Molecular Characterization of Microorganisms in Crude Oil-Polluted Mangrove Swamps of the Niger Delta, Rivers State, Nigeria
Received: 17-Apr-2019 / Accepted Date: 24-May-2019 / Published Date: 31-May-2019
Abstract
The soil microbial community plays a huge role in maintaining soil ecosystem balance and soil biological activity, hence a shift in composition or diversity could affect the ecosystem balance. The aim of this study was to examine the change microbial composition and diversity in oil contaminated soil using both traditional and molecular techniques. Bacterial Isolates were morphologically and biochemically characterized, genomic DNA was extracted from isolate and analyzed using 16s rRNA while molecular identification of fungi was performed by sequences analysis from internal transcribed spacer (ITS) region of nuclear ribosomal RNA genes. The 18S rDNA and 28S rDNA fragmens of fungi were analysed and ITS amplified using PCR, microbial isolates were sequenced using sanger sequencer and affiliation of the isolated samples were analyzed by comparing the ITS sequences with those deposited in the GenBank database. The sequence of the isolate demonstrated at least 99% nucleotide identity with the corresponding sequence in GenBank. The Illumina MiSeq high-throughput sequencing technique was used to study the number of reads in the soil. The results showed significant decline in microbial population, composition and diversity in soils under oil pollution when compared with the unpolluted soil. Dominant bacterial isolates belonged to Phylum Firmicutes and Proteobacteria, fungi isolates which belonged to Phylum Ascomcota was dominant in crude oil polluted sites, while Phylum Basidiomycota was dominant in the unpolluted sites. This study also showed that the number of isolates sequenced molecularly was 0.58% of the total metagenomic sequences of all soil samples. Molecular characterization also revealed 5.5% of the bacterial isolates had no significant similarity when blast in NCBI gene bank and those isolates were isolated from mineral salt agar, hence utilized hydrocarbon as energy source, they could be potential novel isolate. Sequences have been submited in the gene bank with accession numbers KT899779-KT899800.
Keywords: 16s RNA; 18s RNA; Crude oil pollution; Phylogyytenic; Microbial population
Introduction
With a large demand for crude oil as an energy source, oil contamination occurs quite often as a result of exploration, production, maintenance, transportation, storage and accidental release, leading to significant ecological impact [1]. Oil exploration and pipeline sabotage have led to increased amounts of crude oil spills in the Niger Delta. Crude oil spills automatically alters the benefits of the mangroves in negative manners. The adverse effects have led to complete loss or disappearance of this ecosystem in the region [2]. The free-living bacteria, fungi and yeasts were reported to have significant role in formation of detritus in the mangrove ecosystems [3]. Various groups of bacteria like nitrogen fixers, phosphate solubilizers, cellulose decomposers, nitrifiers and denitrifiers, sulphur oxidizers, iron oxidizers and iron reducers were prevalent in this ecosystem [4]. Bacteria and fungi make up about 91% of the total microbial biomass, while algae and protozoa represent 7% and 2%, respectively in these ecosystems. Studies have shown that bacterial role in mangroves is vital for biogeochemical cycles and transformations of most nutrients [5].
Microbial populations with plasmids containing genes for utilization are increased in oil polluted soil [6]. Environmental bacteria is often complex to identify using traditional techniques because of the uncultured bacteria that cannot easily be identified using traditional techniques [7]. As a result of the limitations of traditional techniques, new strategies and approaches are being implemented for rapid, sensitive and specific detection of microorganisms in the environment. The molecular analyses have been found to be more appropriate than the traditional approaches. However, both molecular and traditional techniques are important to characterize novel organisms for bioremediation.
The current culture-dependent methods used can only account for a small subset of the total microbial diversity and underestimates of the microbial communities present in petroleum polluted environments and isolating these organisms pose some challenges [8].
Direct isolation of total community DNA from the environment has been a good tool for evaluating microbial diversity and dynamics. Most of the challenges associated with DNA extraction using readily available and cheap reagents has been circumverted by the use of commercially available DNA extracted kits. Most of these studies using molecular techniques were based on the construction of 16S rRNA clone libraries and subsequent sequencing of individual 16S rRNA clones. Metagenomics is a recently described technique for cloning functional genes. It allows isolation and cloning of large segments (30- 100 kb) from soil [9].
The soil microbial community plays a huge role in maintaining soil ecosystem balance and soil biological activity, hence a shift in composition or diversity could affect the ecosystem balance. The aim of this study was to examine the change in microbial composition and diversity in oil contaminated soil using both traditional and molecular techniques.
Materials and Methods
Sample collection
The top and sub sections of soil samples were collected from three crude oil -polluted sites (Bodo, Bille and Cawthorne channel) in Rivers State, Nigeria. Soil samples from each site were collected randomly from 10 sampling points using soil auger at least 1 metre from each point at a depth of 0-15 cm and 15-30 cm to form composites samples. Collected soil samples were put in amber bottles, stored in ice bags and transported to the laboratory for analysis using standard methods of sample collection, presevation and transportation). Control samples were collected from 2 kilometres away from contaminated sites.
Traditional technique of identification
Sterilization of glassware and media: All glassware was sterilized in a hot air oven at 160°C for 1-3 hrs. Sterilization of all media as well as distilled water used for serial dilutions was carried out in an autoclave at 121°C and 15 ponds per sq inch (psi) pressure for 15 minutes.
Media for isolation: All microbial analyses were carried out within 24 hrs after sample collection in Nutrient agar and Potato dextrose agar for isolation and enumeration of heterotrophic bacteria and fungi, while mineral salt agar was compounded to isolate hydrocarbon utilizing bacteria and fungi. All media were prepared according to manufacturer’s instruction.
Mineral salt medium: The following were compounded for mineral salt.
MgSO4 7H2O - 0.42 g
KCl - 0.29 g
KH2PO4 - 1.25 g
K2HPO4 - 0.83 g
NH4 NO3 - 0.42 g
Nacl - 15 g
Agar - Agar - 15 g
Distilled water 1 litre
20 ml of carbon source (crude oil) was dropped unto a small piece of paper filter paper, placed on the lid of the petridish, hydrocarbon degraders were enumerated after 7-14 days. All the above microbiological media were sterilized by autoclaving at 121°C for 15 minutes.
Enumeration of microbial (Bacterial) population in soil
A standard spread plate method with modification was used [10]. Total viable count of heterotrophic aerobic bacteria.
Determination of survival of indigenous soil microorganism
Inoculum preparation: One gram of homogenized, 2 mm sieved soil sample was aseptically transferred, using a flame sterilized steel spatula, into a sterile test tube containing 9.0 ml of the diluent. This gave 10-1 dilution, subsequently, three-fold (103) serial solution was prepared from 10-1 solution.
Sterile physiological saline i.e., 0.85% (w/v) sodium chloride was used as diluent for inoculum preparation, 0.1 ml aliquot of 10-3 dilution was aseptically removed with a sterile pipette and spread plate with flame-sterile glass spreader on well dried agar plates used for bacteria and mould isolation and enumeration in duplicates. After incubation, plate with about 30-300 colonies was selected for counting. The colonies counted were expressed as colony forming unit (cfu) per gram soil using the formula, bacterial incubation was for 24 hours after which plates that grow between 30 and 300 colonies [11] were counted and recorded.
Colonies expressed as colony forming unit per grams soil and recorded. For fungi, incubation period was 3 days and colonies were also counted. For Hydrocarbon utilizing bacterial and fungi, incubation was for a period of 7-14 days.
