Research Article Open Access
Molecular Assessment of Microbial Species Involved in the Biodegradation of Crude Oil in Saline Niger Delta Sediments Using Bioreactors
| Chioma Blaise Chikere*, Chinedu Christopher Obieze and Phillip Okerentugba | |
| Department of Microbiology, University of Port Harcourt, Rivers State, Nigeria | |
| Corresponding Author : | Chioma Blaise Chikere Department of Microbiology University of Port Harcourt, Rivers State, Nigeria Tel: 2347030912861 E-mail: chioma.chikere@uniport.edu.ng |
| Received July 02, 2015; Accepted August 05, 2015; Published August 07, 2015 | |
| Citation: Chikere CB, Obieze CC, Okerentugba P (2015) Molecular Assessment of Microbial Species Involved in the Biodegradation of Crude Oil in Saline Niger Delta Sediments Using Bioreactors. J Bioremed Biodeg 6:307. doi:10.4172/2155- 6199.1000307 | |
| Copyright: © 2015 Chikere CB, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
| Related article at Pubmed, Scholar Google | |
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
Purpose: At elevated salinities conventional microbiological processes are not very effective, therefore clean up of contaminants using bioremediation strategy will involve the use of halophilic and halo-tolerant bacterial species. This research therefore aimed at isolating and identifying potential halophilic and halo-tolerant bacterial species capable of hydrocarbon degradation during bioreactor based treatment with exogenous nutrients. Methods: The diversity of indigenous bacterial species with potential to degrade hydrocarbons was investigated using both culture-dependent and independent techniques. Bioremediation of hydrocarbon contaminated saline sediments was carried out using seven 2.5 liter bioslurry bioreactors operated over a 64-day period. Physicochemical parameters monitored were pH, nitrate, phosphate, total petroleum hydrocarbon (TPH), polycyclic aromatic hydrocarbon (PAH), temperature, salinity, and total organic carbon (%TOC). Results: The baseline TPH, PAH and pH of the sediments were 19 ppm, 3.1 ppm and 7.0 respectively. The baseline salinity of the sediment was 10% thus the sediment was adjudged moderately saline. TPH ranged from 97 ppm-105 ppm on day zero and decreased to an average of 5.62 ppm on day 64, while PAH ranged from 56 ppm-61 ppm on day zero and decreased to an average of 4.02 ppm on day 64. The bacterial species identified as potential hydrocarbon degraders includes Halomonas lutea, Achromobacter spp, Aquitalea magnusonii, Bacillus sp, Sphingobacterium sp, Shewanella sp, Brevundimonas naejangsanensis, Pseudomonas pseudoalcaligenes, Pseudomonas aeruginosa, unidentified bacterium BH23 and Gordonia sp. The genus Pseudomonas formed majority of the isolates successfully sequenced and exhibited similarity values ranging from 91% to 100% with sequences deposited in GenBank. A combination of both molecular and culture based technique allowed the identification to species level of twelve isolates. One isolate could not be identified while the remaining isolates were identified to their generic level. Treatment BCD recorded highest total culturable heterotrophic bacteria (TCHB) count (7.1 × 108 cfu/g) and total culturable hydrocarbon utilizing bacteria (TCHUB) count (6.7 × 108 cfu/g). There was a significant difference at P<0.05 in TCHUB bacteria counts between the unamended bioreactor slurries and those amended with organic and inorganic nutrients. There were also significant differences in TCHUB counts when the bioaugumented slurry was compared with those amended with NPK, Urea and cow dung using one way ANOVA and Tukey’s multiple comparison tests. Conclusion: This study revealed potentially novel bacterial species and previously described hydrocarbon degrading bacterial species that can be characterized further to determine their role in hydrocarbon degradation as well as their salt tolerance level prior to application in bioremediation of saline environments.
