Review Article Open Access
Modern Approaches into Biochemical and Molecular Biomarkers: Key Roles in Environmental Biotechnology
Paniagua-Michel J1* and Olmos-Soto J21Marine Bioremediation and Bioactive Metabolites Lab, Centro de Investigacion Cientifica y de Educacion Superior de Ensenada, Mexico
2Molecular Marine Microbiology Lab, Centro de Investigacion Cientifica y de Educacion Superior de Ensenada, Mexico
- Corresponding Author:
- J Paniagua-Michel
Marine Bioremediation and Bioactive Metabolites Lab
Centro de Investigacion Cientifica y de Educacion Superior de Ensenada Mexico
Tel: +52-646-1745050
Fax: 526461750569
E-mail: jpaniagu@cicese.mx
Received date: November 26, 2015; Accepted date: December 30, 2015; Published date: January 06, 2016
Citation: Paniagua-Michel J, Olmos-Soto J (2016) Modern Approaches into Biochemical and Molecular Biomarkers: Key Roles in Environmental Biotechnology. J Biotechnol Biomater 6:216. doi:10.4172/2155-952X.1000216
Copyright: © 2016 Paniagua-Michel J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Biotechnology & Biomaterials
Abstract
Biomarkers are characterized by a unique order in their molecular structures and are the first sentinel’s tools for sensitive effect measurements in environmental quality or biotechnological processes assessment. Most of the importance of biomarkers resides in their property to be measurable using different biochemical and molecular approaches. The recent application and or studies of biomarkers and its correlation at the omics era, has revalorized new roles of biomarkers in environmental biotechnology. In this work, some of the common biomarkers actually used, viz, pigments, cytochrome P4501A enzyme induction, acetylcholinesterase inhibition, DNA integrity and metallothiones are analyzed concomitantly to recent applications of Omics technologies to optimize metabolic networks in living biomes. New developments under the umbrella of the culture independent molecular tools applied for the analyses of mixed microbial communities have contributed in understanding catabolism from contaminants in extreme and fragile environments. These approaches open the venue for the new biomarkers for an increased biodiversity expectative of ca. 99% higher than conventional classification. Functional genes by metagenomic arrays, will greatly improve our understanding of microbial interaction and metabolism to facilitate the development of suitable bioremediation strategies for environment clean up.
Keywords
Biomarkers; Bioremediation; Environment; Biotechnology; Omics
Introduction
Contemporary science has been applying biomarkers for more than half a century, but a remarkable explosion has increased since the 21st century [1] and recently due to the development of high-throughput technologies available for generating large-scale molecular-level measurements. These new technological developments have led to an increased interest in the discovery and validation of new biomarkers in environmental and biotechnological studies in the omics era. Nowadays, biomarkers are mostly molecular makers, such as genes, proteins, metabolites, and other molecules, used for disease diagnosis, prognosis, and prediction of response processes in living cells. In recent years, levels of contaminants in different terrestrial and aquatic environments have increased largely because of various anthropogenic activities. The stringent situation caused by xenobiotic damage to the environment has led to develop biomarkers for rapid assessment of the impact of various types of contaminants and thereby help undertake appropriate corrective measures [2,3]. These continuous expositions of organisms and their environment to different anthropogenic pollutants are susceptible to interact with the physiological processes (growth and reproduction) and are important elements for any bioscreening program. That is why, recent developments of biomonitoring aquatic and terrestrial environments are now involving biomarkers, which are measurable parameters at different levels of biological organization (molecular, cellular or physiological). Probably, the major utility of biomarkers, are their capabilities to traduce changes in the metabolic regulatory processes resulting from the effect of anthropogenic stressors (Table 1) [4].
| • Biomarkers can exhibit effect of electron transfer or reception • Biomarkers are effective early warning signals of adverse biological effects. • Biomarkers can indicate biological effects, while chemistry-based surveillance system cannot. • Biomarkers are more effective in revealing overall toxicities of complex mixtures. • Biomarkers are economical. • Biomarker have different degrees of specificity. |
Table 1: Main advantages of Biomarkers (modified after [28]).
The use of biological markers or biomarkers at the molecular and cellular level are considered as the first sentinels tools for sensitive effect in environmental quality or biotechnological processes assessment [5]. Hence, a biomarker can be defined as the measure of the effect of pollutants and or the host response in biochemical, cellular and molecular terms [6,7]. A biomarker also can be useful at the level of the whole animal, which effect is detectable at physiological, behavioural or energetic terms’ [8]. For instance, marine bivalves, such as mussels are effective sentinels of the impact of xenobiotics at level of marine ecosystem [6]. The term exposure biomarkers indicate that the organism had been exposed to pollutants (exposure biomarkers) and/ or the magnitude of the organism’s response to the pollutant are called effect biomarkers or biomarkers of stress [2].
In this work, we analysis the uses of biomarkers with respect to the specific requirements for further implementation of biomarkers in environmental biotechnology monitoring and its significance as diagnostic and prognostic tools for pollution monitoring. New expectative for multiple effectors using omics tools can lead to the development for a new generation of biomarkers.