Total number of cells in cfu/g soil=N/V × Dt,
Where, N=No of colonies,
V=volume of inoculums plated,
DF=Dilution factor used for plating,
Microbial analysis - Two replicates were used for all analysis and the mean values obtained are reported.
Counts of bacteria for the three soil sites were compared statistically using the analysis of variance completely randomized design, using MINITAB for windows V 10 means were compared at the 5% significance level using Duncans multiple range text-DMRT analysis of the three samples revealed significant differences (p<0.05).
Maintenance of pure cultures: Pure cultures of bacteria and fungal isolation were obtained by plating on nutrient agar and potato dextrose agar respectively and incubated at room temperature (28 ± 2°C). Lawns of overnight cultures of the purified isolates were scrapped into 10% (V/v) glycerol thoroughly mixed and stored at -35°C [12]. These glycerol suspensions served as a means for long term storage and a source for weekly working cultures. Working cultures were 24 hours old cultures made from these frozen glycerol suspensions.
Biochemical tests
These were carried out to identify the isolates according to laboratory methods by Cheesbrough [13].
Total heterotrophic moulds
Soil heterotrophic moulds was estimated by the soil dilution plate counts method (IPS, 1990) in which serial dilutions of the soil sample in sterile physiological saline were spread on potato dextrose agar, incubated at 28 ± 2°C for 5 to 7 days after which heterotrophic mould counts was recorded. Colonies were subcultured on to fresh medium for the development of pure isolates which was stored on potato dextrose agar slants for subsequent characterization and identification tests. The composition of the medium was potato 200 g, distilled water, 500 ml glucose 15 g and agar No. 1 20 g. Ampicillin and streptomycin was added to inhibit bacteria growth.
Presumptive identification of mould isolates: The pure isolates were identified using both cultural and microscopic examination in accordance with lactophenol method. Microscopic examination of Fungi by the lactophenol method.
Reagent: Lactophenol cotton.
Constituent of the stain, Phenol (pure crystals)-10 g, Lactic acid - 10 gm, Distilled water - 10 gm.
It is readily prepared by warming the phenol with the water until it dissolved then adding the lactic acid and glycerol.
Method of mounting: This was done by needle mount method. A small portion of the growth was picked with a sterile needle and teased out in a drop of lactophenol cotton blue on a clean microscopic slide. This was covered with a clean cover ship, taking care to exclude air bubble. The prepared slides were examined under the microscope, starting with a low power objective (X10) to a higher power (X40) objective for a better field view and magnification [13].
Identification of fungi
The fungal isolates were characterized based on macroscopic and microscopic appearances which comprised colonial morphology, type of hyphal, presence of sterigma, shape and kind of spore/conidia, presence of special structure such as foot cell, and growth on glucose. Probable identification of the Fungi were determined [14].
Colonies of bacterial and fungi were counted and counted for all soil sites were compared statistically using the analysis of variance completely randomized design using MINTAB for windows v.10, means were compared at 5% significance level.
Molecular microbial techniques
Molecular identification of isolates: Genomic DNA were extracted from pure cultures of all isolates using a total genomic DNA isolation Method or using a fast DNA kit (Bio 101).
DNA extraction: DNA extraction was from a 24 hours growth of soil microbial isolates in BHI broth harvested by centrifugation at 14,000 × g for 10 minutes. The cells were washed three times in 1 ml of Ultra pure water by centrifuging at 12,000 rpm for 5 min. DNA extraction and purification was done using ZR Fungal/Bacterial DNA MiniPrep™ 50 Preps. Model D6005 (Zymo Research, California, USA). 50-100 mg of bacterial cells was resuspended in 200 μl of sterile water. This was transferred into a ZR BashingBead™ Lysis Tube, exactly 750 μl Lysis Solution was added to the tube. The bead containing the solution was Secured in a bead beater fitted with a 2 ml tube holder assembly and process at maximum speed for 5 minutes. The ZR BashingBead™ Lysis Tube was Centrifuged in a microcentrifuge at 10,000 × g for 1 minute. 400 μl of the supernatant was pipeted into a Zymo-Spin™ IV Spin Filter in a Collection Tube and centrifuged at 7,000 × g for 1 minute. This was followed by the addition of 1,200 μl of Fungal/Bacterial DNA Binding Buffer into the filtrate in the Collection Tube. After this 800 μl of the mixture was tansfered into a Zymo-Spin™ IIC Column in a Collection Tube and centrifuge at 10,000 × g for 1 minute. The flow through was discarded from the Collection Tube and the process was repeated to obtain the remaining products. The 200 μl DNA Pre-Wash Buffer was added into the Zymo-Spin™ IIC Column. In a new Collection Tube and centrifuge at 10,000 × g for 1 minute. This was followed by the addition of 500 μl Fungal/Bacterial DNA Wash Buffer into the Zymo-Spin™ IIC Column and centrifuged at 10,000 × g for 1 minute. The Zymo- Spin™ IIC Column was transferred into a clean 1.5 ml microcentrifuge tube and 100 μl of DNA Elution Buffer was then added directly to the column matrix. This was centrifuged at 10,000 × g for 30 seconds to elute the DNA. The Ultra-pure resulting filtrate (DNA) obtained was used as a template during the assay.
Polymerase chain reaction analysis: Amplification of the template DNA was performed using 2 ml volume of the extracted DNA with Bio RAD mini thermal cycler (Mexico). The 50 Nm PCR mixture contained 5 Nl of deoxy nucleoside triphosphates (NDTPs) mixture (2.5 Nm) (Promega, USA), 5 μ of 5X Green GO Taq flexi butter (Promega, USA), 3.5 μ of 25 mm ngcl2 (Promega, USA), 2 μ each of 10 pmol of both forward (primer M) and reverse (primer K) Primers PA8F - GC (5 - CGC - CCG - CCG-CGC -GCG - GCC - GGC - GGG - GCG - GGG - GCA - GGG - GAG - AGT - TTG - ATC - CTG - GCT - CAG - 3’) and KPRUNS518. (5’ ATTACCGCGG’TGCTGG - 3). 0.25 μ of 5 μ hot 20 mg/ml of bovin serum albumin and 27.75 μ of sterile water. A reaction tube without template DNA was included as negative control. The PCR programme was as follows; denaturing step at 95°C for 3 min, followed by 33 cycles of 30 sec at 95°C for 30 sec at 55°C and extension at 72°C for 1 min, followed by a final extension at 72°C for 7 min and held at 4°C. amplified DNA were presented for sequencing. sequencing was done by sanger sequencing blast analysis: clc bio. Sequencing result was FASTA format and corresponding ID after BLAST ANALYSIS on NCBI website (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Metagenomics analysis culture-independent analysis
DNA extraction of bacterial isolates from soil sample: Culture independent analysis of Direct non cultivation based molecular techniques for detecting microorganism in the soil samples were also used to study the population of microbial communities.
DNA extraction from soil sample was performed using ZYMO soil DNA extraction Kit according to the manufacturer’s method. Genomic DNA was extracted by weighing out 0.25 grams of soil sample using an analytical Balance. The sample was the added into a ZR Bashing BeadTM Lysis Tube followed by the addition of 750 μ Lysis solution to the tube. The content of the 2 ml tube disrupted by mixing in a vortex mixer at maximum speed for 5 minutes. The ZR Basing BeadTM Lysis tube was Centrifuge in a micro centrifuge at <10,00 × g for 1 minute. 400 μ of the filtrate was added to a zymo-spinTM IV spin filter in a collection Tube and centrifuge at 7.000 rmp (7,000 × g) for 1 minutes.
