Molecular Characterization of Rice Land Races by SSR Markers
Received: 24-Jun-2021 / Manuscript No. JPGB-22-34625 / Editor assigned: 01-Jan-1970 / PreQC No. JPGB-22-34625(PQ) / Reviewed: 20-Jan-2022 / QC No. JPGB-22-34625 / Revised: 22-Jan-2022 / Manuscript No. JPGB-22-34625(R) / Accepted Date: 27-Jan-2022 / Published Date: 28-Jan-2022 DOI: 10.4172/jpgb.1000112
Abstract
A study was conducted to analysis the molecular characterization of twenty two rice land races at Pandit Jawaharlal Nehru College of Agriculture and Research Institute Karaikal . The SSR marker based diversity analysis grouped the 29 accessions in to 17 clusters which showed its efficiency in grouping the dissimilar individuals. The SSR marker method discriminated the accessions better than the morphological markers. The primer RM 21 identified more number of alleles and based on Polymorphic Information Content (PIC) the primer RM 493 is ascertained as more informative than the rest of the 14 primers used.
Keywords: Rice land races; SSR markers; Clustering; Genetic diversity
Keywords
Rice land races; SSR markers; Clustering; Genetic diversity
Introduction
Land races played an important role in the local food security and sustainable development in agriculture, in addition to their significance as genetic resource for rice genetic improvement [1]. Genetic diversity is conventionally assessed by morphological traits. However, such traits are affected by environment, phenology or development stage of the plant and the type of plant material [2,3]. Further, it requires testing in several replications and environments to establish the genetic contributions, which is highly influenced by several factors. Hence there is a need to go in for a highly reliable and precise method for assessment of genetic variability with no environmental effects. Molecular markers are considered an efficient, powerful tool for the assessment of genetic relationships.
Marker analysis helps to understand the genetic make up of the accessions and also make it possible to analyze the global organization of genetic diversity within a species. In contrast to morphological traits, molecular markers can reveal difference among genotypes at the DNA level, providing a more direct, reliable and efficient tool for germplasm characterization, conservation and management. Several DNA markers are used for molecular diversity anlaysis of which Simple Sequence Repeats (SSR) or microsatellites are a class of repetitive DNA element, and these are the di-,tri- or tetra nucleotide with 5-50 copies per loci, such as (AT)29, (CAC)16, or (GACA)32. Microsatellites are PCR based markers that are technically efficient, cost effective, and common in rice [4]. Compared to RFLP, microsatellite markers detect a significantly higher degree of polymorphism in rice [5] and are especially suitable for evaluating genetic diversity among closely related rice cultivars (or) accessions [6].
A large number of land races are being cultivated which are well adapted to local ecosystems and withstand the adverse effects such as drought, salinity but with a very poor yield [7]. An increase in the yield potential of these land races and at the same time maintaining the desirable traits would be a major objective in rice breeding programme. However assessment of genetic diversity of rice landraces was not given much thrust and the same needs to be studied to identify genetically diverse accession having desired genes which could be better utilised in crop breeding programme.
Materials and methods
The materials used in the study comprise of twenty nine rice land races collected from Department of Plant Breeding and Genetics, Pandit Jawaharlal Nehru College of Agriculture and Research Institute Karaikal, UT of Puducherry. The details of the genotypes are given in Table 1. A field trial was conducted using twenty nine rice land races (Plate.1). These genotypes were sown in raised nursery bed during Kharif, 2008 and 25 days old seedlings were transplanted to the main field in a Randomized Block Design replicated thrice. Each genotype was transplanted in three rows of 3m length adopting a spacing of 30cm between rows and 20cm between plants. All the recommended agronomic package of practices were adopted during the entire crop growth period.
