary compounds are known to be beneficial for humans as pharmaceuticals. Over the last decades, the pharmacological industry had slightly slowed down on the use of natural products and shifted more towards the combinatorial chemistry approach. But recently there has been a renewed interest from the industry to look for active natural products with putative pharmaceutical importance in plants [1]. Success has been limited in the combinatorial-chemistry to deliver novel drugs and consumer demand for natural products have increased, because of their high antioxidant activities while flavonoids are extensively used in human nutrition [2- 4] as anticancer, antimutagenic, antimicrobial, anti-inflammatory, and anti-atherosclerotic agents [4,5]. Current molecular tools are allowing scientists to clone and characterized structural genes from the flavonoid biosynthesis pathway in many plant species to enable the determination of their specific roles.
The structural genes of anthocyanin biosynthesis pathway in Vitis vinifera grapes have been greatly investigated at the molecular level [6,7]. Among them is the flavanone 3-hydroxylase (F3H), which is an important enzyme in the anthocyanin biosynthesis pathway. It catalyzes the stereospecific hydroxylation of (2S) -eriodictyol and (2S)-naringenin to form (2R, 3R)-dihydroquercetin and (2R, 3R)- dihydrokaempferol [8-10]. In other words, it acts at the bifurcation of the anthocyanin and flavonoid branches. The function of F3H was first described from crude extracts of Matthiola incama and illuminated through parsley cell cultures [9,10]. Subsequently, the F3H gene was cloned from Petunia hybrida and functionally expressed in Escherichia coli with high activity [11]. Afterwards, more F3H genes have been cloned and characterized from a variety of plant species: such as Hordeum vulgare [12], Malus [13], Medicago sativa [14], Zea mays [15], Arabidopsis thaliana [16] and Perilla frutescenes [17]. However, there have been no published reports on the molecular cloning and characterization of F3H in muscadine grapes. Nucleotide sequences of anthocyanin biosynthesis genes in muscadine grapes, are lacking in the databases except chalcone synthase (CHS) and dihyroflavonoid 4-reductase (DFR) genes that were reported as cloned by our lab, [18, 19].
In the present study, we report for the first time: (1) the isolation of a full-length cDNA clone of the gene encoding F3H from muscadine grapes (designated VrF3H ), (2) we compare the deduced amino acid sequence of muscadine VrF3H to other published F3H sequences from other plants, and (3) characterize the expression pattern of VrF3H gene in the in vitro grown cell culture and field grown green, véraison and physiologically mature muscadine berries using RT-PCR. In order to unveil the overall biosynthetic pathway of flavonoids in muscadine grapes, it is necessary to identify and characterize each gene involved in the pathway. Together with our previous work on CHS expression [20], these studies add to the molecular understanding of the biosynthesis of flavonoids and anthocyanins in muscadines, with significant practical implications for enhancing the specific nutraceutical contents in muscadine using genetic engineering.
Materials and Methods
Plant materials
Muscadinia rotundifolia (Michx.) Small “Noble” var. berry skins were harvested from the Florida A&M University vineyard at three different development stages (green, véraison and physiologically mature stage). Berries that were free of physical injuries and uniform in size were used. They were washed with distilled water; the skins were peeled and immediately frozen with liquid nitrogen and stored at -80ºC until use. in vitro red cell culture was established from sub-epidermal cells of the same vine [21] (patent publication no. US2011/0054195 A1). These cells were grown in a growth chamber at 23°C under a white light (150 μE m-2S-1) with a 16 h light/8h dark cycle. The developed callus produces anthocyanins.
Sample preparation for phytochemical assays
Two grams fresh tissue (callus and/or skins) were kept frozen at -80°C, then homogenized (5000 rpm, 5 min, Bio Homogenizer, Switzerland) in 10 ml extraction solvent (Methanol: 1% HCL – 1:1) and centrifuged at 11000 rpm for 15 min at 4ºC (Eppendorf 5804R, USA). The supernatant was collected and the residue was homogenized and re-extracted two more times. The combined extract was filtrated by 0.45 μm syringe filter and used for total anthocyanins assay.
Total anthocyanins assay
Total anthocyanins content was measured by using the pH differential spectrophotometric method [22]. Two portions of anthocyanin extracts were diluted (by using pre-determined dilution factor) with potassium chloride buffer (0.025 M, pH 1.0) and sodium acetate buffer (0.4 M, pH 4.5), respectively. After 15 min, the absorbance of both dilutions was measured at 520 nm and 700 nm against water (Ultrospec 3100 Pro UV–Vis Spectrophotometer, GE Healthcare, USA). The corrected absorbance of the diluted sample and the monomeric anthocyanin pigment concentrations were calculated as described elsewhere [22]. The total monomeric anthocyanins concentration was expressed as mg cyanidin-3,5-diglucoside equivalents (molecular weight = 611.5254, molar absorptivity ? = 30175) per g dry weight (DW). Dry weights of muscadine berries skins and callus tissue were determined by using MJ33 Compact Infrared Moisture Analyzer (Mettler Toledo, Switzerland).