| Keywords |
| Bioremediation; Saline; Sediment; Halo-tolerant; Hydrocarbons; Niger-delta |
| Introduction |
| Petroleum is a complex mixture of different hydrocarbons including aliphatic, cycloalkanes, mono and polyaromatics, asphalthenes and resins with majority of these compounds toxic and carcinogenic [1]. The increasing worldwide demand for energy from petroleum and an increase in prospecting for crude oil in the marine environment would mean a higher risk of accidental oil discharge into the marine environment [2,3]. |
| The Niger Delta region of Nigeria has witnessed an intermittent discharge of crude oil into its environment since the inception of crude oil exploration and exploitation, this has led to the destruction of its farmlands, aquaculture, rivers and creeks with hydrocarbon compounds [4]. |
| Recently they have been an increasing agitation to cleanup, reclaim and restore all oil polluted environments within the Niger Delta [5]. Bioremediation is said to be the best approach for environmental clean up because it is a cost effective and an eco friendly strategy. |
| However in moderate to highly saline environments like the marine environment, application of bioremediation is very challenging due to the detrimental effect of salt on microbial life. The salty nature of the environment could even be compounded by produced water, a byproduct or waste associated with oil and gas production which contains high levels of salt (1000-250,000 mg/L), oil and grease, toxic chemicals, heavy metals, and naturally occurring radioactive materials [6,7]. Therefore in other to effectively carryout bioremediation in such environments a different bioremediation approach has to be applied. This approach would involve the use of halophilic and halo-tolerant microbial species capable of effectively degrading hydrocarbons under high salt conditions. Halophiles are classified into three groups according to their optimal salt concentration for growth: slightly halophilic (1-3% w/v), moderately halophilic (3-15% w/v), and extremely halophilic (15-32% w/v) [7-9]. Many hydrocarbon degrading organisms are known [10] and in the marine environment a number of specialist hydrocarbon degrading taxa are also known [1,11]. Aromatic hydrocarbon degraders include Cycloclasticus spp. which utilize biphenyl, naphthalene, anthracene, phenanthrene, toluene, and benzoate [12], and Neptunomonas which can degrade naphthalene, 2-methylnaphthalene and phenanthrene as sole carbon sources, but are unable to degrade 2,6-dimethylnaphthalene, 1-methylnaphthalene, biphenylorace-naphthene [13]. This highlights the drawback caused by the chemical complexity of crude oil which limits the capacity of single microbial specie to efficiently and effectively degrade all crude oil components. A combined effort of mixed bacterial consortia however usually improves hydrocarbon bioremediation. Marine saturated hydrocarbon degrading specialists include Alcanivorax [14,15], Planococcus [16], Oleiphilus [17], Oleispira [18], Thalassolituus [19]. |
| With crude oil production shifting offshore in the Niger Delta region of Nigeria, it is expected that pollution of the marine environment with hydrocarbons will increase. Therefore an understanding of the impact of oil on indigenous microbial communities and the isolation and identification of key oil degrading indigenous microorganisms are prerequisite for directing the management and clean up of oil impacted marine environments. The use of artificial microbial consortia cannot substitute for highly complex and dynamic indigenous microbial population essential for complete and efficient hydrocarbon degradation [11]. Thus the objective of this study was to isolate and identify the bacterial species that are associated with hydrocarbon degradation in saline sediments of the Niger Delta which could be used as model organisms for pollution detection and as specialist organisms for oil spill clean-up. |