| Sl.No. | Genotype | Origin |
|---|---|---|
| 1 | Orumanayoor Anakkodan | Kerala |
| 2 | Vytilla Anakkodan | Kerala |
| 3 | Chettivirippu | Kerala |
| 4 | Chettivirippu Kannamalai | Kerala |
| 5 | Chithyankottai | Tamil Nadu |
| 6 | Chitteani | Kerala |
| 7 | Gopalbhag | Tamil Nadu |
| 8 | Pokkali | Kerala |
| 9 | Jodumani | Tamil Nadu |
| 10 | Kadamakudy Pokkali | Kerala |
| 11 | Kethanur | Kerala |
| 12 | Koorgood | Kerala |
| 13 | Kulavazhai | Tamil Nadu |
| 14 | Vettaikaraniruppu Kulivedichan | Tamil Nadu |
| 15 | Vedaranyam Kulivedichan | Tamil Nadu |
| 16 | Njavara | Kerala |
| 17 | Pallipuram Pokkali | Kerala |
| 18 | Pant Kalanamak 3131 | Utttar Pradesh |
| 19 | Patnai 23 | West Bengal |
| 20 | Cherthallai Pokkali | Kerala |
| 21 | Edavanakad Pokkali | Kerala |
| 22 | Ponnarayan | Kerala |
| 23 | Red Thriveni | Kerala |
| 24 | Sivappu Chitraikar | Tamil Nadu |
| 25 | Sivappu Koompalai | Tamil Nadu |
| 26 | Thulasi Manjari | Bihar |
| 27 | Vattan | Kerala |
| 28 | Vellai Chitraikar | Tamil Nadu |
| 29 | Vellaikurikar | Tamil Nadu |
Table 1: Details of genotypes.
Genetic diversity studies based on Simple Sequence Repeat (SSR) markers
Isolation of Genomic DNA
All the twenty nine genotypes were sown in laboratory condition for the extraction of DNA from leaf samples. DNA was isolated from five days old seedlings from each of the entries for molecular diversity analysis. DNA was extracted by adopting the protocol developed by Dellaporta et al. [8]
Reagents used Cetyl Trimethyl Ammonium Bromide (CTAB) extraction Buffer (100mL):
CTAB 2% (w/v)
Tris HCl (pH8.0) 200 mM
Sodium Chloride 1.4 M
EDTA (pH 8.0) 20 mM
(Tris, sodium chloride and EDTA were autoclaved and 2% CTAB was added after autoclaving and pre heated before using the buffer)
Liquid Nitrogen
β Mercapta ethanol
Ice-cold Isopropanol
Chloroform: Isoamylalcohol (24:1) (v/v)
Ethanol (70 and 100%)
Extraction of Genomic DNA
One gram of fresh and clean leaves were cut into bits with the help of sterile scissors and transferred to prechilled mortar. The leaf tissues were frozen using liquid nitrogen and ground to fine powder. The fine powder was allowed to thaw in the presence of 800μl of preheated extraction buffer in the polypropylene centrifuge tubes, to this 100μl of β-mercaptaethanol was added and incubated for 30-45 minutes at 65ºC in water bath with occasional mixing. The tubes were removed from the water bath, equal volume of 800μl of chloroform: Isoamylalcohol mixture (24:1 v/v) was added and mixed by inversion for 1 hour. It was centrifuged at 10,000 rpm for 20 minutes at room temperature. The clear aqueous phase was transferred to a new sterile tube. Equal volume of Ice-cold isopropanol was added, mixed gently by inversion and then kept in the freezer until DNA was precipitated. The mixture was centrifuged at 13,000 rpm at room temperature for 10 minutes and the supernatant was decanted retaining the pellet. The pellet was washed with 100μl of 70 per cent ethanol and centrifuged at 6000rpm for 5 minutes. The alcohol was discarded and DNA was completely air dried. The dried pellet was resuspended in 100μl of sterile water.
Quantification of DNA
The genomic DNA extracted from the young leaf samples was tested for its intactness by electrophoresis. The DNA was quantified with visually on agarose (0.8%) gel by staining with ethidium bromide. 10μl of each genotype DNA was loaded with tracking dye along with 1 Kb DNA. After the tracking reached one third of the gel, documentation was done using UV transilluminator. The quantity of isolated DNA was determined by its corresponding ladder DNA. Based on this, the dilution of DNA was done for SSR amplification.