RNA extraction, gel electrophoresis and cDNA synthesis
Samples were prepared from different tissues of ‘Noble’ grape as mentioned above. Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen, CA) according to the manufacturer’s protocol. RNA was quantified using Nano drop, and the inactivity was inspected by formaldehyde agarose gel electrophoresis. Purified RNA was treated with RNase-free DNAse 1, and immediately frozen to -20ºC. Formaldehyde gel electrophoresis (1% agarose) was used to evaluate the RNA quality. The gel apparatus (including the gel tray and comb) was treated with RNase Away and rinsed with RNase-free distilled water. Total RNA was used in primary gene expression profiling. The SuperScript First-strand Synthesis System for RT-PCR (Invitrogen) was used to synthesize cDNA in a 20 μL reaction containing 5 μg of DNase I-treated total RNA, 20mM Tris-HCl (pH 8.4), 50mM KCl, 2.5mM MgCl2, 10mM dithiothreitol, 0.5 mg oligo (dT), 0.5mM each of dATP, dGTP, dCTP, and dTTP, and 200U SuperScript II Reverse Transcriptase. RNA, dNTPs, and oligo (dT) were mixed first, heated to 65°C for 5 min, and placed on ice until the addition of the remaining reaction components. The reaction was incubated at 50°C for 50 min, and terminated by heat inactivation at 85°C for 5 min. The cDNA product was treated with 1mL of Rnase H (Invitrogen) for 20 min at 37°C. An identical reaction without the reverse transcriptase was performed to verify the absence of genomic DNA (no-RT control). The cDNA was stored at - 20°C until it was ready for use.
Cloning of VrF3H full-length cDNA by RACE
After RNaseH treatment, the single-strand cDNA mixtures were used as templates for PCR amplification of the core fragment of F3H from V. rotundifolia. Two primers, FF3H (5’-GCGCCTACGACACTGAC-3’) and RF3H (5’-TAGGCCAAAATCTGGTC-3’) were designed based on the conserved regions of F3H and used for the amplification of the core cDNA fragment of VrF3H. The core fragment was amplified at the annealing temperature of 56°C, and sub-cloned into pMDTM 18-T vector (TaKaRa, Japan), transformed into Escherichia coli strain DH5α followed by sequencing. The core fragment was subsequently used to design the primers for the cloning of the full-length cDNA of VrF3H by RACE.
SMARTTM RACE cDNA Amplification Kit (Clontech, USA) was used to amplify the 3’-end and 5’-end of VrF3H cDNA. The firststrand 3’-RACE-ready and 5’-RACE-ready cDNA samples from V. rotundifolia were prepared according to the manufacturer’s protocol and used as templates for 3’-RACE and 5’-RACE, respectively. Two 3’-gene-specific primers were designed for 3’-RACE. For the first cycle of amplification of 3’-end of VrF3H cDNA, VrF3H3-1 (5’-CACCATGGCGCCTACGACACTG-3’) and UPM (Universal Primer A Mix, 5’- CTAATACGACTCACTATA GGGCAAGCAGTGGTA TCAACGCAGAG-3’ and 5’-GGCGCCTACGACA CTGACG-3’) were used with 3’-RACE-ready cDNA as the template. For the nested amplification of 3’-RACE, VrF3H3-2 (5’- TTAGGCCAAAA TCTGGTCAATG-3’) and NUP (Nested Universal Primer A, 5’- AAGCAGTGGTATCAACGCAGAGT-3’) were used with the products of the first amplification as templates. Two 5’-gene-specific primers were designed for 5’-RACE. For the first cycle of amplification of 5’-RACE, VrF3H5-1 (5’-TTTCACC CCTGTTTCCTCC TCTGCTTC-30) and UPM were used with 5’-RACE-ready cDNA as the template. For the nested amplification of 5’- RACE, VrF3H5-2 (5’-GGCCAAAATCT GGTCAAT-3’) and NUP were used with the products of the first amplification as templates. The first and nested PCR procedures were carried out under the same conditions described in the protocol (SMARTTM RACE cDNA Amplification Kit, User Manual, Clontech): 35 cycles of amplification (30 s at 94ºC, 30 s at 68ºC, 2 min at 72ºC).The nested 3’-RACE and 5’-RACE products were purified and sub-cloned into pGEM-T easy vector, and then transferred into E.coli strain JM109. The pGEM-VrF3H plasmid was sequenced from both ends at Eurofins mwg/Operon (Huntsville, Alabama U.S.A). The PCR amplification and sequencing for the full-length cDNA of VrF3H was repeated three times to avoid PCR errors. The full-length cDNA of VrF3H was subsequently analyzed for the homology to other F3H genes.