| Methodology |
| Site description |
| The examined sediments contaminated with crude oil originated from Bodo creek, in Bodo community of Gokana Local Government Area, in the lower Niger Delta Basin of Nigeria. Bodo creek is characterized by brackish water system and has undergone varying degrees of crude oil pollution due to exploration, exploitation and artisanal refining activities. |
| Sample collection |
| Sediment and water samples were collected in July 2013. Sediment was collected using Eckman grab, while water was collected using sterile 10 L Jerry can. All samples were transported to the Environmental Microbiology laboratory, University of Port Harcourt within 6 h for analysis. |
| Phco-chemical analysisysi |
| The parameters analysed were the pH, nitrate, phosphate, temperature and total organic carbon (%TOC). These parameters were analysed as described in ref. [20]. Salinity was determined using refractometer (S/mill-E Range 0-100%) after extracting water from sediment by centrifuging at 1000 rpm for 15 minutes. Gas chromatographic analyses were carried out as stated below. |
| Total petroleum hydrocarbons (TPH): Dried sediment samples were powder sieved and cold-extracted in conical flask for a total of two hours in each case using 100% dichloromethane. The solvent from the resultant solution was removed by means of rotary evaporator under vacuum (pressure not greater than 200 mbar) and finally by a flow nitrogen at not more than 30°C to yield the extracted organic matter (EOM). |
| The semi volatile compounds were introduced into the GC/MS by injecting the sample extract into a gas chromatograph (GC) equipped with a narrow-bore fused silica capillary column. The GC column was temperature-programmed to separate the analytes, which were then detected with a mass spectrometer (MS) connected to the gas chromatograph. |
| Analyte eluted from the capillary column were introduced into the mass spectrometer via a jet separator. Identification of target analytes was accomplished by comparing their mass spectra with the electron impact spectra of authentic standards. Quantitation was accomplished by comparing the response of a major (quantitation) ion relative to an internal standard using an appropriate calibration curve for the intended application. |
| Chromatograhic condition: The GC/MS system comprised of Agilent 6890GC, (Agilent technologies, Wilmington, USA) with 5975B MSD and MSD chemstation (version D. 03.00). Helium gas was used as the carrier gas at a constant flow rate of 1 mL/min at a pressure of 75 kpa. The injector temperature was set at 250°C. The program used was; 2 min hold time, ramp to 240°C at 7°C/min and a final ramp to 285°C at 12°C with an 8 min hold time. Column—30-m × 0.25-mm ID × 0.25 μm film thickness silicone-coated fused-silica capillary column. |
| MSD condition: Solvent delay: 4 min, Mode-Scan at 3.54, Solvent delay: 3 min, Quard temp: 150°C, Source temp: 230°C, Transfer line temp: 280°C, Sampling: 2, Low mass: 45.0 amu, High mass: 450 amn, Threshold: 150. |
| Polycyclic aromatic hydrocarbons (PAH): Solvents: pentane (Sigma-Aldrich, Germany) methylene chloride (99.50% Sigma-Aldrich Germany) acetone (BDH, chemicals, poole England), sodium sulphate anhydrous (May and Baker, Dagenham, England), silica gel 60/120 mesh (Hopkins and Williams, Essex England) distilled water. |