PCR Amplification
Sequence of micro satellite primer pairs were downloaded from Genome data bases, Rice genes micro satellite markers (http://www. gramene.org/markers/microsat/ssr.html) and 15 primers were chosen randomly covering all the 12 chromosomes and the details of the primer sequences and the product size are given in Table 2. These primer sequences synthesized by Sigma Aldrich Inc., Bangalore were utilized for amplification.
| Sl. No. | Oligo Name | Chromosome location |
Sequence (51-31) | Product Size (bp) |
|---|---|---|---|---|
| 1 | RM493 | 1 | TAGCTCCAACAGGATCGACC* GTACGTAAACGCGGAAGGTG** |
211 |
| 2 | RM110 | 2 | TCGAAGCCATCCACCAACGAAG* TCCGTACGCCGACGAGGTCGAG** |
156 |
| 3 | RM53 | 2 | GCCTCGAGCATCATCATCAG* ATCAACCTGCACTTGCCTGG** |
182 |
| 4 | RM489 | 3 | ACTTGAGACGATCGGACACC* TCACCCATGGATGTTGTCAG** |
271 |
| 5 | RM261 | 4 | CTACTTCTCCCCTTGTGTCG* TGTACCATCGCCAAATCTCC** |
125 |
| 6 | RM131 | 4 | TCCTCCCTCCCTTCGCCCACTG* CGATGTTCGCCATGGCTGCTCC** |
215 |
| 7 | RM403 | 5 | GCTGTGCATGCAAGTTCATG* ATGGTCCTCATGTTCATGGC** |
241 |
| 8 | RM343 | 6 | CCACGAACCCTTTGCATC* GTGATGATGCGTCGGTTG** |
233 |
| 9 | RM429 | 7 | TCCCTCCAGCAATGTCTTTC* CCTTCATCTTGCTTTCCACC** |
159 |
| 10 | RM481 | 7 | TAGCTAGCCGATTGAATGGC* CTCCACCTCCTATGTTGTTG** |
169 |
| 11 | RM515 | 8 | TAGGACGACCAAAGGGTGAG* TGGCCTGCTCTCTCTCTCTC** |
211 |
| 12 | RM524 | 9 | TGAAGAGCAGGAACCGTAGG* TCTGATATCGGTTCCTTCGG** |
198 |
| 13 | RM222 | 10 | CTTAAATGGGCCACATGCG* CAAAGCTTCCGGCCAAAAG** |
213 |
| 14 | RM21 | 11 | ACAGTATTCCGTAGGCACGG* GCTCCATGAGGGTGGTAGAG** |
157 |
| 15 | RM17 | 12 | TGCCCTGTTATTTTCTTCTCTC* GGTGATCCTTTCCCATTTCA** |
184 |
| * Forward primer **Reverse primer |
||||
Table 2: Details of SSR primers used for PCR amplification.
The total reaction volume was 15μl, the cocktail for the amplification was prepared as follows in 0.2ml PCR tubes.
DNA 2.00μL
dNTPs (2.5mm) (Bangalore Genei Ltd., India) 1.50μL
Primer (Sigma Aldrich Inc., USA) 2.00μL
PCR buffer 1.50μL
Taq Polymerase (3 units/μL) (Bangalore Genei Ltd., India) 0.2μL
Magnesium chloride 0.3μL
Sterile distilled H2O 7.50μL
The reaction mixture was given a short spin for thorough mixing of the cocktail components. Then the 0.2mL PCR-tubes were loaded on to a thermal cycler (MJ Research Inc. USA). The thermal cycler was programmed as follows.