Relative-quantitative real-time PCR using SYBR green assay
Relative-quantitative real-time PCR reactions were performed in a 96-well plate with an iCycler iQ Multicolor Real-Time PCR Detection system (Bio-Rad; [23]), using an iQ SYBR Green Supermix (Bio-Rad) to monitor cDNA amplification, according to the manufacturer’s protocol. Three independent experiments were performed for each sample. After the real-time PCR had been performed, the absence of unwanted byproducts was confirmed by an automated melting curve analysis and agarose gel electrophoresis of the PCR products. In all the experiments, three replicates for each RNA sample were included; averages were calculated, and differences in the threshold cycle (Ct) were evaluated. The comparative Ct method was used, which mathematically transforms the Ct data into the relative transcription-level genes. When comparing the expression of VrF3H in different tissues, the relative quantification of VrF3H expression was achieved by calibrating its transcription level to that of the reference gene, Actin. The expression level calculated by the formula 2- ΔΔCt represents the x-fold difference from the calibrator.
Search for muscadine F3H-related sequences
F3H sequence was retrieved through Basic Local Alignment Tool (BLAST), homology, and domain searches in public domains, namely GenBank (www.ncbi.ncbi.nlm.nih.gov). The GenBank submitted F3H protein sequence of muscadine (Accession no. KF040970) was used for BLAST and homology searches against other plants.
Multiple sequence alignments and phylogenetic tree construction
Multiple alignment of the putative amino-acid sequence of muscadine F3H was performed using the T-Coffee program [24]. The alignment of 20 F3H proteins was summarized using the Plotcon sequence similarity graph (http://bioweb2.pasteur.fr/docs/ EMBOSS/plotcon .html), which represents the similarity along the set of aligned sequences. The molecular phylogenetic tree for F3H was built with Neighbor Joining, using p-distance as a substitution model, and Maximum Parsimony methods in MEGA Version 5.0, with 5000 iterations for calculating boostrap confidence levels [25]. The phylogenetic tree construction included the sequences for the muscadine F3H protein and/or putative F3H proteins reported in the NCBI database for 20 plants.
Protein three-dimensional structure prediction
The Muscadine F3H structural model was obtained from its amino-acid sequence by using the SWISS MODEL (http://swissmodel. expasy.org) [26] and Protein Homology/analogy Recognition Engine (PHYRE) (www.sbg.bio.ic.ac.uk/phyre) prediction servers [27,28]. The model obtained was classified according to identity percentage.
Results
Cloning of full-length VrF3H cDNA and sequence analysis
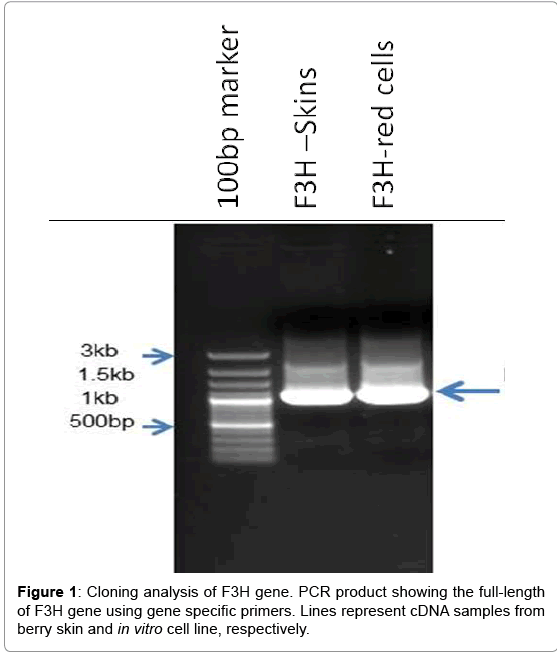
Following PCR amplification, an approximately 985 bp product was obtained and sequenced. A BLAST search revealed that this 985 bp cDNA fragment had high homology to F3H genes from Vitis species (99% identity to Vitis vinifera). Thus, this fragment was used to design gene specific primers for both 5’-RACE and 3’-RACE.