This was followed by the addition of 1,200 μl of soil DNA Binding buffer to the filtrate in the collection Tube. 800 μl of the mixture from above was added to a Zymo-spinTM IIC Columu in a collection tube and centrifuge at 10,000 × g for 1 minute. Flow through from the collection tube was discarded and this particular step was repeated with the remaining filtrate. 200 μl of DNA pre-Wash Buffer was thereafter added to the Zymo-spinTM IIC Column in a new collection tube and centrifuge at 10,000 × g for 1 minute after which 500 μ soil DNA wash Buffer was added to the Zymo-spinTM IIC Column and centrifuge at 10,000 × g for 1 munite. Flow through from the Collection Tube was discarded and this particular step was repeated with the remaining filtrate. 200 μl of DNA Pre-wash Buffer was thereafter added to the Zymo-Spin TM IIC Column in a new Collection tube and centrifuge at 10,000 × g for I minute after which 500 μ Soil DNA Wash Buffer was added to the Zymo-Spin TM IIC Column and centrifuge at 10,000 × g for 1 minute. The Zymo-Spin TM IIC Column was transferred into a clean 1.5 ml micro centrifuge tube and add 100 μ DNA Elution Buffer directly to the column matrix. This was centrifuge 10,000 × g for 30 seconds to elute the DNA. The elute DNA from was transferred into a filter unit of Zymo-Spin TM IV-HRC Spin Filter in a cleanb 1.5 ml micro centrifuge tube and centrifuge at exactly 8,000 × g for 1 minutes. The filtered DNA was then used for PCR and DNA sequencing.
Identification of fungi isolates by DNA sequencing and corresponding codon
Fungal cultures were harvested from Potato dextrose agar after 72 hours of inoculation and culture. Molecular identification was performed by sequences analysis from internal transcribed spacer (ITS) region of nuclear ribosomal RNA genes. This DNA fragment included the 3' end of the 18S rDNA, ITS1, and the 5' end of the 28S rDNA. Genomic DNA was isolated from monosporic fungal cultures by conventional procedures [15]. The ITS was amplified using primers ITSl and ITS4 [16]. The amplification reactions were performed in a 50 μl reaction volume under the following PCR cycling conditions: one cycle of denaturation at 95°C for 3 min, followed by 34 cycles of denaturation at 95°C for 1 min, annealing at 52°C for 30 sec, and elongation at 72°C for 1 min, with a final extension step of 72°C for 10 min. The PCR products of approximately 550 bp length were resolved in 1% agarose gels stained with ethidium bromide. The PCR products were sequenced by mean of the mentioned primers in an Applied Biosystem 3130 sequencer (www.appliedbiosystems.com) based on the procedure described by Sanger et al. [17]. Sequencing result was in FASTA format and corresponding ID was got after BLAST ANALYSIS on NCBI website (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Results
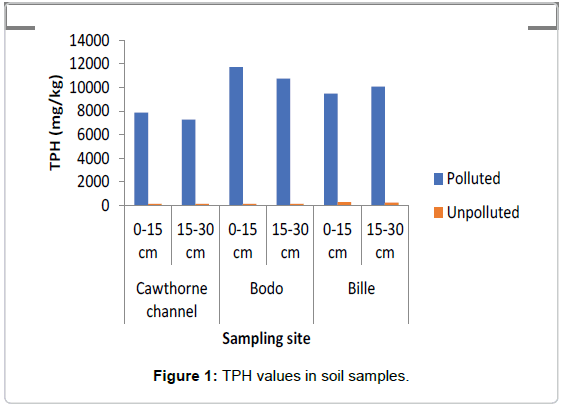
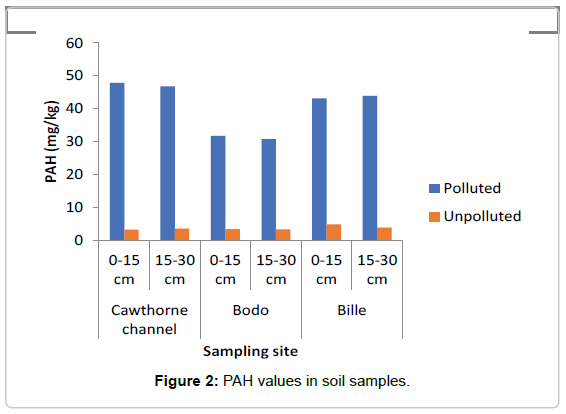
The concentrations of TPH and PAH as represented in Figures 1 and 2 showed that the swamp was heavily polluted with crude oil as the TPH values in the polluted soils ranged between 7000 mg/kg to 12000 mg/kg while in the unpolluted soil the values ranged between 145 mg/kg to 150 mg/kg The values of PAH ranged between 30 mg/ kg and 48 mg/kg in the polluted soil, while in the the unpolluted soil, values ranged between 3.35 mg/kg to 4.2 mg/kg.
Microbial counts in polluted and unpolluted sites
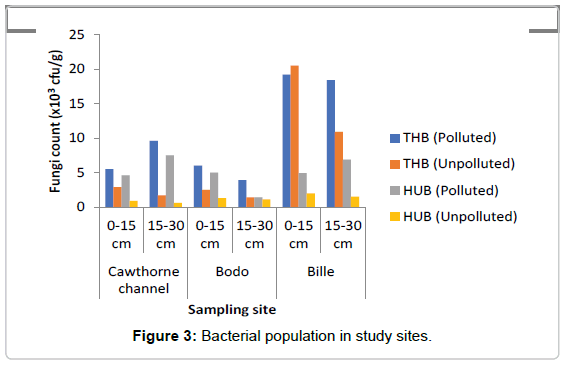
Total Heterotrophic Bacteria (THB) count: It was observed that the total heterotrophic bacteria (THB) counts as represented in Figure 3 ranged from 3.9-18.2 × 104 cfu/g in the polluted soil and 1.4- 20.4 × 104 cfu/g in the unpolluted soil, In this study total microbial population of crude oil in polluted soil ranged between 5.5-19.2 × 104 cfu/g at soil depth 0-15 cm. At depth 15-30 cm the count ranged from 3.9-18.4 × 104 cfu/g. The results showed that bacteria count increased by 3.9% at depth of 0-15 cm when compared to count at depth of 15- 30 cm in the oil polluted soils. The total heterotrophic bacteria in the unpolluted soils ranged from 2.5-20.5 × 104 cfu/g in the top layer of the soil (0-15 cm), while at depth of 15-30 cm total heterotrophic bacteria count ranged from 1.4-10.9 × 104 cfu/g. Result showed that bacteria count decreased by 45.9% at depth of 15-30 cm when compared to microbial count at depth of 0-15 cm in the unpolluted soil. The mean count value in the polluted and unpolluted sites were 4.77 ± 0.14 cfu/g and 5.26 ± 0.06 cfu/g respectively. THB decreased in the polluted soil when compared to the unpolluted soil. A two-way ANOVA showed that there was a significant difference in the THB values between the polluted and unpolluted soil (p=0.005). No significant difference was observed between depths (p=0.06).