Profile 1: 95ºC for 1 minute Initial denaturation
Profile 2: 94ºC for 45 seconds denaturing
Profile 3: 55ºC for 1 minute Annealing 34 cycles
Profile 4: 72ºC for 1 minute 30 seconds Extension
Profile 5: 72ºC for 10 minutes Final Extension
Profile 6: 4ºC for infinity to hold the samples
Agarose Gel Electrophoresis
After PCR amplification, products were separated by electrophoresis on agarose gels visualized by ethidium bromide staining.
Materials
Loading dye
Bromophenol Blue 0.5% (w/v)
10 x TBE (Tris Borate EDTA) Buffer
Trisbase – 10.76 gm
Boric acid – 5.5 gm
EDTA – 4 ml
(Dissolved in 800mL and made upto 1000mL and stored at 4ºC)
Protocol
The open ends of the Pyrex gel casting plate were sealed with cello tape and placed on a perfectly horizontal platform. Agarose (2.5 %) was added to 1X TBE, boiled until the agarose dissolved completely and then cooled to lukewarm temperature. Ethidium bromide was added. It was then poured in to the gel mould and then comb was placed properly and allowed to solidify. After solidification of the agarose, the comb and the cello tape were removed carefully. The casted gel was placed in the electrophoresis unit with wells towards the cathode and submerged with 1X TBE to a depth of about 1 cm.
Loading the DNA samples
10μl of DNA samples were pipetted on to a parafilm and mixed well with 2μl of loading dye by pipetting up and down for several times. The gel was run at 90 volt until the tracking dye was one third of the gel and bands were visualized and documented in gel documentation system (Model Alpha Imager 1200, Alpha Innotech Corp., USA). The viewed picture was photographed and saved for further scrutiny.
Scoring
Clearly resolved, unambiguous bands were scored visually for their presence or absence with each primer. The scores were obtained in the form of matrix with ‘1’ and ‘0’ which indicate the presence and absence of bands in each variety respectively.
Data Analysis
Polymorphic Information Content (PIC) values were calculated for each of the SSR loci using the formula developed by Nei [9]
PIC = 1 - Σx2k
Where, x2k represents the frequency at the kth allele. The data of microsatellite markers were analysed using NTSYS-pc statistical package, version 2.02 i [10]. The bands on the gel were scored for each of the SSR primer pairs in each genotype based on the presence or absence of bands generating a matrix of 1 and 0. Informative bands were used to calculate the genetic distance based on Jaccard’s similarity coefficient using SIMQUAL procedure. The DNA data of SSR markers for 29 rice land races were clustered using an unweighted pair group method (UPGMA) with the module of SHAN in the NTSYS-pc package.
Genetic Divergence Based On SSR Marker
Polymorphism of SSR markers
A total of 15 microsatellite markers was used to assess the extent of genetic diversity in the 29 rice land races and all of them were found to be polymorphic. As much as 49 alleles were detected among the genotypes. The number of alleles per locus ranged from 2 (RM 110 and RM 343) to 5 (RM21) with an average of 3.3 per locus.(Plate 4 and 5). The PIC values derived from allelic diversity and frequency among the genotypes were not uniform for all the SSR loci tested. The PIC value for 15 primers varied between 0.19 (RM 429 and RM 524) and 0.69 (RM 493) with a mean of 0.38 (Plate 6). In a set of the 15 primers used in this study there were no null alleles. The details on PIC values are given in Table 3.
| Sl.No. | Primer name | Chromosome location | Number of alleles | PIC values |
|---|---|---|---|---|
| 1 | RM 493 | 1 | 4 | 0.69 |
| 2 | RM 110 | 2 | 2 | 0.37 |
| 3 | RM 53 | 2 | 3 | 0.44 |
| 4 | RM 489 | 3 | 4 | 0.36 |
| 5 | RM 261 | 4 | 3 | 0.25 |
| 6 | RM 131 | 4 | 3 | 0.38 |
| 7 | RM 403 | 5 | 3 | 0.30 |
| 8 | RM 343 | 6 | 2 | 0.37 |
| 9 | RM 429 | 7 | 3 | 0.19 |
| 10 | RM 481 | 7 | 4 | 0.45 |
| 11 | RM 515 | 8 | 3 | 0.47 |
| 12 | RM 524 | 9 | 3 | 0.19 |
| 13 | RM 222 | 10 | 4 | 0.35 |
| 14 | RM 21 | 11 | 5 | 0.53 |
| 15 | RM 17 | 12 | 3 | 0.34 |
Table 3: Allele variation and PIC values for SSR markers identified in 29 genotypes.