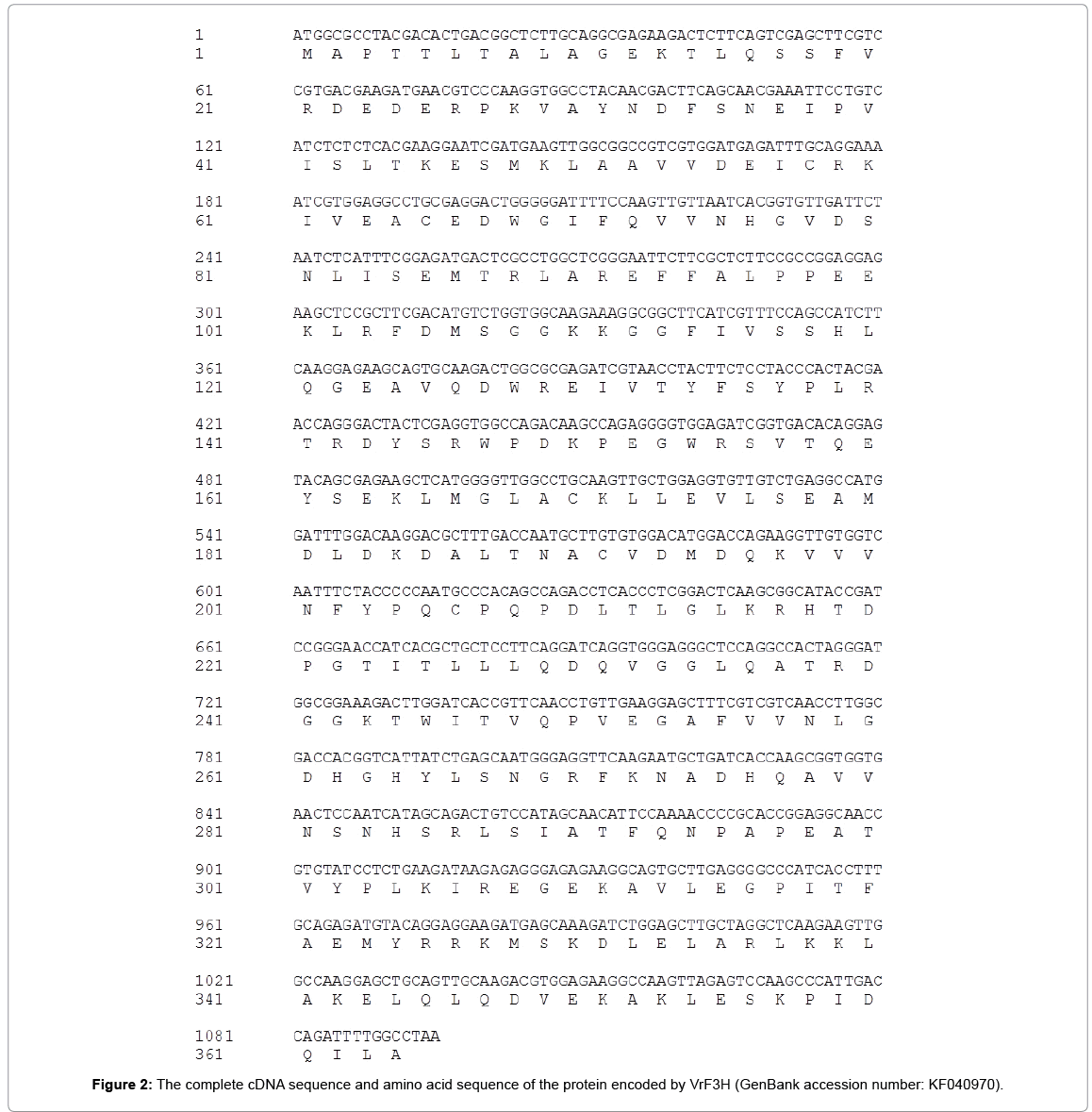
Using 3’-RACE and 5’-RACE, two PCR products sized about 500 and 315 bp were obtained, respectively. The products were sequenced and confirmed to be the 364 bp 3’-end and 255 bp 5’-end. By aligning and assembling the sequences of 3’-RACE, 5’-RACE and the core fragment on Contig Express (Vector NTI Suite 8.0), the full-length cDNA sequence of VrF3H with 1081 bp (Figure 1) was deduced and subsequently confirmed by sequencing (Figure 2). ORF Finder program analysis on NCBI showed that the VrF3H contained a 1081 bp ORF encoding a protein of 364 amino acids (Figure 2) with a calculated molecular mass of 40.8 kDa and an isoelectric point of 5.60.