Hydrocarbon Utilizing Bacteria (HUF): The range of HUB in the polluted soil as represented in Figure 3 ranged from 1.4-7.5 × 102 cfu/g, while in the unpolluted soil the count ranged from 0.6-2.0 × 102 cfu/g, increase in HUB was observed, similarly, the count for hydrocarbon utilizing bacteria (HUB) ranged from 4.6-5.0 × 102 cfu/g at depth of 0-15 cm. At depth of 15-30 cm the HUB count ranged from 1.4-7.5 × 102 cfu/g. Percentage of HUB increased at depth 15-30 cm by 8.9% when compared to the depth of 0-15 cm in the polluted soil. Study also showed that the hydrocarbon utilizing bacteria count in the unpolluted soils ranged between 0.9-2.0 × 102 cfu/g at depth of 0-15 cm, while at depth of 15-30 cm HUB ranged from 0.6-1.5 × 102 cfu/g. The result showed HUB decreased by 23.8% at depth of 15-30 cm when compared to a depth of 0-15 cm. HUB were slightly higher at the subsection (15- 30 cm) of polluted soils when compared to the unpolluted soils, where HUB were higher at depth of 0-15 cm in the unpolluted soil. The mean value of hydrocarbon utilizing bacteria (HUB) count in the polluted soil was 8755 ± 8249 cfu/g, while the mean value of HUB count in the unpolluted soil was 123.30 ± 19.90 cfu/g. A two-way ANOVA showed that there was a statistically significant difference in the value of HUB count between the polluted and unpolluted soil (p=0.030). While across the depth there was no significant difference observed (p=0.264). The result showed that crude oil pollution greatly increased the number of HUB in the soil. HUB were found more at the subsection of the polluted soil and more at the top section of the unpolluted soil. It was observed that age of spill may have effect on HUB population. The older the spill, the more the hydrocarbon utilizing bacteria population. Site T (50 weeks)>G (14 weeks)>B (3 weeks).
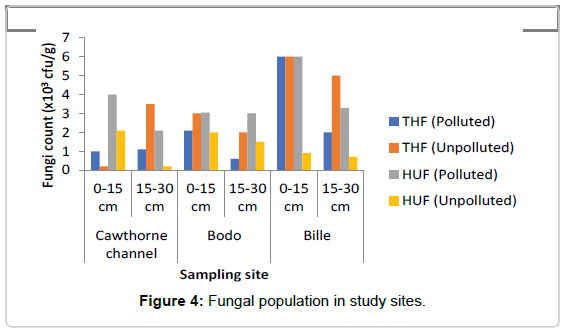
Total Heterotrophic Fungi (THF): Total fungi isolates ranged as represented in Figure 4 ranged from 0.6-6.0 × 103 sfu/g in the polluted soil while in the unpolluted soil the count ranged from 0.2-6.8 × 103 sfu/g. The microbial population for total fungi ranged from 1.0-6.0 × 103 sfu/g at depth of 0-15 cm in the oil polluted soil, while the count at depth of 15-30 cm ranged from 1.1-2.0 × 103 sfu/g, in the polluted soil, decrease was observed at depth of 15-30 cm when compared to the depth of 0-15 cm. In the unpolluted soil total fungi count at depth of 0-15 cm ranged from 2.0-6.8 × 103 sfu/g while the count at depth 15-30 cm ranged from 2.0-5.0 × 103 sfu/g. A decrease was observed in THF in the polluted soil when compared to the unpolluted soil. The mean value of THF count in the polluted site was found to be about 2133.0 ± 810.0 cfu/g, while the mean value of THF count in the unpolluted site was 3333.0 ± 882.0 cfu/g. A two-way ANOVA showed that there was no significant difference in the value of THF count in polluted soil when compared with the value in the unpolluted soil (p=0.602). There was also no significant difference across the depths of the study sites (p=0.99).
Hydrocarbon Utilizing Fungi (HUF): In contrast, HUF was generally found at the top layer (0-15 cm) in the spill soil as represented in Figure 4. Study did not really ascertain their abundance with spillage because there was no variation between 50 week and 3 week spill except for 14 week spill that had highest HUF as was also observed in heterotrophic fungi count in all sites.
The least number of Total heterotrophic bacteria count was observed at freshly spill site B (3 weeks), while sites G and T had more counts, though site G with 14 weeks had the highest count. Total Heterotrophic Bacteria also showed that age of spills might affect heterotrophic bacteria count as was observed in 14 week and 50 week Cawthorne channel (T), having 50 weeks of pollution history had the least heterotrophic fungi count, followed by Bodo (B) with fresh spill. Though Bille (G) with 14 weeks spill recorded the highest microbial count at both sections of soil. It also showed that both bacteria and fungi were more populated at the top layer (0-15 cm) of the control soil, this varied in the oil polluted environment. Crude oil pollution affected community profile, microbial population across the top and sub soil section for bacteria and not much variation in fungal population.
Molecular characterization of hydrocarbon utilizing isolates
A Total of 55 bacteria isolates were characterized in this study from polluted and unpolluted soils. The study showed that 3.9% showed no amplification with universal primers used. This means that the primers used could not amplify the genes, more studying should look at other primers design that can amplify the genes (5.5%) of the isolates had never been identified in any study, there was no sequence corresponding to this isolates. This study had been able to prove that molecular study discovers novel isolates microorganism.
About 90.2% isolates were identified to specie level this had shown that molecular technique remain the best tool for characterization of microorganism. Bacteria species characterized in this study include Bacillus (32.1%), Pseudomonas (24.8%) PaeniBacilluslactis, Alkaligens faecalis (5.9%), LysiniBacillus sphaericus (3.9%), Ralstonia sp (3.9%), Serratia sp (3.9%) other species include Ochrobacterium oryzae, (1.96%), Stenotrophormonas maltophilia (1.96%). The study showed that Bacilli 32.1% was mostly isolated, followed by Pseudomonas sp and Alkaligens (5.9%) and the potential unclassified isolates 5.9%.. The strains of bacteria molecularly identified showed that dominant strains belonged to Bacilli, Gamma Proteobacteria and Beta Proteobacteria classes and the Firmicutes and Proteobacteria phyla.
Molecular and traditional characterization of fungi cultures
A total of 23 fungal cultures were characterized in this study, 8.7% of the isolates showed no amplification with universal primers used, other primers should be designed that can amplify these genes. 65.2% of the fungal cultures were Aspergillus spp. The dorminant cultures belonged to phylum Ascomycota in the polluted soil while the dorminant organisms in the unpolluted soil belonged to the phylum Basiomycota as represented in Table 1.
| Sites | Phylogenic group | Class | Closest bank relative | Accession number | Strain | Max. ID | Evaluation |
|---|---|---|---|---|---|---|---|
| Polluted | Ascomycota | Eurotiomycetes | Aspergillus Fumigates | K317993.1 | SH5.EGY | 100% | 0 |
| Aspergillus norminus | KP100549.1 | F20 | 100% | 0 | |||
| Aspegillus flavus | KM284800.1 | ALCOO1 | 99% | 0 | |||
| Aspergillus niger | KRO7579.1 | SCTCC323 | `99% | 0 | |||
| Aspergillus fumigates | KP689196.1 | FR18 | 99% | 0 | |||
| Penicillium janthinelium | KM284800.1 | ACC001 | 99% | 0 | |||
| KM46103631 | WHAD9 | 99% | 0 | ||||
| Ascomycota | Saccharomycetes | Pichia kudriavzevii | KP132507.1 | CNRMA9-WASHAM-ITS | 100% | 0 | |
| Unpolluted | Basidiomycota | Tremetomycetes | Cryptococcus laurentii | KP337904.1 | 2-b | 99% | 0 |
| Tremetes lactinea | |||||||
| Agaricomycetes | Peyronellaea pomorun | JX082368.1 | BRF | 99% | 0 | ||
| Dothideomycetes | FJ839846 | IBT4176 | 99% | 0 | |||
| Ascomycota | Eurotiomycetes | Aspergillus niger | KRO75079.1 | SCTCC323 | 100% | 0 | |
| Aspergillus nomilus | TQ781730.1 | AT5A | 100% | 0 | |||
| Penicillium janthinelium | KM46103631 | WHAD9 | 99% | 0 |
Table 1: Molecular characterization of Fungi isolates from crude oil polluted site.