Biomarkers and Environmental Biotechnology
The use of biomarkers at molecular or cellular level have been proposed as sensitive ‘early warning’ tools for biologic effect measurement in environmental quality assessment [8]. These early warning biomarkers can be used in a predictive way, allowing the application of strategies before irreversible environmental damage of ecological consequence occurs, viz short term indicators of long-term biological effects (Table 2).
| •Discrimination between contaminants and the organisms. •Detection of the presence contaminants. •Early detection of effects (signals for preventive action). •Integration of measure of bioavailable pollutants. •Attribution to exposure and risks to environmental pollutants •Detection of changes in ecosystems due to pollutants. •Prevention of irreversible environmental damage •Identification of main routes of exposure at trophic levels •Toxicity of chemicals and or effluents undetected (chemicals speciation, absorption and uptake effects). •Detection of toxic effects of parent compounds and metabolisable metabolites including contaminants (PAHs and organophosphate). •Integration of toxicological interactions of mixtures of contaminants (various congeners of PCBs, PAHs, metals) measurable t a particular molecular, cellular or tissue target. •Short-term predictors of long-term ecological effects at different levels of contamination Sarkar et al 2006 (Behrens and Segner, 2005). •Versatility of application (to both laboratory and field studies). |
Table 2: Functional Roles of Biomarkers in Marine Environmental Applications [modified after [2].
In view of the continuous increase of xenobiotic of anthropocentric activity at planetary levels, the use of molecular biomarkers has drawn worldwide attention for monitoring programs of large ecosystems, such as the aquatic and marine environments. Hence, commonly used biomarkers such as cytochrome P450 1A enzyme induction, DNA integrity, acetylcholinesterase activity, metallothioneins and Pigments, are downlines explained (Table 3).
| Biomarker | Detection/Application | Analytical Tools |
|---|---|---|
| Pigments | Presence of photosynthesisSpecificantioxidants, Vit A | Fluorescence, HPLC, Mass spectra |
| CytochromeP4501A | Presence of organic contaminationcertain PAHs, PCBs, PCDDs, etc. | Enzyme activity (fluorometry andspectroscopy) and amount (Elisa) |
| DNA—damage | Presence/damage of organicxeno-biotics (PAHs, PCBs and PCDDs) | DNA strand breaks measurement, |
| AChE inhibition | Indicator of exposure to organo-phosphorous Carbamate, toxins,metals (Cd, Pb, Cu, etc.,). | Enzyme activity (spectrophometer pH) |
| Metallothioneins | Indicator of exposure to metals(Zn, Cu, Cd and Hg).In: vertebrates (mammals) andAndinvertebrates (molluscs, fish)Algae. | Proteins (pulse polarography, liquidchromatography |
Table 3: Particular molecular biomarkers of biological effects of contaminants on aquatic organisms and respective analytical technique of their measurements (modified after [2]).
Cytochrome P4501A Induction
The need of environmental markers for contaminants, like dioxins, furans, polychlorinated biphenyls and polycyclic aromatic hydrocarbons recently have found in the cytochrome P4501A (CYP1A) a sensible biomarker detecting the biotransformation of such contaminants [2,3]. In this action, the presence of aryl hydrocarbon receptor from cytosol enhances the induction of CYP1A due to exposure of organisms to such pollutants, which has been used as a biomarker in pollution monitoring in fish. For instance, in the case of marine bivalves, interest in the use of biomarkers has led to the application of ethoxyresorufin dealkylation (EROD) [3,8]. In the case of the Zebra mussel (Dreissena polymorpha), a significant induction of EROD activity occurred when mussels were exposed to 100 ng/l of PCB mixture of Arochlor 1260 and dioxin-like CB-126, but this congener showed also a clear competitive inhibition after 48 h of exposure. The CYP1A biomarker was able to discriminate the pollution status, of small streams receiving different levels of contamination by AhR-binding PAHs and PCBs [9]. The results of the enzymatic CYP1A activity by measuring the EROD activity, pointed-out as a good biomarker of pollutants in marine and estuarine environments [3,9].
DNA integrity as a Biomarker of Pollution
The susceptibility of the integrity of DNA can be greatly affected by genotoxic agents due to DNA strand breaks, loss of methylation and formation of DNA adducts [2,3]. It is known that the double strandedness with respect to total DNA exhibit the effect of various genotoxic chemicals on the integrity of DNA. The exogenous agents induces the breaking of DNA strand breaks due to direct DNA damage, or may be produced during DNA repair process or other physiologic responses in the cell. Members of the PAH such as Benzo(a)pyrene (BaP), is oxidized to chemically reactive diolepoxide (BaPDE), which subsequently interact with DNA to form both stable and unstable adducts with DNA [10]. Each type of adduct may contribute towards the eventual transformation of the cell [10]. Single strand breaks can be produced in cells by direct chemical damage of DNA [2,3]. The chemical reactions that are known to generate single strand breaks proceed by ionizing radiation, oxidation-reduction reaction or photoreaction.
Studies on the status of integrity of DNA in marine snail (Planaxis sulcatus) have shown the impact of pollution at different sampling sites, as evidenced from measurements of the DNA strand breaks [2]. It the case of marine snail P. sulcatus, DNA integrity decreased significantly at the contaminated sites, which was attributed to the extent of contamination of these sites by petroleum hydrocarbons of waste materials into the coastal water.
Metallothioneins (MTs)
MTs are cytosolic proteins of low molecular weight and rich in cysteine. Its ubiquitous occurrence has been associated to the inactivation of toxic metal ions by binding them to sulfur atoms of the peptide cysteine residues [11]. In fact, it has repeatedly been shown that concentrations levels of MT can be correlated to accumulated fractions of toxic metal ions such as copper or cadmium in animals’ tissues [12]. The presence and role of metallothioneins (MTs) allowing mollusk bivalve species to accumulate and tolerate high amounts of heavy metals are based on their metal handling by metallothioneins (MTs). In addition, metallothionein concentrations in molluscs may also vary due to influence by nonmetallic pollutants. That is why the idea that MTs might be used as biomarkers for environmental pollution has emerged by measuring their concentrations in bivalves from contaminated habitats.