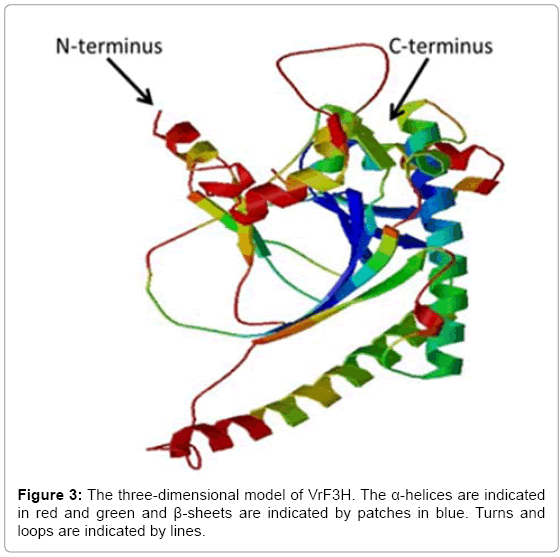
The deduced amino acid sequence of VrF3H was submitted to the NCBI for PSI-BLAST search and the result showed that VrF3H was homologous to F3H sequences from other plant species, with 97% identity to F3H from V. vinifera. VrF3H was also similar to F3H from P. lactiflora (96% identities), P. suffruti (95% identities), which suggests that VrF3H belongs to the F3H family. The search for a conserved domain database in the GenBank revealed that VrF3H belongs to the 2OG-FeII_Oxy superfamily. This family contains other genes like flavonol synthase (FLS) and anthocyanidin synthase (ANS). Our analysis indicated that VrF3H contains conserved domains from 2-ODD superfamily similar to other members of the 2OG-Fe (II) dioxygynase family. A comparative modeling of 3-D structure of VrF3H (Figure 3) was performed using SWISS-MODEL software. This was done to enhance an understanding of VrF3H. The 3-D model showed the presence of a conserved undisturbed helix on the surface containing a noticeable motif with amino acids resembling a putative leucine zipper.
Phylogenetic relationships of F3Hs
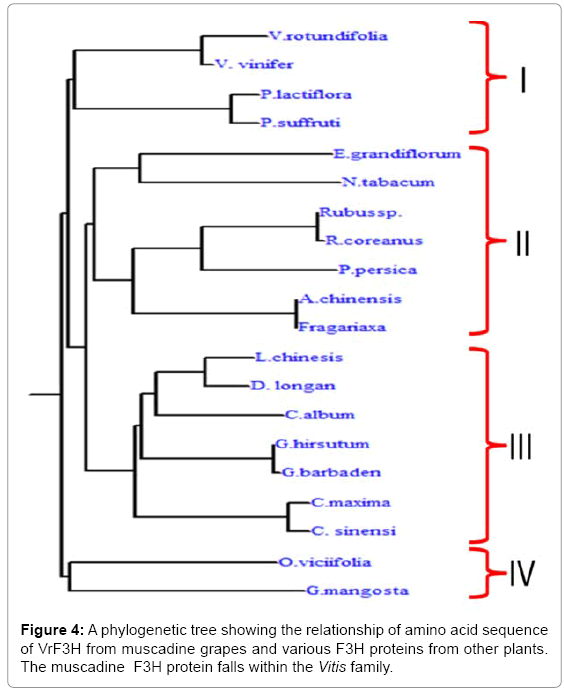
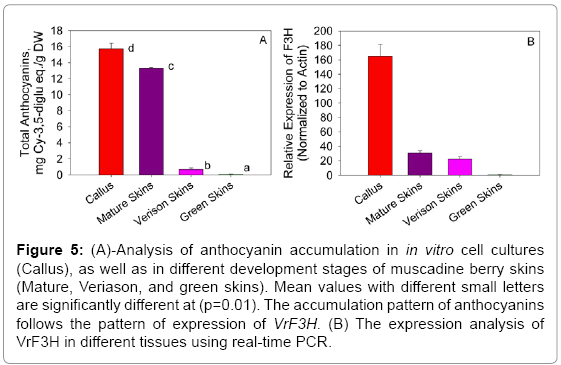
Phylogenetic tree was constructed using the predicted amino sequence of the putative F3H protein from muscadine as well as other plant species (Figure 4). The F3Hs from different plant species were divided into four subgroups: I, II, III, and IV (Figure 5). Our cloned VrF3H belongs to I subgroup, close to V. vinifera. In fact, part of the branch forming subgroup I are members of the Vitis spp. Phylogenetic tree confirmed that there is a close relationship between muscadine grapes F3H and V. vinifera F3H. Our results concur with those by [15], which suggest that F3H is well conserved among plants of different classifications and has distinct species specificity.
Figure 5: (A)-Analysis of anthocyanin accumulation in in vitro cell cultures (Callus), as well as in different development stages of muscadine berry skins (Mature, Veriason, and green skins). Mean values with different small letters are significantly different at (p=0.01). The accumulation pattern of anthocyanins follows the pattern of expression of VrF3H. (B) The expression analysis of VrF3H in different tissues using real-time PCR.