Discussion
Bacterial counts were found to be higher than the fungal counts and this corroborates with the findings of Olukunle [18], he studied oilpolluted soil and water at Awoye, Mese and Oluwa villages in Ondo State and three different flow stations in River State. In this study, the isolates characterized in the crude oil polluted soil were similar to the findings of Olukunle and Boboye et al. [18,19] who worked on degradation activity of bacteria obtained from crude oil polluted sites. He obtained Micrococcus, Aerococcus, Closridium, Staphylococcus, Streptococcus, Lactobacillus, Bacillus, Enterobacter, Pseudomonas, Acinetobacter, while Olukunle [18] isolated Bacillusfirmis, B. Sphaericus, B. pumilus, Staphylococcus aureus, Micrococcus sp, Pseudomonas stutzeri. Though there were variation in species such as Clostridium, Stapylococcus aureus, Pseudomonas sp being one of the most extensively studied oil degrader was also isolated and this agrees with the report by Wackett [20]. Bacillus spp and Pseudomonas sp were the most dominant bacterial isolates in both contaminated and non-contaminated soil this agrees with the report of Boboye et al. [19] also isolated Pseudomonas, Bacillus and Micrococcus from crude oil polluted soils. Oil-polluted sites harbour a vast array of microbial flora, hence diversity in the types of bacteria and fungi identified in this. The isolation of Bacillus from Minerals salt agar showed its ability to utilize hydrocarbon as an energy source, Bacillus species are known as r-strategists [21] they are not good competitors, they use rapid growth as survival strategists when the nutrients are in abundant quantities, they grow well if there is a great quantity available nutrients in low competition. Friedrich et al. [22] reported that traditional cultivation strategies are often selective and favour faster-growing population, whose role in situ may not be relevant, this could account for the abundance of Bacillus isolates from mineral salt media. Thus, it is possible that such isolates were obtained only because the cultivation techniques favoured their selection. the cultivation of numerous hydrocarbon-degrading isolates from soil environments might overestimate their importance in situ without confirmation using culture-independent molecular analyses.
It is possible that these isolates were not involved in the degradation of crude oil components under the conditions used in the current different environmental conditions with different niches [22].
Effect of crude oil pollution on hydrocarbon degraders
The increase observed in hydrocarbon utilizing bacterial counts in the polluted soils when compared with the unpolluted soil agrees with the work of Tyagi et al. [23], that hydrocarbon degrading population increase and certain degrading taxa become dominant in the soil that is polluted with crude, this could be accounted for by natural selection. John et al. [24] also observed an increase of 10% in HUB when compared to the unpolluted soil. Chikere et al. [25] also observed an increase in THB and HUB when they examined population dynamics during bioremediation of an African soil contaminated with Arabian light crude oil and nutrient amendment. The findings of this work showed total heterotrophic bacteria counts and total heterotrophic fungi were higher in the unpolluted soil when compared to the polluted soil. Though HUF was higher in the polluted soils but they had no statistical significant differences. The above findings were similar to the works of Olukunle, John et al., Saadoun et al. [18,24,26].
Bacterial counts were also observed to be more at the top soil 0-15 cm when compared to the sub soil (15-30 cm), this agrees with the work of Saadoun et al. [26], where it was observed that bacteria counts declined to 65-95% in the depth of 10-20 cm when compared to 0-10 cm. Study also showed more microbial diversity at the top soil when compared to the sub soil. This findings also agree with the work of Saadoun et al. [26]. This could be as a result of the concentration of the oil at the lower soil, HUB were also found to be more at the lower strata of the polluted soil, the above reason could account for more HUB at the lower soil strata. Crude oil contamination reduced fungal count by 63% in the subsoil and 44.56% in the top soil about 55% decrease in heterotrophic fungi was observed in the polluted soil by Saadoun et al. [26] and tha corroborates with the findings of this work.
The study also observed a general increase in total heterotrophic bacteria counts than the fungal counts, this agrees with the study of Onifade et al. and Olukunle et al. [27,28]; the nutrient (hydrocarbon) status of the soil could be responsible for increase in bacteria counts. A high significant difference at P=0.014 was observed between TPH and heterotrophic bacteria counts as well as hydrocarbon utilizing bacteria P=0.02, this agrees with the reports of Yuting et al., Rodrigo et al. [29,30], between the polluted and unpolluted soil as well as a highly positive relationship exist between TPH and THB as well as between TPH and HUB and thus agrees with the work of Yuting et al. [29].
A decrease in the number of bacteria counts in polluted site when compared to the unpolluted sites was observed corrobrates with the findings of Saadoun et al. [26] he observed a declined of bacteria counts in the fresh spill soil when compared to old spill sites. Total heterotrophic count decreased by 27-47.8% when compared to their control soils thus findings agrees with Saadoun et al. [26] he observed a decrease of about 92% in heterotrophic bacteria count when compared to the unpolluted soil. Various environments conditions and microbial activities contribute to degradation of hydrocarbon pollutant. More counts were observed in the older spill when compared to the fresh spill. A decrease of 66.1% in bacterial count at soil depth of 15-30 cm when compared to soil depth of 0-15 cm was observed and this was similar to the findings of Saadoun et al. [26] he observed between 65- 95% decrease in bacteria count at 15-30 cm when compared to the depth of 1-15 cm. fungal counts were also observed to be more at 0-15 cm than 15-30 cm by 16-19%, decrease was also observed at the top soil (0-15 cm) when compared to (15-30 cm) sub soil in the polluted soil this findings agree with the work of Saadoun et al. [26].
The Study revealed a positive correlation between hydrocarbon degrading bacteria counts and TPH, this agrees with the work of Saadoun et al. [26] in his study on the effect of crude oil pollution on nitrogen fixing bacteria, he observed a positive relationship between TPH and HUB [29], also observed a significant relationship between TPH and HUB however, the findings is in contrast to the work of Margesin et al. [31], that observed no correlation between HUB and TPH.
Results in Table 2 showed that percentage number of sequences from cultured THB when compared with the metagenomic reads was 0.289% in the polluted site and 0.295% in the unpolluted site, and that of HUB in the polluted and unpolluted sites were 0.109% and 0.032% respectively. The percentage of cultured bacteria when compared to the metagenomics reads as represented in Table 2 showed a total of 0.58% and 0.141% of THB and HUB counts respectively, this corroborates with the reports of Tyson et al., Hugenholtz et al. [8,32] that <1 to 10% can be cultivated on laboratory media since the growth requirement for most strains are not known therefore plate count technique underestimates the true microbial population. Microbial enumeration is carried out but it is not an effective measure of soil microbial population. Tyson et al. [8] also reported that culture-dependent methods used can only account for a small subset of the total microbial diversity and underestimates of the microbial communities present in petroleum polluted environments and isolating these organisms pose some challenges. Cultural methods of cultivation recover less than 1% of the total bacterial species present in the polluted site sample, about 1% bacteria can be detected from many environments when traditional techniques are used [7,33-35] the culturable portion of bacteria is not a representative of the total phylogenetic diversity. Indeed, most of the cultivated microorganisms are those that grow quickly and are capable to grow in nutrient-rich media [36] and the dominant and key players in the environmental system.
| Polluted sites | Number of culturable THB | Number of culturable HUB | Total number of reads (metagenomics) | Percentage of culturable THB | Percentage of culturable HUB |
|---|---|---|---|---|---|
| T (0-15 cm) | 55 | 46 | 1309 | 0.042 | 0.035 |
| T (15-30 cm) | 96 | 75 | 1940 | 0.049 | 0.039 |
| B (0-15 cm) | 60 | 50 | 4471 | 0.013 | 0.011 |
| B (15-30 cm) | 39 | 14 | 4456 | 0.01 | 0.003 |
| G (0-15 cm) | 192 | 49 | 2977 | 0.06 | 0.016 |
| G (15-30 cm) | 184 | 69 | 1590 | 0.115 | 0.005 |
| Unpolluted site | |||||
| T (0-15 cm) | 229 | 9 | 1818 | 0.126 | 0.005 |
| T (15-30 cm) | 117 | 6 | 1622 | 0.072 | 0.009 |
| B (0-15 cm) | 125 | 13 | 6273 | 0.02 | 0.002 |
| B (15-30 cm) | 84 | 11 | 5970 | 0.014 | 0.006 |
| G (0-15 cm) | 295 | 20 | 6700 | 0.044 | 0.003 |
| G (15-30 cm) | 109 | 15 | 5547 | 0.02 | 0.007 |
| Total Percentage cultured | 0.584 | 0.141 | |||
Table 2: Percentage of culturable bacteria at 0-30cm at study sites when compared to metagenomics reads.