Genetic diversity pattern
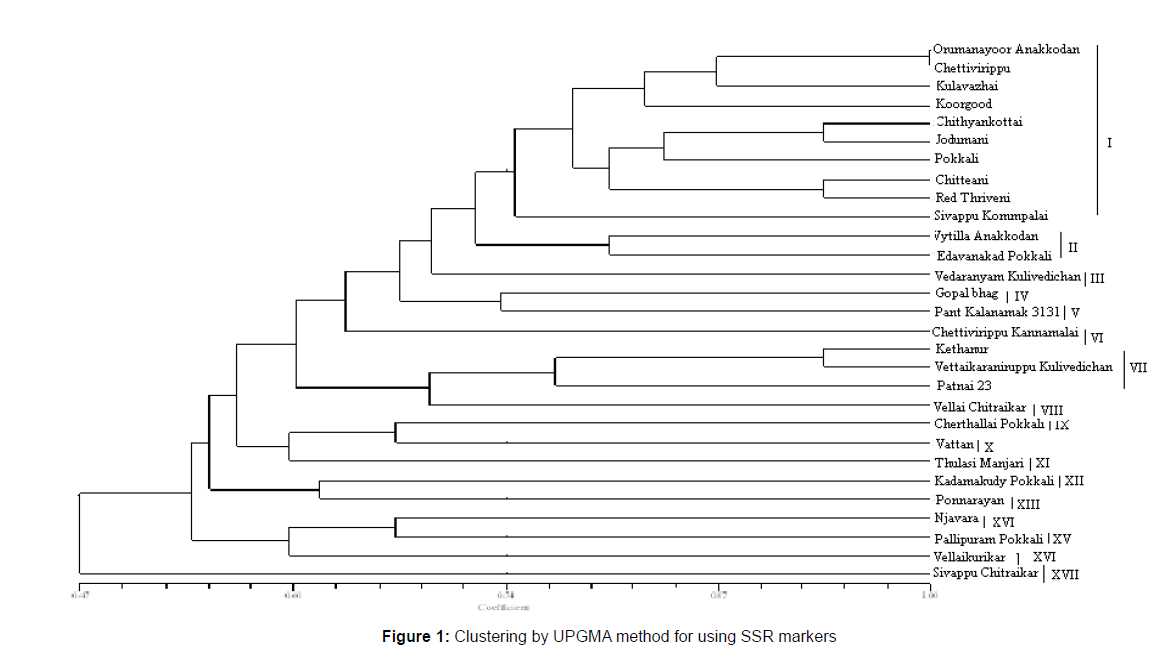
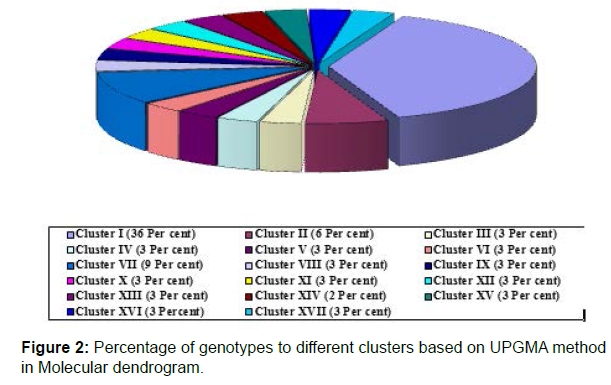
Dendrogram based on UPGMA analysis grouped the 29 genotypes in to different clusters that were demarcated at a similarity coefficient of 0.74. At this level of 74 per cent genetic similarity, the genotypes were grouped into 17 clusters each clusters distinguishes the genotypes clearly from the other. Cluster I had a maximum of ten land races of which five were originated from Kerala (Figure 1). Cluster II has all the two land races which were collected from Kerala. The remaining clusters except cluster VII are menogenotypic. The cluster VII had three land races (Table 4).