Expression profile analysis of VrF3H gene in berry skin and in vitro cell lines and total anthocyanin assay
The expression of VrF3H was investigated in samples taken throughout the development of muscadine grape berry skin tissues (Figure 5B). Real-time PCR analysis indicated that VrF3H gene expression occurred in two phases. It was expressed at veraison, the start of the berry ripening and the expression continued to physiological maturity stage. VrF3H was highly expressed in in vitro cell lines starting from the 12th day of cultivation. There was no expression of VrF3H at the green stage of berry development. Following this period of little or no expression (green berries), there was a coordinated increase in expression of VrF3H gene. Expression of VrF3H gene then continued throughout the remainder of berry development. Thus, the expression of VrF3H gene coincided with other genes previously investigated in the anthocyanin biosynthetic pathway [7,17,29]. The expression of VrF3H, coincided precisely with the accumulation of anthocyanin pigments in the berry skin as indicated in Fig. 5A. The accumulation of total anthocyanin was monitored in all the samples, and the results showed that in vitro red cells had the highest accumulation followed by mature red skins (Figure 5A) with significant difference between the in vitro red cell cultures and physiologically mature skins.
Discussion
Results from this study further support that, functions of different plant species and their relationship in genome structure can be determined by comparative genomics [30]. Previous research indicates that F3H proteins from different species are highly conserved [13-15,17], and that the genes encoding F3H protein have been characterized at the genetic, chemical, and proteomic levels [16,29,31- 34]. However, cloning of an F3H gene from muscadine grapes has not yet been reported. In this study, VrF3H gene from muscadine grapes was isolated, cloned and characterized. The isolated VrF3H cDNA was verified to have 1081bp encoding 364 amino acids. Comparison of its amino acid sequence showed high homologies (>90%) with the proteins of seven species: V. vinifera, P. lactiflora, P. suffruti, E. grandiflorum, N. tabacum, P. persica, and A. chinensis. Our results indicate that VrF3H is highly conserved and this is in agreement with [16]. From this study we determined that VrF3H gene is highly expressed in in vitro red cell cultures, as compared to véraison or physiologically mature berry skins (Figure 5B). The expression of VrF3H is controlled by transcriptional regulators, which have been studied in other plants including for the regulation of F3H. Quattrochio et al., [35] reported that F3H gene is independently expressed in Petunia, however, according to Martin et al., [36], F3H expression in Antirrhinum is coordinately controlled with downstream genes such as dihydroflavonol reductase. In Arabidopsis, F3H gene is coordinately controlled with the upstream genes for chalcone isomerase and chalcone synthase [16]. In another study, Quattrochio et al., [37] demonstrated deferential control of F3H expression in Z. mays and P. hybrid, which indicated that regulatory anthocyanin genes were conserved between species and that divergent evolution of the target gene promoters were responsible for the speciesspecific differences in regulatory networks.
However, the appearance of anthocyanins in muscadine grape berry skins at the onset of ripening coincides with increased expression of VrF3H gene encoding F3H enzyme in anthocyanin biosynthetic pathway. This means that VrF3H is one of the rate-determining enzymes in anthocyanin biosynthesis pathway of V. rotundifolia. But, further investigations are required to characterize the F3H enzyme activity and fully understand its structural role in anthocyanin biosynthesis. The tissue-specific expression analysis revealed higher levels of F3H expression in in vitro cell lines than in véraison and physiologically mature berry skins. This indicates that in vitro cultivation of pigmentspecific tissues can be an improved model of increasing the production of anthocyanin for pharmaceuticals. According to Jeong et al., [38], the expression of F3H gene can also be induced by abiotic and biotic stresses. For example, Shinozaki and Yamaguchi-Shinozaki [39] demonstrated that F3H genes not only showed increased expression under drought stress, but also under salt, cold, and hormonal stresses. Since different hormones were used in our in vitro cultivation processes in this study, the cells likely underwent a certain level of stress, which could have led to increased expression of the VrF3H gene (Figure 5). However, studies have also shown that plants that contain gene families from F3H show different transcription patterns [40]. According to Khlestkina et al., [40] the transcript level of RtF3H2 was significantly higher than that of RtF3H1 in the root of R. trigyna. However, both genes were highly expressed in the stem and not in the leaves. But in a separate study, F3H was highly expressed in the roots and stems of alfalfa seedlings [41]. Therefore, the expression of VrF3H suggests tissue specificity, which allows anthocyanin accumulation in the skins, as well as in the in vitro cell lines of muscadine grapes.
In Shiraz grape berries, anthocyanins have been determined to accumulate in the skin but not in the flesh [42]. The pattern of expression seen in the muscadine berry skin was similar to that in the berry skin of Shiraz grape, where anthocyanins begin to accumulate at about véraison, and this coincides with the increase in expression of F3H. This suggests that there is coordinated regulation of F3H gene during the development of grape berry skin. According to Sparvoli et al. [7], as anthocyanins accumulate in dark-grown grape seedlings subsequently exposed to light, there is a coordinated induction of the genes from the committed steps of the anthocyanin biosynthetic pathway. This is similar to the control of the anthocyanin biosynthetic pathway in maize aleurone, which is regulated by the R and C1 gene families [43].