MTs as biomarkers of oxidative stress
MTs act also as molecular biomarkers for oxidative stress and are used extensively in aquatic organisms, especially for toxic metal pollution [13]. MT act as metal-chelating agents for the excess of toxic metals in the cells, thus playing important roles in metal metabolism in aquatic organisms and particularly in the detoxification mechanisms, through oxygen free radical scavenging actions and metal binding [14]. A huge increase of research and applications into the field of oxidative stress has been stimulated by free radical reactions and ROS in the physiological processes of living organisms and in the mechanisms of toxicity by exposure to a variety of environmental pollutants. The resulting oxidative damage to lipids, DNA, and proteins and the adverse effects on the antioxidant, enzymatic and non-enzymatic, defense mechanisms of aerobic organisms have been used in recent years as biomarkers for monitoring environmental pollution. The current knowledge that such processes of oxidative damage occur in aquatic organisms gave the impetus to extend environmental and ecotoxicological studies to aquatic organisms as sentinels of environmental contamination by toxic chemicals [15]. All these studies indicate that oxidative biomarkers in combination with other types of biomarkers (hepatic, genotoxic, hematoimmunotoxic) in aquatic organisms can be useful in large-scale environmental monitoring programs [16].
Metallothionein induction
MTs are ubiquitous in eukaryotes, and have unique structural characteristics to give potent metal-binding and redox capabilities [17]. One of the characteristics of the thiol groups (-SH) of cysteine residues enable MTs to bind particular heavy metals. MT-like proteins have been reported in many vertebrates including many species of fish [18] and aquatic invertebrates mainly molluscs [19] and crustaceans. MTs can be induced by the essential metals Cu and Zn and the non-essential metals Cd, Ag and Hg in both vertebrates and invertebrates. The exposure of aquatic organisms to toxic metals and associated changes in biochemical processes are indicatives of early warning signals of the presence of these particular contaminants, which enhances its uses as biomarkers. Effective response to induction of MTs has been detected in more than 50 different species of aquatic invertebrates, viz, in the oyster Crassostera virginica [20] and the mussel (M. galloprovincialis, Mullus barbatus) [21,22]. That is why; MTs are good candidates to monitor metal contamination as well as a safe and useful biomarker for evaluation of pollution in aquatic environments. Several organs of marine organisms are the first target of pollution and respective sequestration of metals by MT such as in the gills, digestive gland and kidney, indicating the significance of these tissues in uptake, storage and elimination of metals [23]. Actually, studies incorporating the quantification of several biomarkers and MTs allow more accurate in situ applications.
Pigments as Biomarkers
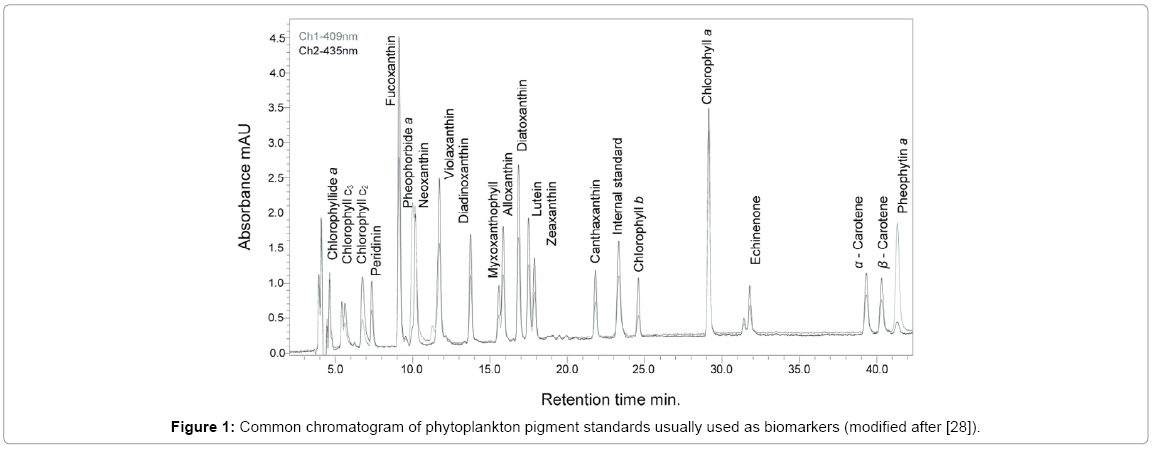
Algae and plant biomarkers are molecules whose carbon skeleton can unambiguously be linked to that of a known biological precursor compound. In 1961, Jeffrey [24] recommended the use of pigments as biomarkers in studies of distribution and abundance of phytoplankton in the oceans, but was until the 80’s that phytoplankton markers were recognized based on the precise separation and quantification of most algal species. Pigments are present in all photosynthetic organisms, which main primary function is as light harvesting agents for photosynthesis and photo protection. In plants and algae, there are three basic classes of pigments. Chlorophylls are greenish that contain a porphyrin ring and is present in all plants, algae and cyanobacteria. The most important is chlorophyll "a", which makes photosynthesis possible, by passing its energized electrons on to molecules, which will manufacture sugars. Carotenoids are usually red, orange, or yellow pigments, which gives plants and algae their characteristic color. These compounds are composed of two small six-carbon rings connected by a "chain" of carbon atoms. Both phycocyanin and phycoerythrin fluoresce at a particular and distinctive wavelength, which are reliable to be used as chemical "tags" with extensive use in cancer research, for "tagging" tumor cells. Chlorophyll (the primary photosynthetic pigment is ubiquitous for all autotrophic organisms cells) are related to biomass of phytoplankton and macrophytes in marine and freshwater ecosystems [25]. The distribution of accessory pigments in plants and algal divisions is unique, that is why pigments are useful and relevant biomarkers representing taxonomic specificity [26], and hold the representation of the entire phototrophic community and overall primary production. For instance, chemotaxonomy is often used as a commonly accepted and as a standard method in oceanographic studies. Carotenoids unlike chlorophylls are often broken down to colorless compounds, by destruction of the long chain of alternating double bonds that cannot be detected by regular pigment analysis methods [27]. As biomarkers, pigments have been used as antioxidants, to characterize marine organic matter and to assess rates of key oceanographic processes, as well as to determine the phytoplankton community structure in water samples as a supplement or alternative to microscopically counts (Figure 1) [28].