| Cluster No | Number of genotypes | Name of genotypes | Origin |
|---|---|---|---|
| I | 10 | Orumanayoor Anakkodan | Kerala |
| Chettivirippu | Kerala | ||
| Kulavazhai | Tamil Nadu | ||
| Koorgood | Kerala | ||
| Chithyankottai | Tamil Nadu | ||
| Jodumani | Tamil Nadu | ||
| Pokkali | Kerala | ||
| Chitteani | Kerala | ||
| Red Thriveni | Kerala | ||
| Sivappu Koompalai | Tamil Nadu | ||
| II | 2 | Vytilla Anakkodan | Kerala |
| Edavanakad Pokkali | Kerala | ||
| III | 1 | Vedaranyam Kulivedichan | Tamil Nadu |
| IV | 1 | Gopal bhag | Bangladesh |
| V | 1 | Pant Kalanamak 3131 | Uttar Pradesh |
| VI | 1 | Chettivirippu Kannamalai | Kerala |
| VII | 3 | Kethanur | Kerala |
| VettaikaraniruppuKulivedichan | Tamil Nadu | ||
| Patnai 23 | West Bengal | ||
| VIII | 1 | Vellai Chitraikar | Tamil Nadu |
| IX | 1 | Cherthallai Pokkali | Kerala |
| X | 1 | Vattan | Kerala |
| XI | 1 | Thulasi Manjari | Bihar |
| XII | 1 | Kadamakudy Pokkali | Kerala |
| XIII | 1 | Ponnarayan | Kerala |
| XIV | 1 | Njavara | Kerala |
| XV | 1 | Pallipuram Pokkali | Kerala |
| XVI | 1 | Vellai kurikar | Tamil Nadu |
| XVII | 1 | Sivappu Chitraikar | Tamil Nadu |
Table 4: Distribution of genotypes to different clusters based on UPGMA method in genotypic dendrogram.
Genetic divergence studies based on SSR marker
Characterization and quantification of genetic diversity have been a major goal in evolutionary biology. The analysis of genetic variation within and among the elite breeding materials is of fundamental interest to plant breeders. Diversity based on phenological and morphological characters usually varies with environment and evaluation of these traits requires growing the plant with full maturity prior to identification. To have rapid and easy analysis of a large number of loci distributed throughout the genome of plants, use of molecular marker is increasingly common method and also serves as a powerful tool in the assessment of genetic variation and elucidation of genetic relationships within and among the species. Information regarding genetic variability at molecular level could be used to help, identify and developed genetically unique germplasm that complements existing cultivars.
Cluster analysis of dendrogram constructed using SSR markers
The dendrogram based on UPGMA grouped the 29 genotypes in to 17 clusters that were demarcated at a similarity coefficient of 0.74. Cluster I was the largest and included 10 land races followed by cluster VII and II each of which had three and two land races respectively (Figure 2). The remaining clusters had one genotype each. This dendrogram revealed that the land races that are originated from different places are not grouped together which is indicative of non parallelism exist between geographical origin and genetic diversity.