Due to the nutritional and physiological roles of flavonoids, it is essential to understand the flavonoid pathway in muscadine grapes. Therefore, cloning and characterization of genes encoding key enzymes or transcriptional factors is the first step to understanding the regulatory mechanisms controlling flavonoid biosynthesis in muscadine grapes. It is clear that much more work is required before the regulation of flavonoid biosynthesis pathway and its biological significance are fully understood in muscadine grapes. We have isolated a muscadine F3H gene and performed some necessary analysis. But further investigations are required to characterize enzymatic activities of VrF3H to fully understand and deduce its regulatory role in anthocyanin biosynthesis.
Acknowledgements
The research has been done with the financial support of USDA/NIFA/AFRI Plant Biochemistry Program Grant # 2009-03127 and USDA/NIFA/1890 Capacity Building Grant #2010-02388.
References
- Müller-Kuhrt L (2003) Putting nature back into drug discovery. Nat Biotechnol 21: 602.
- Potapovich AI, Kostyuk VA (2003) Comparative study of antioxidant properties and cytoprotective activity of flavonoids. Biochemistry (Mosc) 68: 514-519.
- Middleton E Jr, Kandaswami C, Theoharides TC (2000) The effects of plant flavonoids on mammalian cells: implications for inflammation, heart disease, and cancer. Pharmacol Rev 52: 673-751.
- Ananga A, Georgiev V, Tsolova V (2013) Manipulation and engineering of metabolic and biosynthetic pathway of plant polyphenols. Curr Pharm Des 19: 6186-6206.
- Georgiev V, Ananga A2, Tsolova V3 (2014) Recent advances and uses of grape flavonoids as nutraceuticals. Nutrients 6: 391-415.
- Ford CM, Boss PK, Hoj PB (1998) Cloning and characterization of vitisviniferaUDP-Glucose: flavonoid 3-O-glucosyltransferase, a homologue of the enzyme encoded by the maize bronze-1Locus that may primarily serve to glucosylateanthocyanidins in vivo. JBiolChem 273: 9224-9233.
- Sparvoli F, Martin C, Scienza A, Gavazzi G, Tonelli C (1994) Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitisvinifera L.). Plant MolBiol 24: 743-755.
- Britsch L, Grisebach H (1986) Purification and characterization of (2S)-flavanone 3-hydroxylase from Petunia hybrida. Eur J Biochem 156: 569-577.
- Forkmann G, Heller W, Grisebach H (1980) Anthocyanin biosynthesis in flowers of Matthiolaincanaflavanone 3-and flavonoid 3'-hydroxylases. Zeitschrift fur Naturforschung. Section C: 691-695.
- Britsch L, Heller W, Griesbach H (1981) Conversion of flavanone to flavone, dihydroflavonol and flavonol with an enzyme system from cell cultures of parsley. ZeitschriftfuerNaturforschung, Section c 36: 742-750.
- Britsch L, Ruhnau-Brich B, Forkmann G (1992) Molecular cloning, sequence analysis, and in vitro expression of flavanone 3 beta-hydroxylase from Petunia hybrida. J BiolChem 267: 5380-5387.
- Meldgaard M (1992) Expression of chalcone synthase, dihydroflavonolreductase, and flavanone-3-hydroxylase in mutants of barley deficient in anthocyanin and proanthocyanidin biosynthesis. TheorAppl Genet 83: 695-706.
- Davies KM (1993) A cDNA clone for flavanone 3-hydroxylase from Malus. Plant Physiol 103: 291.
- Charrier B, Coronado C, Kondorosi A, Ratet P (1995) Molecular characterization and expression of alfalfa (Medicago sativa L.) flavanone-3-hydroxylase and dihydroflavonol-4-reductase encoding genes. Plant MolBiol 29: 773-786.
- DebooGB,Albertsen MC, Taylor LP (1995) Flavanone 3-hydroxylase transcripts and flavonol accumulation are temporally coordinate in maize anthers. Plant J 7: 703-713.
- Pelletier MK, Shirley BW (1996) Analysis of flavanone 3-hydroxylase in Arabidopsis seedlings. Coordinate regulation with chalcone synthase and chalconeisomerase. Plant Physiol 111: 339-345.
- Gong Z, Yamazaki M, Sugiyama M, Tanaka Y, Saito K (1997) Cloning and molecular analysis of structural genes involved in anthocyanin biosynthesis and expressed in a forma-specific manner in Perillafrutescens. Plant MolBiol 35: 915-927.
- Samuelian SK, Simova EP, Colova VM (2009)Chalcone synthase gene (CHS1) expressed in the North American 'Noble' muscadinia var. during ripening.