Effect of crude oil on microbial composition and diversity
The result of bacterial isolates characterized molecularly from crude oil polluted soils belonged to eight (8) bacteria genera: Bacillussp, Pseudomonas aeruginosa, Bacilluscereus, Bacillusthurisgiensis, Lysinibacillus sphaericus, Bacillusflexus, Bacilluscirculans, serratia marcescers, Bacillusspharicus, Strenotrophomonas maltophila, PaeniBacilluslactis, Alkaligens faecalis, Bacillu pulmilus, Enterobacter cloaca, as represented in Table 3, while in the unpolluted soils bacterial isolates characterized molecularly from unpolluted soils belonged to Eleven (11) bacteria genera: Enterobacter saccari, Bacillus sp, Herbaspirillium huttiense, Lysini Bacillus sphaericus, Bacillusthioparans, Ochromobacterium oryzae, Pseudomonas aeruginosa, Ralstonia manitolytica, Alkaligens faecalis, Bacillusflexus, Paeni Bacilluslactis. Similarly, the unpolluted soil in Table 1 shows five (5) genera of fungi (Tremetes sp, Peyronellaea sp, Cryptococcus sp and Aspergillus sp and Penicillium sp) belonging to four (4) classes: Eurotiomycetes, Dothideomycetes, Agaricomycetes and Tremetomycetes when compared to the polluted soils which had three genera of fungi (Aspergillus spp, Penicillium sp and Pichia sp) belonging to two classes: Eurotiomycetes and Saccharomycetes. In this study, fungal isolates in polluted soil belonged to phylum Ascomycota when compared to the fungal isolates characterized in the unpolluted soil which belonged to two phyla: Ascomycota and Basidiomycota. Similarly, 18 genera of fungal cultures were characterized traditionally, Six (6) genera were identified from the polluted sites: Aspergillus sp, Chrysosporium sp, Trichoderm sp, Penicillium sp, Verticillium sp and Fusarium sp, while 12 genera where identified in the unpolluted sites: Fusoma sp, Gliocladium sp, Aspergillus sp, Geotrichum sp, Fusarium sp, Cladosporium sp, Basidiobotrys spp, Monilla sp, Botrodiplodia sp, Alternaria, Botrydia, Nitrospora, Rhizobium sp and Trichoderma sp. Result showed more fungi cultures in the unpolluted soil when compared to the polluted soil.
| Polluted | Phylogenic group | Class | Closest gene bank relative organisms | Accession number | Strain | Max. identity | E. Value |
|---|---|---|---|---|---|---|---|
| T | Firmicutes | Bacilli | PaeniBacilluslactis | KF607091.1 | LG2 | 0.99 | 0 |
| Bacilli | Bacilluspumilus | KRO55041.1 | IHBB11166 | 0.99 | 0 | ||
| Bacilli | Bacillusflexus | KT226113.1 | PJS5 | 1 | 0 | ||
| Bacilli | Bacillus sphaericus | DQ923492.1 | D45 | 1 | 0 | ||
| Proteobacteria | Betaproteobacteria | Alkaligenes faecalis | KT356811.1 | GC | 0.99 | 0 | |
| Gammaproteobacteria | Pseudomonas aeruginosa | KF977860.1 | PA5-2-2 | 0.99 | 0 | ||
| Gammaproteobacteria | Strenotrophomonas maltophilia | KR856199.0 | L77 | 1 | 0 | ||
| Gammaproteobacteria | Serratia marcescens | KJ877667.1 | MSSRFQS38 | 1 | 0 | ||
| Unknown | No hit | No significant similarity | - | - | - | ||
| B | Proteobacteria | Gamma proteobacteria | Enterobacter cloacae | CPO12167.1 | 34978 | 99% | 0 |
| Pseudomonas aeruginosa | NCO025162 | PA01 | 100% | 0 | |||
| Pseudomonas aeruginosa | KF977860.1 | PA5-2-2 | 99% | 0 | |||
| Bacilli | Bacilluscirculans | KRO55041.1 | PJS5 | 100% | 0 | ||
| Firmicutes | Bacilli | Bacillusflexus | KJ226113.1 | HB12266 | 99% | 0 | |
| Bacilli | Bacillus sp | KJ200599.1 | HB1226 | 99% | 0 | ||
| Bacilli | LysiniBacillus sphaericus | EU869266.1 | BG-111 | 99% | 0 | ||
| Beta proteobacteria | Alkaligenes faecalis | KT356811.1 | GC | 99% | 0 | ||
| Unknown | No hit | No significant similarity | - | - | - | - | |
| G | Firmicutes | Bacilli | Bacillus sp | HQ916741.1 | Bg-1 | 100% | 0 |
| Proteobacteria | Gamma proteobacteria | Pseudomonas aeruginosa | NCO025162 | PA01 | 100% | 0 | |
| Firmicutes | Bacilli | Bacilluscereus | NCO04722-1 | ATCC14579 | 99% | 0 | |
| Bacilli | Bacillusthuringiensis | KT216618.1 | DD179 | 99% | 0 | ||
| Unpolluted soils | |||||||
| T | Proteobacteria | Gamma proteobacteria | Enterobacter saccari | CPO07215.2 | SPI | 97% | 0 |
| Firmicutes | Bacilli | Bacillus sp | KC94319.1 | G3-2-20 | 100% | 0 | |
| Proteobacteria | Gamma proteobacteria | Pseudomonas aeruginosa | KF977860.1 | RZS9 | 100% | 0 | |
| Firmicutes | Bacilli | Lysinibacillus sphaericus | KP980616.1 | JXRH19 | 100% | 0 | |
| Bacilli | Bacillus thioparans | NR04762.1 | BMP | 100% | 0 | ||
| Proteobacteria | Beta proteobacteria | Ochromobacterium oryzae | KM017981.1 | R16 | 7.40% | 3.00E-77 | |
| Gamma proteobacteria | Pseudomonas aeruginosa | KP866815.2 | RZS9 | 100% | 0 | ||
| Beta proteobacteria | Bordetella trematum | KJ604707.1 | NN | 93% | 0 | ||
| B | Proteobacteria | Beta proteobacteria | Ralstonia manitolytica | KJ806364.1 | JNT | 100% | 0 |
| Beta proteobacteria | Herbaspirillium huttiense | KM272770.1 | LAMA1106 | 99% | 0 | ||
| Firmicutes | Bacilli | Bacillus sp | KJ200599 | HB12266 | 99% | 0 | |
| Gamma proteobacteria | Bacillusflexus | KR140376.1 | OU91 | 100% | 0 | ||
| Beta proteobacteria | Ralstonia mannitolytica | KF751574.1 | Faro7 | 86% | 0 | ||
| Firmicutes | Betaproteobacteria | Alkaligens faecalis | KP823043.1 | Unwasa 32 | 79% | 0 | |
| Proteobacteria | Bacilli | PaeniBacilluslactis | FN429978.1 | IB-188B | 100% | 0 | |
| Gamma proteobacteria | Pseudomonas aeruginosa | KR007310.1 | 364 | 100% | 0 | ||
| B | Proteobacteria | Beta proteobacteria | Ralstonia manitolytica | KJ806364.1 | JNT | 100% | 0 |
| Firmicutes | Beta proteobacteria | Herbaspirillium huttiense | KM272770.1 | LAMA1106 | 99% | 0 | |
| Bacilli | Bacillus sp | KJ200599 | HB12266 | 99% | 0 | ||
| Gamma proteobacteria | Bacillusflexus | KR140376.1 | OU91 | 100% | 0 | ||
| Beta proteobacteria | Ralstonia mannitolytica | KF751574.1 | Faro7 | 86% | 0 | ||
| Firmicutes | Beta proteobacteria | Alkaligens faecalis | KP823043.1 | Unwasa 32 | 79% | 0 | |
| Proteobacteria | Bacilli | PaeniBacilluslactis | FN429978.1 | IB-188B | 100% | 0 | |
| Gamma proteobacteria | Pseudomonas aeruginosa | KR007310.1 | 364 | 100% | 0 | ||
| G | Bacilli | PaeniBacilluslactis | KF607091.1 | LG2 | 100% | 0 | |
| Gamma proteobacteria | Serretia liquefaciens | KJ781948.1 | B7 | 99% | 0 | ||
| Bacilli | LysiniBacillus sphaericus | KP980616.1 | JXH19 | 100% | 0 | ||
| Unknown | No hit | No significant similarity | - | - | - | - | |
Table 3: Molecular Characterization Hydrocarbon Utilizing Isolates.