Biomarkers Exemplified for Environmental and Aquatic Pollution
Selected bio markers in different organisms can represent sensitive indexes, or early warning signals of effectors, viz, measuring stress by contaminants on the organism, and are effective sentinels for biomonitoring, both in the marine and freshwater environment [29]. Among the aquatic organisms susceptible of research, fish have become an interesting subject because the effect factors on fish health temperature changes, habitat and water quality deterioration as well as aquatic pollution which may result in mortalities and population decline [30]. That is why the innate immune responses in fish is considered as suitable biomarkers for monitoring biological effects of pollution induced by xenobiotics on fish immune responses [29-31].
Metals are binding molecules with biologically active constituents of the organisms body such as lipids, amino acids and proteins [32], that under stress conditions induced by excess of certain heavy metals, like nickel, exhibit a significant decrease in total glycogen content of gill, digestive gland and whole body freshwater bivalve, Parreysia cylindrical [33]. On the other hand, increases in the level of pollutants increases also the levels of cortisol, which have a marked modulatory potential on immune functions [30], and hence promissory to be used as a biomarker. Several agencies have addressed the importance of tracing sources and fates as well as the effects of contaminants into aquatic ecosystems, such as estuaries where effects have been most strongly observed [29,34]. Effective role of biomarkers was developed from analysis of DNA of fish from polluted sites revealed the biomarker presence of DNA adducts containing aromatic or bulky hydrophobic moieties not present in DNA of fish from an unpolluted reference site [35]. The authors [35] suggested that the amount of DNA adducts in eel and earthworm may be a suitable and sensitive indicator and biomarker for the exposure to carcinogenic PAH from contaminated sediments or soils, respectively, and therefore useful in human exposure risk assessment.
Omics for New Biomarkers
Unculturable microbial communities can find scaling possibilities by high-throughput sequencing, DNA cloning and amplification technology, coupled with genomic tools, which allow holistic approaches of the predominantly species and respective biomes (Figure 2) [36]. Genomic studies into the biodiversity of different environments are promissory tools leading to the biotechnological exploitation from basic applications to industrial developments purposes. In recent years, 16S ribosomal RNA has been overshadowed by the application of high throughput shotgun sequencing to environmental DNAs (metagenomics) [37,38]. Metagenomic sequencing randomly samples of all genes present in a habitat rather than just 16S, thereby providing clues to the functional capacity of a community rather than just its phylogenetic composition. Community composition profiling by 16S is now often used as a preliminary step before metagenomic analysis and can be of great value in guiding decisions regarding sequencing technology to be used (Figure 2). However, 16S is reemerging as a stand-alone molecular tool owing to a confluence of methodological advancements.
The Great Plate Count Anomaly’, established that almost 99 percent of the microbial communities are not yet discovered neither cultivated. The advent of culture-independent high-resolution technologies, have solved the problem of identification. Moreover the “omics” technologies (genomics, transcriptomics, proteomics, and metabolomics) and the next generation sequencing platforms can sequence the genomes of single isolates as well as those belonging to a complex mixture of organisms via shotgun metagenomic sequencing [39,40]. This massive parallel sequencing has also enhanced the depth of 16S ribosomal RNA (rRNA) gene surveys much greater than Sanger sequencing [41-43] and the ability to increase the sequencing depth of phylogenetic markers broadened our knowledge of the diversity of a microbial community in various environments.
Decoding What is There (16S rRNA)
Advances in modern genomic technologies provide a variety of possibilities to access marine bio diversity otherwise hard to achieve by traditional techniques. Sequencing and other techniques for identifying DNA from environmental samples can yield a far more complete analysis of the role of organisms from a particular community. In this sense, phylogenetic markers genes becomes as a priority tool for Identification and classification of novel organisms, considering their common cell tasks such as transcription and translation [44,45]. Although alternative techniques, such as metagenomics, provide insight into all of the genes (and potentially gene functions) present in a given community, 16S rRNA-based surveys are extraordinarily valuable given that they can be used to document unexplored biodiversity and the ecological characteristics of either whole communities or individual microbial taxa. Perhaps because 16S rRNA phylogenies tend to correspond well to trends in overall gene content (8), the ability to relate trends at the species level to host or environmental parameters has proven immensely powerful [45]. These approach may result from sequenced rRNA genes directly [5], from scan rRNA genes in largeinsert clones or by computational identification of the 16S rRNA gene. Additionally, these studies have revealed that the diversity of different communities can vary significantly up to the level of thousands species [46]. For example, actually is possible to discover novel clades of dominant members of bacterioplankton communities in the ocean by using the 16S rRNA gene [47], but which had escaped detection by other approaches [47]. But if the interest is on the functionality of the identified microorganisms, DNA sequencing of environmental samples can be used as biomarkers. However, challenges associated to the complexity of the sample of interest such as the comprehensiveness of the sequencing, and the length of the fragments themselves needs to be overcome. Some authors pointed out that environmental DNA sequences are not sufficient to determine gene function, because of the large number of both conserved and non-conserved genes with unknown function in individual bacterial genomes and metagenomes. According to [36], It is clear that omics level technologies derived from primary sequence information are necessary to make the transition from gene and genome catalogues to functional significance.