| The samples were dried, crushed and sieved using 0.5 mm sieve. 2.0 ± 0.1 g of samples was weighed into a clean extraction container. 10 ml of extraction solvent (pentane) was added into the sample and mixed thoroughly and allowed to settle. The mixture was carefully filtered into clean solvent rinsed in extraction bottles using filter paper fitted into Buchner funnels. The extracts were concentrated to 2 ml and then transferred for clean up/separation. |
| 1 cm of moderately packed glass wool was placed at the bottom of 10 mm long chromatographic column. Slurry of 2 g agitated silica gel in 10 ml methylene chloride was prepared and placed into the chromatographic column. To the top of the column was added 0.5 cm of sodium sulphate. The column was rinsed with addition of 10 ml methylene chloride. The column was pre-eluted with 20 ml pentane; this was allowed to flow through the column at the rate of about 2 min until the liquid in the column was just above the sulphate layer. Immediately 1 ml of the extracted sample was transferred into the column, the extraction column was rinsed with 1 ml pentane and added to the column as well. |
| The stop-cock of the column was opened and the eluent was collected with a 10 ml graduated cylinder. Just prior to exposure of the sodium sulphate layer to air. Accurately measured volume of 8-10 ml of the eluent was collected and labeled Aliphatic. |
| Following recovery of the aliphatic fraction and just prior to exposure of the sodium sulphate layer, the column was eluted with 1:1 mixture of acetone and methylene chloride in 1-2 ml increments. Another accurately measured 8-10 ml of the eluent was collected and labeled ‘’Aromatics”. The aromatic fraction was concentrated to 1 ml for PAHs analysis using gas chromatography. |
| Bioreactor design and operation |
| Bioremediation of hydrocarbon-contaminated sediments from Bodo creek was carried out using seven 2.5 L bioslurry bioreactors operated over a 64-day period. Two reactors served as controls (unamended) and (heat-killed), four out of the remaining five as nutrient amended bioreactors while one was amended with 108 cfu/g consortium of indigenous hydrocarbon utilizing bacteria. The bioreactors were designated as BPD, BCD, BUR, BNPK, BAUG, BUNa and BHK (Poultry dropping, Cow dung, Urea, NPK, consortium of indigenous hydrocarbon utilizing bacteria, unamended control and heat killed control respectively). Each of the 7 bioreactors received 1 kg (wet weight) of sediments, 20 ml of crude oil and 20 mg of anthracene. For the controls, the unamended treatment was spiked with hydrocarbons without nutrient addition to determine whether the indigenous bacteria have the natural ability to degrade petroleum hydrocarbons, whereas the heat-killed treatment (killed by autoclaving sediments at 121°C for 15 min at 15 psi on 2 consecutive days) served to measure the role of abiotic factors in the loss of petroleum hydrocarbons. The bioreactors were continuously stirred (by 2 impellers) at 150 rpm throughout the 64-day experimental period. Filtered air was supplied to the bioreactors from the air compressor through hoses running in and out of them. The reactors were sealed with Teflon to prevent the ingress of atmospheric air and egress of the slurry. Throughout the 64-days of experimentation the reactors were operated at room temperature (30°C). |
| Microbiological analysis of samples |