Some of the identified fungi spp were similar to the ones identified [18] in a crude oil polluted soil. Gomes et al. [37], also isolated and identified 50 species of filamentous fungi from the mangrove sediment of Barra das Jangades Jaboatoo dos Guarrapes Pernambuco in the North East of Brazil. Penicillium and Aspergillus were the dominant genera they isolated, their work agreed with the findings of this work where Aspergillus sp was the dominant, Fusarium, Trichoderma, Phoma, Talaromyces, Cladosporium, Gongronella, Microspheropsis and Mucor were also characterized in their study. Fusarium, Aspergillus, Botrytis, Alterneria sp, Vertcillium sp, Botryoplodis, Chrysosporium, Geotrichium sp, Monillia sp, Penicillium sp, Rhiopus sp, Nitrospora sp, Basidiobotrys, Torulopsis sp, Pichia sp, Rhodotorula sp, Saccharomyces sp and Candida sp were similarly isolated. Fungi cultures belonging to Ascomycota and Basidiomycota were dominant in the sites of study, this agrees with the work of Sridhar [38] who reported the presence of Ascomycetes, Mitosporic fungi, Basidiomycetes, Chitridiomycetes, Myxocycetes, Oomycetes, Thraustochikids and Zygomycetes in the mangrove swamp. Yeasts, Saccharomyces, Candida and Rhodotorule species were isolated in this study and these fungi have been found to have the ability to degrade hydrocarbons. Liu et al., Emtiazi et al., Hassanshahian et al. [39-41] reported molds which belong to the genera of Aspergillus, Penicillium, Fusarium, Yeasts, Candida, Yanonia, Pichia to be involved in hydrocarbon degradation. Santos et al. [42], isolated fungi from estuarine sediments near Santos (São Paulo) belonging to the following groups: Ascomycetes, Basidiomycetes, Zygomycetes and mitosporic, which were found to be tolerant to phenanthrene and pyrene. Cyclothryrium sp., Penicillium simplicissimum and Psilocybe sp. were able to degrade these compounds and Cyclothryrium sp. was the most effective [43].
This study has shown that culture-dependent method though can account for a small subset of the total microbial diversity and underestimates of the microbial communities present in petroleum polluted environments but revealed that crude oil pollution affected microbial compositin and diversity and isolating these organisms pose some challenges.
The study showed that 3.9% of isolates showed no amplification with universal primers used and 5.5% amplified bacterial isolates belonded to could not be classified, this findings corroborates with that of Rodrigo et al. [30] on molecular characterization of bacteria in different soils in brazil. In their studies, of bacteria diversity they reported different quantities of bacteria that cannot be classified within a known phylum present in the soil.
Successful colonization and high fungal biodiversity is favored by moist conditions, environments rich in organic matter, aeration and low pH amongst other factors. Though potentially ubiquitous, some fungal species are restricted to very specific niches and are endemic, especially the symbiotic and parasitic forms [44]. Marine sediments are inhabited by fungi.
The presence of bacteria and fungi in contaminated soils of study indicates that microorganisms are everywhere, diverse in nature and are able to adapt in extreme environments [45]. The ability of the bacteria to grow in hydrocarbon source indicates that they can utilize crude oil as energy and carbon source. Das et al. [46] hence, their ability to grow in mineral salt agar. The higher population of hydrocarbon utilizing bacteria observed in the polluted soils could be attributed to their ability to break down part of the oil and use it to grow; low population of hydrocarbon utilizing bacteria in the unpolluted soil could account for low carbon or energy in the environment. Similar report were published by Boboye et al., Onifade et al., Okerentugba et al. [19,27,47]. Abundant numbers of oil degrading microorganisms in an environment implies that those organisms are active degraders of the oil. The increase in HUB indicates that the indigenous microbes in the polluted soil may have catabolized part of the oil and used it to grow this is similar to the findings of Ojo [48], the activities of the indigenous microbes could be responsible for the bioremediation of the polluted sites [19,27,47].
Conclusion
The presence of bacteria and fungi in contaminated soils of study indicates that microorganisms are everywhere, diverse in nature and are able to adapt in extreme environments. The methods of characterization revealed significant decline in microbial population, composition and diversity in soils under oil pollution when compared with the unpolluted soil. Though, microbial enumeration is carried out traditionally, it is not an effective measure of soil microbial population. Molecular characterization also revealed the presence of hydrocarbon utilizing isolates which could not be classified. Sequences have been submited to the gene bank with accession numbers KT899779- KT899800.
References
- Allen JP, Atekwana EA, Duris JW, Werkema DD, Rossbach S (2007) The microbial community structure in petroleum-contaminated sediments corresponds to geophysical signatures. Appl Environ Microbiol 73: 2860-2870.
- Orji FA, Ibiene AA, Ugbogu OC (2012) Petroleum Hydrocarbon Pollution of Mangrove Swamps: The Promises of Remediation by Enhanced Natural Attenuation. Am J Agric Biol Sci7: 207-216.
- Maria GL, Sridhar KR (2002) Richness and diversity of filamentous fungi on woody litter of mangroves along the west coast of India. Current Science83: 1573-1580.
- Holguin G, Bashan Y, Vazquez P (2001) The role of sediment microorganisms in the productivity, conservation, and rehabilitation of mangrove ecosystems: an overview. Biol Fert Soils 33: 265-278.
- Kathiresan K, Bingham BL (2001) Biology of mangroves and mangrove ecosystems. Adv Mar Biol 40: 81-251.
- Atlas RM (1995) Bioremediation of petroleum pollutants. Int Biodeteriorat Biodegradat 35: 317-327.