- Ananga AO, Acheampong K, Zheng C, Kambiranda D, Georgiev V, et al.(2013) Vitisrotundifolia cultivar Noble dihydroflavonol-4-reductase mRNA, complete cds.
- Davis G, Ananga A, Krastanova S, Sutton S, Ochieng JW, et al. (2012) Elevated gene expression in chalcone synthase enzyme suggests an increased production of flavonoids in skin and synchronized red cell cultures of North American native grape berries. DNA Cell Biol 31: 939-945.
- Colova V (2009) Synchronized strains of subepidermal cells of muscadine (muscadine sp.) grapevine pericarp for use as a sourse of flavonoids (nutraceuticals). Google Patents.
- Giusti MM, Wrolstad RE (2001) Characterization and measurement of anthocyanins by UV-visible spectroscopy. Current Protocols in Food Analytical Chemistry.
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3: RESEARCH0034.
- Notredame C, Higgins DG, Heringa J (2000) T-Coffee: A novel method for fast and accurate multiple sequence alignment. J MolBiol 302: 205-217.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. MolBiol and Evol 28: 2731-2739.
- Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37: D387-392.
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389-3402.
- Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA (2008) Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins 70: 611-625.
- Shen G, Pang Y, Wu W, Deng Z, Zhao L, et al. (2006) Cloning and characterization of a flavanone 3-hydroxylase gene from Ginkgo biloba. Biosci Rep 26: 19-29.
- Hardison RC (2003) Comparative genomics. PLoSBiol 1: E58.
- KoesRE, Spelt CE, van den Elzen PJ, Mol JN (1989) Cloning and molecular characterization of the chalcone synthase multigene family of Petunia hybrida. Gene 81: 245-257.
- Ursula Niesbach-Klösgen, Ellen Barzen, Jürgen Bernhardt, Wolfgang Rohde, Zsuzsanna Schwarz-Sommer, et al. (1987)Chalcone synthase genes in plants: a tool to study evolutionary relationships. Journal of Molecular Evolution 26: 213-225.
- Ferrer JL, Jez JM, Bowman ME, Dixon RA, Noel JP, et al. (1999) Structure of chalcone synthase and the molecular basis of plantpolyketide biosynthesis. NatStructBiol 6: 775-784.
- Jez JM, Noel JP (2000) Mechanism of chalcone synthase. pKa of the catalytic cysteine and the role of the conserved histidine in a plant polyketide synthase. J BiolChem 275: 39640-39646.
- Quattrocchio F, Wing JF, Leppen H, Mol J, Koes RE (1993) Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 5: 1497-1512.
- Martin C, Prescott A, Mackay S, Bartlett J, Vrijlandt E (1991) Control of anthocyanin biosynthesis in flowers of Antirrhinum majus. Plant J 1: 37-49.
- Quattrocchio F, Wing JF, van der Woude K, Mol JN, Koes R (1998) Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J 13: 475-488.
- Jeong ST, Goto-Yamamoto N, Kobayashi S, Esaka M (2004) Effects of plant hormones and shading on the accumulation of anthocyanins and the expression of anthocyanin biosynthetic genes in grape berry skins. Plant Science 167: 247-252.
- Shinozaki K, Yamaguchi-Shinozaki K (1996) Molecular responses to drought and cold stress. CurrOpinBiotechnol 7: 161-167.
- Khlestkina EK, Röder MS, Salina EA (2008) Relationship between homoeologous regulatory and structural genes in allopolyploid genome - a case study in bread wheat. BMC plant biol 8: 88.
- Shen X, Martens S, Chen M, Li D, Dong J, et al. (2010) Cloning and characterization of a functional flavanone-3ß-hydroxylase gene from Medicagotruncatula. MolBiol Rep 37: 3283-3289.
- Boss PK, Davies C, Robinson SP (1996) Analysis of the Expression of Anthocyanin Pathway Genes in Developing Vitisvinifera L. cv Shiraz Grape Berries and the Implications for Pathway Regulation. Plant Physiol 111: 1059-1066.
- Martin C, Gerats T (1993) Control of Pigment Biosynthesis Genes during Petal Development. Plant Cell 5: 1253-1264.
Relevant Topics
- Agricultural biotechnology
- Animal biotechnology
- Applied Biotechnology
- Biocatalysis
- Biofabrication
- Biomaterial implants
- Biomaterial-Based Drug Delivery Systems
- Bioprinting of Tissue Constructs
- Biotechnology applications
- Cardiovascular biomaterials
- CRISPR-Cas9 in Biotechnology
- Nano biotechnology
- Smart Biomaterials
- White/industrial biotechnology
Recommended Journals
Article Tools
Article Usage
- Total views: 15338
- [From(publication date):
August-2015 - Jul 15, 2025] - Breakdown by view type
- HTML page views : 10651
- PDF downloads : 4687