| Enumeration of total culturable heterotrophic bacteria (TCHB): 1 g (wet weight) of sediment was homogenized in 0.85% of normal saline. Decimal dilutions (tenfold) of the suspensions was plated out in duplicate on plate count Agar (PCA) modified with 10% NaCl and incubated at 30°C for 24 h for the THB counts [15]. |
| Enumeration of total culturable hydrocarbon utilizing bacteria (TCHUB): For hydrocarbon utilizing bacteria counts, appropriate dilutions of sediment suspensions (1 g wet weight of sediment homogenized in 0.85% of normal saline) was plated out in duplicate on Bushnell-Haas agar (Sigma-Aldrich, USA) modified with 10% NaCl. Hydrocarbons were supplied through the vapour phase transfer to putative hydrocarbon utilizers by placing sterile Whatmann No. 1 filter papers impregnated with 5 ml Okono medium crude oil in the lids of the inverted Petri plates. Plates were then incubated at 30°C for 7 days [15]. |
| Biochemical and phenotypic analysis |
| Individual cultures were identified by morphological and biochemical techniques using Colour Atlas of Medical Bacteriology [21]. |
| Molecular characterizations |
| Bacterial DNA extraction was done with Zymo Research Fungal/Bacteria DNA kit™. The DNA samples were quantified using NanoDrop ND-2000 spectrophotometer. A portion of the 16S rRNA gene sequence extracted from each bacterial species was subjected to polymerase chain reaction using the primer set pA8f-GC (59-CGCCCG- CCG-CGC-GCG-GCG-GCG-GGC-GGGGCG-GGG-GCAGGG- GAG-AGT-TTG-ATC-CTGGCT-CAG-39) and KPRUN518r (59-ATTACCGCGGCTG CTGG-39) [22]. Amplification of the template DNA was performed using 2 μl volume of the extracted bacterial DNA with Eppendorf thermal cycler. The 50 μl PCR mixture contained 5 μl of deoxy nucleoside triphosphates (dNTPs) mixture (2.5 μM) ( Promega, USA), 5 μl of 5X Green Go Taq Flexi buffer (Promega, USA), 3.5 μl of 25 mM MgCl2 (Promega, USA), 2 μl each of the primer set pA8f-GC (59-CGC-CCG-CCG-CGC-GCG-GCG-GCG-GGCGGGGCG- GGG-GCA-GGG-GAG-AGT-TTG-ATC-CTGGCTCAG- 39) and KPRUN518r (59-ATTACCGCGGCTG CTGG-39), 0.25μl of 5U/μl hot start Go Taq DNA polymerase, 2.5μl of 20 mg/ml of bovine serum albumin and 27.75 μl of sterile water. A reaction tube without template DNA served as negative control. |
| Amplified DNAs were examined by electrophoresis in 1.4% agarose gel with 2 μl aliquots of PCR products in 1X Tris-Acetate-EDTA buffer using a UV-transilluminator. PCR products of the bacterial DNA were sequenced using GATC Sanger sequencer. (GATC Biotech AG European Custom Sequencing Centre Koln). Sequence identification was performed using the BLAST-N facility of the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih. gov/BLAST/). Each 16S rRNA sequence was subjected to a basic local alignment search tool (BLAST) analysis on the GenBank database and matching hits, with e-values closest to 0.0 indicating a statistically plausible match, was selected. |
| Okono medium crude oil degradation screening |
| Pure isolates obtained from Bushnell-Haas agar were inoculated into Bushnell-Haas broth amended with Okono medium crude oil (9 ml broth culture-1 ml crude oil) as sole carbon source and incubated at room temperature for 9 days. Biodegradation was determined by monitoring the change in turbidity of liquid culture (OD600nm) using a spectrophotometer (Optima SP-300 spec. Japan). |
| Statistical analysis of data |
| Statistical analysis was performed on the data generated from the microbial counts, using one way ANOVA and Tukey’s Multiple Comparison Test. The software Graph-Pad Prism (GraphPad Software, CA, USA) for Windows version 5.04 was used for the analysis. |
| Results |
| Baseline analysis of sediment sample |