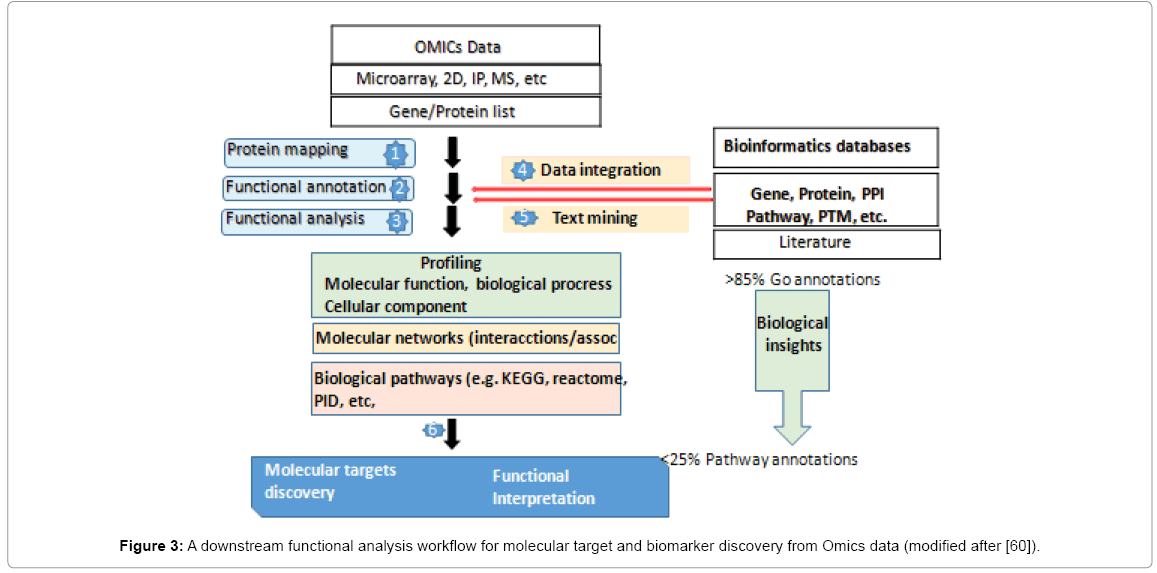
In natural microbial communities, microarray technology can be applied both as a tool to monitor critical gene activities across a diverse spectrum of genomes [48,49] or to access the transcriptome of single microbial strains in a complex community. Probably the most useful approach of the microarray-based gene expression is the quantitative assessment of transcript abundance that provides, which can be used to predict gene function based on the hypothesis that functionally related genes are more likely to be transcriptionally coregulated. Metagenomic sequencing randomly samples all genes present in a habitat rather than just 16S, thereby providing clues to the functional capacity of a community rather than just its phylogenetic composition. ‘Classical’ community composition profiling by 16S is now often used as a preliminary step before metagenomic analysis and can be of great value in guiding decisions regarding sequencing technology to be used (454 vs. Sanger, shotgun vs. large-insert clones) and amount of sequencing necessary [46]. Actually, a new reemerging of 16S is occurring as a platform-alone molecular tool owing an integration of methodological advancements, such as in Omics tools (Figure 3).
Almost all of the 16S sequence data in the public repositories to date are the products of PCR amplification of the 16S rRNA gene using 15– 25 nucleotide primers broadly targeting bacteria or archaea. However, such primers are known to miss some organisms owing to target mismatches [50,51] and the recent application of short (10 nucleotide) ‘miniprimers’ suggests that a considerable amount of diversity may be overlooked in environmental samples using standard 16S primers [52]. Pyrosequencing of cDNAs prepared from environmental RNAs may be the way of the future. This approach not only bypasses any potential primer bias, but simultaneously also provides a community profile of all three domains of life and functional information in the form of expressed messenger RNAs [53]. Actually, the revolution in sequencing technology, such as high-throughput sequencing technologies combined with the development of advanced computational tools have made important advances in the identification of cost-effective of microbial phylotypes using metadata which corroborate the excellence of the 16S rRNA phylogenetic marker for taxa identification purposes [41].
Genomics and Proteomics: Toward Functional
Functional genomics and proteomicsare extremely useful for understanding the dynamic aspects of gene expression and protein function at the level of transcription, translation, protein–DNA, and protein–protein interactions [54]. Moreover, these tools contribute to identify which genes and metabolic pathways are involved in the studied processes. But, nevertheless, functional genomics and proteomics studies give a more robust understanding of the origin of the particular function selected from the information encoded in an organism’s genome [55]. In principle, when studying the genome of a community, also the transcriptome, or proteome may contribute to the precise understanding of the role, as in the case of the subfields, metatranscriptomics, or metaproteomics. In particular studies aiming apply new developments of massive sequencingand high-throughput screening technologies have allowed to establish the function of genomes at the levels of DNA, RNA transcripts, and protein products. In this way, is possible to understand the association or differences on the genomic structure of particular organisms and its phenotypic response to environmental changes [56]. The application of functional genomics allow to corroborate in deep the expression of all the genes of a biological system from the study of a single gene and contribute to validate in deep the early verdict of any particular of organisms in an ecosystem [54].