- Kamagata Y, Tamaki H (2005) Cultivation of Uncultured Fastidious Microbes. Microbes and Environments - Microbes Environments 20: 85-91.
- Tyson GW, Banfield JF (2005) Cultivating the uncultivated: a community genomics perspective. Trends Microbiol 13: 411-415.
- Rondon MR, August PR, Bettermann D, Brady SF, Grossman TH, et al. (2000) Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol 66: 2541-2547.
- Pelczar JM, Chan LEA, Krieg NR (1993) Microbiology, Concept and Application. McGraw Hill Inc, New Jersey, USA, p: 847.
- Wellington EMH, Williams ST (1978) Preservation Of Actinomycete inoculum in frozen glycerol. Microbios Letters 6: 151-157.
- Cheesbrough M (2006) District Laboratory Practice in Tropical Countries. Cambridge University Press, Cambridge, UK.
- Harrigan WF, McCance ME (1990) Laboratory Methods in Food and Diary Microbiology. Academic Press, London, UK.
- Sambrook J, Russell DW (2001) Molecular Cloning.: A Laboratory Manual. Cold Spring Harbor Laboratory Press, New York, USA.
- White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: A Guide to Methods and Applications.Academic Press, London, UK, pp: 315-322.
- Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences 74: 5463-5467.
- Olukunle OF (2013) Characterization of Indigenous Microorganisms Associated with Crude Oil-polluted Soils and Water Using Traditional Techniques. Microbiol J 3: 1-11.
- Boboye B, Olukunle OF, Adetuyi FC (2010) Degradative activity of bacteria isolated from hydrocarbon-polluted site in Ilaje, Ondo State, Nigeria. African J Microbiol Res 4: 2484-2491.
- Wackett LP (2003) Pseudomonas putida – a versatile biocatalyst. NatureBiotechnol 21: 136-138.
- Atlas RM (1998) Microbial Ecology: Fundamentals and Applications. 4th edn.
- Friedrich M, Grosser RJ, Kern EA, Inskeep WP, Ward DM (2000) Effect of model sorptive phases on phenanthrene biodegradation: molecular analysis of enrichments and isolates suggests selection based on bioavailability. Appl Environ Microbiol 66: 2703-2710.
- Tyagi M, da Fonseca M, de Carvalho C (2011) Bioaugmentation and biostimulation strategies to improve the effectiveness of bioremediation processes. Biodegradation 22: 231-241.
- John R, Itah A, Essien J, Ikpe DI (2011) Fate of Nitrogen-Fixing Bacteria in Crude Oil Contaminated Wetland Ultisol. Bulletin of Environmental Contamination and Toxicology 87: 343-53.
- Chikere CB (2010) Bacterial diversity and community dynamics during the bioremediation of crude oil-polluted soil. Microbiology Department, University of Port Harcourt.
- Saadoun I, Mohammad MJ, Hameed KM, Shawaqfah M (2008) Microbial populations of crude oil spill polluted soils at the Jordan-Iraq Desert (The Badia Region). Braz J Microbiol 39: 453-456.
- Onifade AK, Abubakar FA (2007) Characterization of hydrocarbon-degrading microorganisms isolated from crude oil contaminated soil and remediation of the soil by enhanced natural attenuation. Res J Microbiol 2: 149-155.
- Olukunle OF, Boboye B, Ikuomola OT (2012) Indigenous bacteria and fungi responsible for bioremediation of oil-polluted soils in Ondo soils in Ondo State, Nigeria. Environtropica 8: 138-148.
- Yuting L, Joy D, Van Nostrand YD, Zhihi H, Liyon W, et al. (2011) Functional gene diversity of soil microbial communities from five oil contaminated fields in China. ISME Journal 5: 403-413.
- Jacques RJ, Okeke BC, Bento FM, Teixeira AS, Peralba MC, et al. (2008) Microbial consortium bioaugmentation of a polycyclic aromatic hydrocarbons contaminated soil. Bioresour Technol 99: 2637-2643.
- Margesin R, Labbé D, Schinner F, Greer CW, Whyte LG (2003) Characterization of Hydrocarbon-Degrading Microbial Populations in Contaminated and Pristine Alpine Soils. Appl Environ Microbiol 69: 3085-3092.
- Hugenholtz P, Goebel BM, Pace NR (1998) Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol 180: 4765-4774.
- Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59: 143-169.
- Pace NR (1997) A molecular view of microbial diversity and the biosphere. Science 272: 734-740.
- Hugenholtz P, Goebel BM, Pace NR (1998) Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol 180: 4765-4774.
- Leadbetter JR (2003) Cultivation of recalcitrant microbes: cells are alive, well and revealing their secrets in the 21st century laboratory. Current Opinion Microbiology 6: 274-281.
- Gomes NC, Flocco CG, Costa R, Junca H, Vilchez R, et al. (2010) Mangrove microniches determine the structural and functional diversity of enriched petroleum hydrocarbon-degrading consortia. FEMS Microbiology Ecology 74: 276-290.
- Sridhar KR (2005) Diversity of fungi in mangrove ecosystems. In: Microbial diversity: Current perspectives and potential applications. IK International Publishing House Pvt Ltd., New Delhi, pp: 129-148.
- Liu C, Zongze S (2005) Alcanivorax dieselolei. A novel alkane-degrading bacterium isolated from sea water and deep-sea sediment. Int J Syst Evolutionary Microbiol 55: 1181-1186.
- Emtiazi G, Saleh T, Hassanshahian M (2009) The effect of bacterial glutathione Stransferase on morpholine degradation. Biotechnol J 4: 202-205.
- Hassanshahian M, Emtiazi G, Cappello S (2012) Isolation and characterization of crude-oil degrading bacteria from the Persian Gulf and the Caspian Sea. Marine Pollution Bulletin 64: 7-12.
- Santos D, Cury JC, Do Carmo FL, Dos Santos AL, Tiedje J (2011) Mangrove Bacterial Diversity and the Impact of Oil Contamination Revealed by Pyrosequencing: Bacterial Proxies for Oil Pollution. PLoS ONE 6: e16943.
- Santos HF, Carmo FL, Paes JES, Rosado AS, Peixoto RS (2010) Bioremediation of mangroves impacted by petroleum. Watter Air Soil Pollut 216: 329-350.
- Watanabe K, Kodama Y, Kaku N (2002) Diversity and abundance of bacteria in an underground oil-storage cavity. BMC Microbiol 2: 10.
- Das K, Ashis K, Mukherjee F (2006) Crude petroleum-oil biodegradation efficiency of Bacillussubtilis and Pseudomonas aeruginosa strains isolated from a petroleum-oil contaminated soil from North-East India. Bioresource Technology 98: 1339-1345.
- Okerentugba PO, Ezeronye OU (2003) Petroleum degrading potentials of single and mixed microbial cultures isolated from rivers and refinery effluent in Nigeria. African Journal Biotechnol 2: 288-292.
- Ojo OA (2006) Petroleum-hydrocarbon utilization by native bacterial population from a wastewater canal Southwest Nigeria. African J Biotechnol 5: 333-337.
Citation: Nwankwo CC, Okpokwasili GC (2019) Molecular Characterization of Microorganisms in Crude Oil-Polluted Mangrove Swamps of the Niger Delta, Rivers State, Nigeria. J Bioremediat Biodegrad 10: 463.
Copyright: © 2019 Nwankwo CC, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Usage
- Total views: 3852
- [From(publication date): 0-2019 - Dec 20, 2025]
- Breakdown by view type
- HTML page views: 2907
- PDF downloads: 945