| Gas Chromatographic analysis of TPH detected the presence of C10-C40 carbons as shown in supplementary material. The PAHs detected includes Acenaphthene, Acenaphthylene, Anthracene, Benzo(a)pyrene, Benzanthracene, Chrysene, Fluoranthene, Fluorene, Naphthalene, Phenanthrene and Pyrene. The values for the total heterotrophic bacteria count and hydrocarbon utilizing bacteria count and other physicochemical parameters measured are shown in Table 1. |
| TPH and PAH analysis of bioreactor treatments |
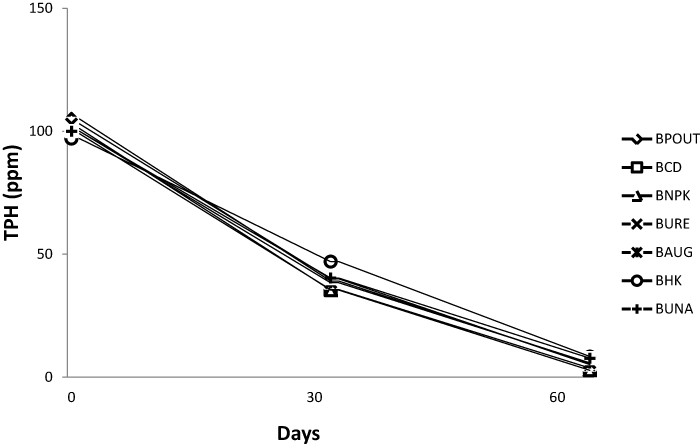
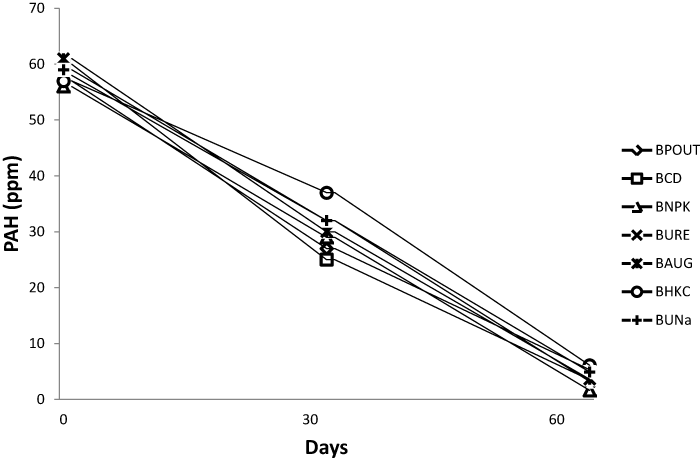
| The TPH and PAH value for all bioreactor treatment on day 1 was 105 and 61 ppm respectively. On day 64 PAH content decreased to an average of 4.02 ppm while TPH also decreased to an average of 5.62 ppm. Figures 1 and 2 represents the TPH and PAH degradation profile for each bioreactor treatment. |
| Bacterial enumeration from bioreactor treatments |
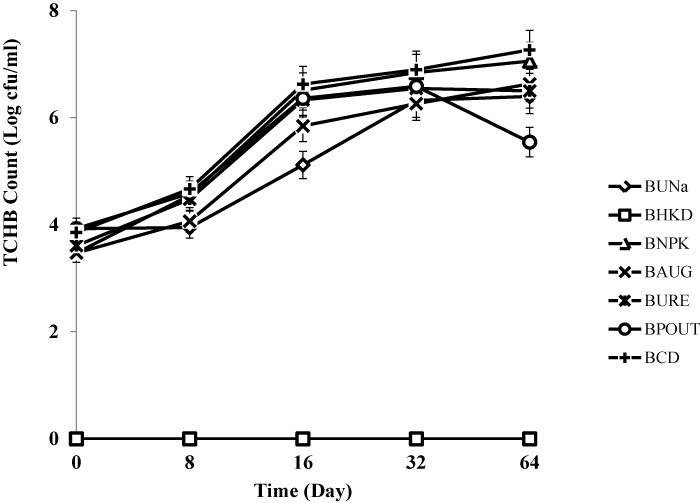
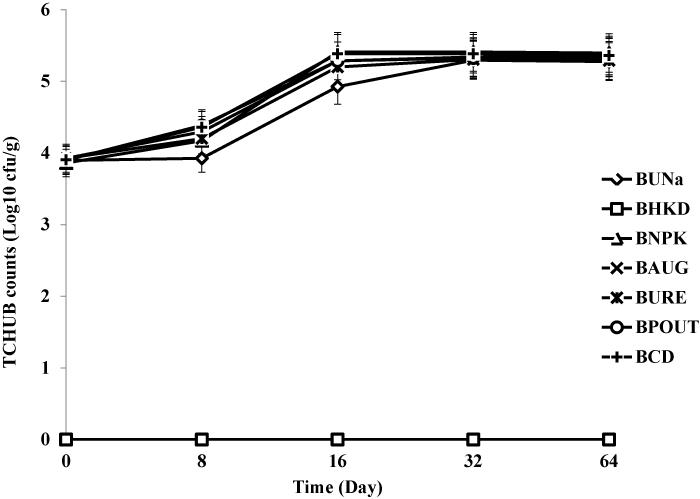
| Treatment BCD recorded highest heterotrophic bacteria count (7.1 × 108 cfu/g) and hydrocarbon utilizing bacteria count (6.7 × 108 cfu/g) respectively. The heat killed control BHKC showed no growth for TCHB and TCHUB throughout the 64 days of experimentation. There was no significant difference in TCHB between the un-amended bioreactor slurry and those amended with organic and inorganic nutrients, but there were statistically significant differences for TCHUB counts when BUNa was compared with other treatments. There were also significant differences in TCHUB counts when the bioaugumented slurry was compared with those amended with NPK, Urea and cow dung. Figures 3 and 4 represent microbial counts across all bioreactor treatments and controls during the 64 days of bioremediation. |
| Characteristics of bacterial isolates |
| After performing a preliminary phenotypic characterization that included the colony morphology, gram reaction and relevant biochemical analysis to eliminate duplicate strains, 25 isolates recovered from the different bioreactor treatments were selected. In an attempt to identify as many bacterial isolates as possible, DNA sequencing results were combined with biochemical, macroscopic and microscopic morphological data as shown in Table 2. Amplification of bacterial DNA samples with primer pair pA8f-GC (5′-CGCCCG- CCG-CGC-GCG-GCG-GCG-GGC-GGGGCG-GGG-GCAGGG- GAG-AGT-TTG-ATC-CTGGCT-CAG-3′) and KPRUN518r (5′-ATTACCGCGGCTG CTGG-3′) resulted in fragments of approximately 550bp. PCR products were checked on 1% agarose gel in 1x TAE buffer. All PCR products obtained with primer pair pA8f- GC-KPRUN518r were sequenced with primer KPRUN518r and were identified using the BLASTn program. |
| Comparative sequence analysis of the 16S rRNA of the selected isolates was performed. The sequences were aligned to the most similar 16S rRNA sequences in the data base. Table 2 shows the 16S rRNA closest match and accession numbers for the different isolates. |
| The genus Pseudomonas formed majority of the isolates successfully sequenced and exhibited similarity values with those deposited in GenBank ranging from 91% to 100%. One isolate (A59) could not be identified as there was no sequence match available in GenBank database. The remaining isolates were identified as organisms related to Gordonia sp (98% sequence similarity), Brevundimonas naejangsanensis (99% sequence similarity), Shewanella sp. (99% sequence similarity), Achromobacter sp. (96% sequence similarity), Sphingobacterium sp. (97% sequence similarity), Bacillus sp. (98% sequence similarity), Aquitalea magnusonii (96% sequence similarity), Achromobacter sp. (100% sequence similarity) and Chromohalobacter salexigens (94%) sequence similarity). |