Information related to the quantification of the gene expression to a pollutant in a cell provide information of genes regulation to the mechanisms involved in several cell functions to the different responses of the cells to the environment, viz, defense, detoxification, or adaptation. For instance, the effect of aromatic hydrocarbons on alterations of microbial cells was studied involving the expressions of regulatory proteins/enzymes in response to external stimuli through the techniques of functional genomics and proteomics, which revealed the complexity of the pollutant degradation [53]. Much work in the field of cellular and molecular approach on the degradation of pollutants, demand for integrative approaches, including functional genomics of the study of several proteins and genes working in the process, which is a hard and complex task in current progress [56]. Recently, metagenomics is contributing into the study of the whole genetic material from an environmental sample without culturing the microorganisms. These approaches allow the access direct to the genome of an entire microbial community including the former missing unculturable fraction of microorganisms, and allow more advanced programs of bioremediation.
Metagenomics commonly refers to construction and analysis (screening) of metagenomic libraries. However, metagenome isolation from a chemically complex, polluted environment represents a challenging task because all the interfering compounds from the inhibitory contaminants. Metagenomic libraries are important because may contribute to the function-driven screening for an expressed trait, an aspect often based on heterologous expression of the desired trait and selection is dependent on successful expression of the functional gene at a detectable level [56]. Accuracy of the sequence-driven approach is also very much dependent on the reliability of the information available in various sequence databases. In practice, a combination of functiondriven and sequence-driven approaches may be the best solution for understanding a complex and dynamic microbial ecosystem of a site undergoing bioremediation treatment [54-56].
A biomarker represents a substance indicator of a biological state. In the case of a diseased condition, a structural and functional change occurs at the level of proteins or enzymes. The measurable changes between the normal and altered diseased condition (changes in genes and proteins expression profiles) may be considered a strategic approach looking for biomarkers [1]. Microarrays-RNA, DNA, protein or antibody, play an important role in analyzing a biomarker. Omics is an emerging and exciting area in the field of science. In the postgenome era, efforts are focused on biomarker discovery and the early diagnosis of different processes through the application of various omics technologies. Numerous promising developments have been elucidated using omics, transcriptomics, proteomics, meabolomics, peptidomics, glycomics, lipidomics, on biology tissue samples and body fluids in different pathologies.
The omics technologies that has driven these new areas of research consist of DNA and protein microarrays, mass spectrometry and a number of instruments that enable high-throughput analysis of the relative low-cost (Figures 1 and 3).
Until now, and according to the ´The Great Plate Count Anomaly’, most marine bacteria have not yet discovered neither cultured, mainly their uniqueness in genes and metabolites of the biomes living in such environments [57]. This has imposed a limit on our present capabilities to develop biotechnological processes. Using whole genome shotgun sequencing´ of 16SrDNA of water samples, Venter et al identified nearly 1800 genomic sequences of bacteria in Sargasso Sea waters, of which 148 phylotypes were presumably new taxa [58]. Complementarily, the ship based pyrosequencing of the environmental metagenome has the potential of accessing almost all the genes in a given environmental sample, from which one may select the useful ones [59]. In this sense, metagenomics is particularly helpful when useful genes are present in uncultured organisms. Ensuring access to novel organisms or their genes is fundamental for sustaining biotechnology. Creation of metagenome libraries of new organisms and environment samples will aid future researches in the discovery of useful genes and biosynthetic pathways, which in turn should lead to the discovery of new and novel biomarkers. Active and orientated applications need to be actively sought (Figure 3).
A practical biomarker should satisfy a number of criteria [4], as described downlines. 1. The biomarker response should be sensitive enough to detect early stage of the process of toxicity; 2. The biomarker should be specific to a particular contaminant or for a class of contaminants; 3. The biomarker should respond in a concentration dependent manner to change in ambient levels of the contaminant; and 4. Identification of the non-toxicological variability identified in particular variations linked to biotic factors.
Methods to rapidly screen or select clones of interest from the thousands in a standard metagenome library are required to bring this technology to the average research laboratory lacking high-throughput infrastructure [60]. The key to this approach will be the integration of gene expression, proteomics, and metabolic data into working cellular models that can accurately predict the response of the organism to a given environment [61].
The advent of recent development of DNA microarrays has enabled researchers simultaneously analyze thousands of phylogenetic or functional genes in order to characterize microbial communities involved in biotechnology and bioremediation.
Conclusion
The increasing performances of next generation sequencing and metadata analyzes on engineered systems especially for complex aquatic systems and pollutants, together with meta-omicstechnologies could be exploited simultaneously to develop stable and reliable biomarkers of sustainable energy and resource recovery. The study of most of the omics methods simultaneously to particular features query of microbiomes under different conditions are promising issues of future of applying ´Omics approaches to optimize metabolic networks of classical and new biomarkers. It is expected that a suite of biomarkers should be confirmed in order to corroborate the sentinel results for a more precise verdict of the nature of the toxicological effects or mitigation measures. The progress in the development of proteomics by examining changes in the protein expression of hundreds to thousands of genes by organisms exposed to multiple stressors may supply new biomarker based technology that might permit diagnosis of complex stressors based on key proteins.
References
- Rapley R, Whitehouse D (2015) Molecular Biology and Biotechnology. Royal Society of Chemistry, London.
- Sarkar A, Ray D, Shrivastava AN, Sarker S (2006) Molecular Biomarkers: their significance and application in marine pollution monitoring. Ecotoxicology 15: 333-340.