Polymorphic information content
The term Polymorphic Information Content (PIC) was originally introduced in to human genetics by Botstein et al. [11] and it refers to the value of a marker detecting polymorphism within a population depending on the number of detectable alleles and their distribution of their frequency. In the present investigation, 15 SSR markers revealed 49 alleles in the 29 land races, the number of alleles per locus varied among these markers ranges from 2 to 5 with an average of 3.3 per locus. These kind of less number of alleles per locus was already reported by Ram et al. [12] where an average of 4.86 alleles were detected, indicating less magnitude of diversity among the genotypes. Out of the 15 SSR primers derived from genes, none of them were found monomorphic and however showed average PIC value lesser than 0.50. Lower PIC value may be the result of closely related genotypes and higher PIC values might be the result of diverse genotype. Low PIC values for some other primers were earlier reported by Juneja, et al. [13].
The study also further revealed that the primer RM 21 is ascertained to be showed more number of alleles with PIC value more than 0.50 which indicates the efficiency of this primer in detecting the most heterogenus accession. This is in corabaration with the findings of Giarrocco, et al. [14]. However, the present study indicated that use of more number of markers would be efficient to characterize the land races than used, which highlighted the presence of diversity at genotypic level among the accessions studied.
References
- Tang SX, Jiang YZ, Wei XH, Li ZC, Yu HY (2002) Genetic diversity of isozymes of cultivated rice in China. Acta Agron Sin 28:203-207.
- Winter P, Khal G (1995) Molecular marker technologies for plant improvement. World J Microbiol and Biotechnol 11:438-448.
- Smith JSC, Smith OS (1989) The description and assessment of distance between inbred lines of maize II The utility of morphological, biochemical and genetic descriptors and a scheme for testing distinctness of inbred lines. Maydica 34:151-164.
- Termnykh S, Park WD, Ayres N, Cartinhour S, Hauck N, et al. (2000) Mapping and genome organization of microsatellite sequences in rice (Oryza sativa L). Theor Appl Genet 100:697-712.
- Yang GP, Maroof MAS, Xu CG, Zhang Q, Biyanshev RM (1994) Comparative analysis of microsatellite DNA Polymorphism in land races and cultivars of rice. Mol Gen Genet 245:187-194.
- Akagi H, Inagaki YA, Fujimura T (1997) Highly polymorphic microsatellites of rice consist of AT repeats, and a classification of closely related cultivars with these microsatellite loci. Theor Appl Genet 94:61-67.
- Wenzel WG, Agishi KK, Mogashoa A, Donaloson G, Mohammed R, et al. (2001) Improved sorghum varieties for small holder farmers. J Appl Bot 75:207-209.
- Dellaporta SL, Woode J, Hicks JB (1983) A plant DNA preparation: Version 2. Plant Mol Biol Rep 1:10-22.
- Nei J, Pewter M, Mackill BJ (2002) Evaluation of genetic diversity in rice sub species using microsatellite markers. Crop Sci 42:601-607.
- Rohlf FJ (1998) NTSYS-pc: Numerical taxonomy and multivariate analysis system, version 2:02 Setauket, New York: Exeter software.
- Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphism. Am J Hum Genet 32:314-331.
- Ram SG, Thiruvengadam V, Vinod KK (2007) Genetic diversity among cultivars, land races and wild relatives of rice as revealed by microsatellite markers. J Appl Genet 48:337-345.
- Juneja H, Inagaki YA, Fujimura T (2006) Highly polymorphic microsatellites of rice consist of AT repeats and a classification of closely related cultivars with these microsatellite loci. Theor Appl Genet 94:61-67.
- Giarrocco LE, Marassi MA, Salerno GL (2007) Assessment of the genetic diversity in Argentine rice cultivars with SSR markers. Crop Sci 47:853-860.
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Citation: Rajesh T, Paramasivam K, Thirumeni S, Raja Rajan D, Suresh Kumar M (2022) Molecular Characterization of Rice Land Races by SSR Markers. J Plant Genet Breed 6: 112 DOI: 10.4172/jpgb.1000112
Copyright: © 2022 Rajesh T, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Open Access Journals
Article Tools
Article Usage
- Total views: 2691
- [From(publication date): 0-2022 - Nov 24, 2024]
- Breakdown by view type
- HTML page views: 2261
- PDF downloads: 430