| Biodegradation screening |
| All bacterial isolates recovered using Bushnell Haas Agar (BHA) were further subjected to biodegradation screening for their ability to utilize Okono medium crude oil as their sole source of carbon and energy. The results obtained showed that all the isolates were significant for crude oil degradability assay evidenced by biomass increase, increase in turbidity, emulsification and a concomitant visual gradual decrease in the oil layer after 9 days of incubation when compared with isolates on day zero and controls. |
| Discussion |
| In the last two decades, research in the marine environment has increased because of the prospects of isolating novel microbial species within the marine/moderately saline environment that possess enzymes adapted to crude oil degradation or that may be of general importance to the biotechnology industry [23]. In the Niger Delta region of Nigeria where crude oil exploration and exploitation activity is shifting offshore, there is a paucity of research in this regard. |
| The examined sediments contaminated with crude oil originated from a moderately saline creek (Bodo) in the Niger Delta region of Nigeria. Total heterotrophic bacterial counts, hydrocarbon utilizing bacterial counts, as well as gas chromatography and other physicochemical analysis were carried out on the sediment in order to investigate the role of the autochthonous bacterial population in crude oil degradation. |
| There was a general increase in microbial population in the amended sediment slurries with attendant reduction in TPH and PAH. The hydrocarbon losses recorded in the bio-stimulated sediment slurries can be attributed to microbial activities which resulted in consumption of nitrogen and phosphorus added in the form of urea, NPK fertilizer, poultry droppings, and inorganic sources of nitrogen and phosphorus. Several researchers have reported that nutrient amendment with both organic and inorganic fertilizers significantly affected crude oil biodegradation [11,15,24]. Statistical analysis of the various bio-stimulated sediment slurries did not show significant differences for TPH and PAH. The significant differences in TCHUB counts recorded when the bio-augmented slurry was compared with those amended with NPK, Urea and cow dung is also consistent with previous research. |
| The reduction in TPH and PAH concentration in the contained system and the ability to culture the various species of bacteria recovered during the period of bioremediation using okono medium crude oil as the sole carbon source implies that the isolates are potential hydrocarbon degraders and could play important roles in bioremediation of pollutants. |
| The study carried out suggests the proliferation of potential halophilic and halotolerant bacterial species capable of Okono medium crude oil degradation in a contained system. The recovery of Halomonas is consistent with previous research carried out in saline habitats. The Genus is known to contain no non halophilic species [25] and constitute the most commonly cultured inhabitants in saline environments. Halomonas sp. and Marinobacter sp. have been recovered from hydrocarbon contaminated saline soil collected from several regions in Iran [26]. Halomonas have also been previously suggested for use as model organism for the study of degradation of aromatic compounds at high salt concentrations, as this group of microorganisms have few nutritional requirements, can withstand a wide range of salinities, degrade a wide range of aromatic compounds and are easily cultured [23]. |