- Sarkar A (2006) Biomarkers of marine pollution and bioremediation. Ecotoxicology 15: 331-332.
- Hamza-Chaffai A (2014) Usefulness of Bioindicators and Biomarkers in Pollution Biomonitoring. International Journal of Biotechnology for Wellness Industries 3: 19-26.
- de la Torre FR, Ferrari L, Salibia´n A (2005) Biomarkers of a native fish species (Cnesterodondecemmaculatus) application to the water toxicity assessment of a peri-urban polluted river of Argentina. Chemosphere 59: 577-583.
- Bodin N, Burgeot T, Stanisie`re JY, Bocquene´ G, Menard D, et al. (2004) Seasonal variations of a battery of biomarkers and physiological indices for the mussel Mytilusgalloprovincialistransplanted into the northwest Mediterranean Sea. CompBiochemPhysiol C ToxicolPharmacol 138: 411-427.
- Magni P, De Falco G, Falugi C, Franzoni M, Monteverde M, et al. (2005) Genotoxicity biomarkers and acetylcholinesterase activity in natural populations of Mytilusgalloprovincialis along a pollution gradient in the Gulf of Oristano (Sardinia, western Mediterranean) Environ. Pollut. 142: 65-72.
- Binelli A, Ricciardi F, Riva C, Provini A (2006) New evidences for old biomarkers: effects of several xenobiotics on EROD and AChE activities in Zebra mussel (Dreissenapolymorpha). Chemosphere 62: 510-519.
- Behrens A, Segner H (2005) Cytochrome P4501A induction in brown trout exposed to small streams of an urbanised area: results of a five-year-study. Environ Pollut 136: 231-242.
- Everaarts JM, Sarkar A (1996) DNA damage as a biomarker of marine pollution: strand breaks in sea stars (Asteriasrubens) from the North Sea. WatSciTechnol 34: 157-162.
- Amiard JC, Cosson RP (1997) Les métallothionéins: Biomarqueurs en Ecotoxicologie, Aspects Fondamantaux. Masson editors, Paris.
- Roesijadi G (2000) Metal transfer as a mechanism for metallothionein-mediated metal detoxification. Cell MolBiol (Noisy-le-grand) 46: 393-405.
- Valavanidis A, Vlahogianni T, Dassenakis M, Scoullos M (2006) Molecular biomarkers of oxidative stress in aquatic organisms in relation to toxic environmental pollutants. Ecotoxicol Environ Saf 64: 178-189.
- Andrews GK (2000) Regulation of metallothionein gene expression by oxidative stress and metal ions. BiochemPharmacol 59: 95-104.
- Kalpaxis DL, Theos C, Xaplanteri MA, Dinos GP, Catsiki AV, et al. (2003) Biomonitoring of Gulf of Patras, N. Peloponnesus, Greece. Application of a biomarker suite including Evaluation of translation efficiency in Mytilusgalloprovincialis cells. Environ Res 37: 1-8.
- Monserrat JM, Geracitano LA, Bianchini A (2003) Current and future perspectives using biomarkers to assess pollution in aquatic ecosystems. Commun. Toxicol. 9: 255-269.
- Coyle P, Philcox JC, Carey LC, Rofe AM (2002) Metallothionein: the multipurpose protein. Cell Mol Life Sci 59: 627-647.
- Roeva NN, Sidorov AV, Yurovitskii YG (1999) Metallothioneins, proteins binding heavy metals in fish. Biol Bull 26: 617-622.
- Isani G, Andreani G, Kindt M, Carpenè E (2000) Metallothioneins (MTs) in marine molluscs. Cell MolBiol (Noisy-le-grand) 46: 311-330.
- Roesijadi G, Hansen KM, Unger ME (1997) Concentration–response relationships for Cd, Cu, and Zn and metallothionein mRNA induction in larvae of Crassostreavirginica. CompBiochemPhysiol C PharmacolToxicolEndocrinol 118: 267-270.
- Petrovic S, Ozretić B, Krajnović-Ozretić M, Bobinac D (2001) Lysosomal membrane stability and metallothioneins in digestive gland of Mussels (MytilusgalloprovincialisLam.) as biomarkers in a field study. Mar Pollut Bull 42: 1373-1378.
- Lionetto MG, Caricato R, Giordano ME, Pascariello MF, Marinosci L, et al. (2003) Integrated use of biomarkers (acetylcholinesterase and antioxidant enzymes activities) in Mytilusgalloprovincialis and Mullusbarbatus in an Italian coastal marine area. Mar Pollut Bull 46: 324-330.
- JEFFREY SW (1961) Paper-chromatographic separation of chlorophylls and carotenoids from marine algae. Biochem J 80: 336-342.
- Jeffrey SW,Vesk M (1997) Introduction to marine phytoplankton and their pigment signatures: Phytoplankton pigments in oceanography. UNESCO publisher, Paris.
- Paniagua-Michel J, and Subramanian V (2016)Omics-based Advances in biosynthetic pathways of isprenoids production in microalgae: Marine Omics: Principles and Applicatiòns. CRC Press, Boca Raton, Fl.
- Leavitt PR, Hodgson DA (2001) Sedimentary pigments: Developments in paleoenvironmental research, Tracking environmental changes using lake sediments: terrestrial algal and siliceous indicators. Kluwer Academic Publishers, Dordrecht.
- Adedeji OB, Okerentugba PO, Okonko IO (2012) Use of molecular, biochemical and cellular biomarkers in monitoring environmental and aquatic pollution. Nature and Science 10: 83-104.