| Pseudomonas and Bacillus, which are commonly isolated in physiologically normal environments, have species recognized as halotolerant and with potential for hydrocarbon degradation [25]. These organisms are often referred to as generalist hydrocarbon degraders, yet they seem to constitute a significant proportion of hydrocarbon degraders and play significant roles in hydrocarbon degradation in the marine environment. The Genus Pseudomonas formed the majority of the species recovered during the period of bioremediation; this could be as a result of their effective competitive capabilities in a wide range of environments [25] with saline environment not being an exception. Pseudomonas, Acinetobacter and Alcanivorax have been previously recovered from Niger-Delta marine sediments [15]. Previous scientific investigations have constantly reported Pseudomonas to be effectively involved in hydrocarbon degradation [4,15,27,28]. They are very versatile and have been reported to produce biosurfactants which aid in hydrocarbon emulsification and in reduction of hydrophobicity [29]. |
| Halo-tolerant Achromobacter xylosoxidans capable of degrading multiple PAH have been isolated from a crude oil polluted saline site [30]. Bacillus sp has been reported as part of the diverse halophilic hydrocarbon utilizing bacteria from Kuwaiti coast of the Arabian Gulf [31]. The significance of the presence of the genus Brevundimonas and Aquitalea as part of the diversity of bacteria recovered would require further screening. Though it was able to grow on Bushnell Haas agar with crude oil as sole carbon source, there is paucity of information on its role in hydrocarbon degradation and thus would require further screening for the presence hydrocarbon degradative genes alongside other isolates recovered. The unidentified bacterium (strain BH23) can be said to be potentially novel species and will be investigated further for its role in hydrocarbon degradation. |
| The incubation of marine sediment in the presence of phenanthrene and bromodeoxyuridine revealed Shewanella as part of the diverse microorganisms capable of PAH degradation [32]. In another study of the influence of crude oil on changes in bacterial communities in Arctic sea-ice, the dominant phylotypes recovered were Pseudomonas spp., Shewanella sp and Marinobacter spp [33]. |
| Three strains of Gordonia sp have been reported to be capable of degrading pyrene, benzo[a] pyrene, anthracene, naphthalene, phenanthrene, and fluoranthene as the sole source of carbon and energy [34]. Sphingobacterium multivorum have previously been reported as part of bacteria consortia capable of utilizing used lubricant [35]. |
| Conclusion |
| This study revealed potentially novel bacterial species and previously described hydrocarbon degrading bacterial species that can be characterized further to determine their role in hydrocarbon degradation as well as their salt tolerance level prior to application in bioremediation of saline environments. |
| Acknowledgements |
| We acknowledge the International Foundation of Science (IFS) Stockholm, Sweden, for funding this research through grant W4263-2 given to Dr. (Mrs.) Chioma Blaise Chikere as well as staff members of Molecular Biology and Biotechnology Division, Nigerian Institute of Medical research (NIMR) Yaba, Lagos Nigeria. |
References
|
Tables and Figures at a glance
| Table 1 | Table 2 |
Figures at a glance
 |
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 14951
- [From(publication date):
September-2015 - Apr 03, 2025] - Breakdown by view type
- HTML page views : 10239
- PDF downloads : 4712