- Tamm M, Freiberg R1, Tõnno I1, Nõges P1, Nõges T1 (2015) Pigment-based chemotaxonomy--a quick alternative to determine algal assemblages in large shallow eutrophic lake? PLoS One 10: e0122526.
- Skouras A, Broeg K, Dizer H, von Westernhagen H, Hansen PD, et al. (2003) The use of innate immune responses as biomarkers in a programme of integrated biological effects monitoring on flounder (Platichthysflesus) from the southern North Sea. Helgol Mar Res 57: 190-198.
- Bols NC1, Brubacher JL, Ganassin RC, Lee LE (2001) Ecotoxicology and innate immunity in fish. Dev Comp Immunol 25: 853-873.
- Vutukuru SS (2005) Acute effects of hexavalent chromium on survival, oxygen consumption hematological parameters and some biochemical profiles of the Indian major carp, Labeorohita. Int J Environ Res Public Health 2: 456-462.
- Azmat R, Farha A, Madiha Y (2008) Monitoring the Effect of Water Pollution on four Bioindicators of Aquatic Resources of Sindh Pakistan. Research Journal of Environmental Sciences 2: 465-473.
- Livingstone DR, Chipman JK, Lowe DM, Minier C, Pipe RK (2000) Development of biomarkers to detect the effects of organic pollution on aquatic invertebrates: recent molecular, genotoxic, cellular and immunological studies on the common mussel (MytilusedulisL.). International Journal of Environment and Pollution 13: 56-91.
- vanSchooten FJ, Maas LM, Moonen EJ, Kleinjans JC, van der Oost R (1995) DNA dosimetry in biological indicator species living on PAH-contaminated soils and sediments. Ecotoxicol Environ Saf 30: 171-179.
- Deutschbauer AM, Chivian D, Arkin AP (2006) Genomics for environmental microbiology. CurrOpinBiotechnol 17: 229-235.
- Tringe SG, Hugenholtz P (2008) A renaissance for the pioneering 16S rRNA gene. CurrOpinMicrobiol 11: 442-446.
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, et al. (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66-74.
- Gude J (2015) A New Perspective on Microbiome and Resource Management in wastewater Systems. J BiotechnolBiomater 5: 1-7.
- Hess M, Sczyrba A, Egan R, Kim TW, Chokhawala H, et al. (2011) Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331: 463-467.
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, et al. (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. ProcNatlAcadSci U S A 108 Suppl 1: 4516-4522.
- Pedrós-Alió C (2007) Ecology. Dipping into the rare biosphere. Science 315: 192-193.
- Olmos Soto J (2009) Caracterización molecular de bacteriaspatógenas de organismosmarinoscultivados: BiotecnologíaMarina. AGT Editores, Mexico.
- Konstantinidis KT, Tiedje JM (2005) Genomic insights that advance the species definition for prokaryotes. ProcNatlAcadSci U S A 102: 2567-2572.
- Hamady M, Knight R (2009) Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res 19: 1141-1152.
- Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, et al. (2005) Comparative metagenomics of microbial communities. Science 308: 554-557.
- Morris RM, Rappé MS, Connon SA, Vergin KL, Siebold WA, et al. (2002) SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420: 806-810.
- Dennis P, Edwards EA, Liss SN, Fulthorpe R (2003) Monitoring gene expression in mixed microbial communities by using DNA microarrays. Appl Environ Microbiol 69: 769-778.
- Rhee SK, Liu X, Wu L, Chong SC, Wan X, et al. (2004) Detection of genes involved in biodegradation and biotransformation in microbial communities by using 50-mer oligonucleotide microarrays. Appl Environ Microbiol 70: 4303-4317.
- Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, et al. (2002) A new phylum of Archaea represented by a nanosizedhyperthermophilicsymbiont. Nature 417: 63-67.
- Baker BJ, Tyson GW, Webb RI, Flanagan J, Hugenholtz P, et al. (2006) Lineages of acidophilic archaea revealed by community genomic analysis. Science 314: 1933-1935.
- Isenbarger TA, Finney M, Ríos-Velázquez C, Handelsman J, Ruvkun G (2008) Miniprimer PCR, a new lens for viewing the microbial world. Appl Environ Microbiol 74: 840-849.
- Urich T, Lanzén A, Qi J, Huson DH, Schleper C, et al. (2008) Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One 3: e2527.
- Sana B (2015) Bioresources for control of environmental pollution. AdvBiochemEngBiotechnol 147: 137-183.
- Zhao B, Yeo CC, Poh CL (2005) Proteome investigation of the global regulatory role of sigma 54 in response to gentisate induction in Pseudomonas alcaligenes NCIMB 9867. Proteomics 5: 1868-1876.
- Zhao B, Yeo CC, Tan CL, Poh CL (2007) Proteome analysis of heat shock protein expression in Pseudomonas alcaligenes NCIMB 9867 in response to gentisate exposure and elevated growth temperature. BiotechnolBioeng 97: 506-514.
- Raghukumar S (2011) Marine Biotechnology: An approach based on components, levels and players. Indian Journal of Geo Marine Sciences 40: 609-619.
- Giovannoni SJ, Stingl U (2005) Molecular diversity and ecology of microbial plankton. Nature 437: 343-348.
- Hu ZZ, Hongzhan H, Wu CH, Jung M, Dritschilo A, et al. (2011) Omics-Based Molecular Target and Biomarker Identification. Methods MolBiol 719: 547-571.
- Isenbarger TA, Finney M, Ríos-Velázquez C, Handelsman J, Ruvkun G (2008) Miniprimer PCR, a new lens for viewing the microbial world. Appl Environ Microbiol 74: 840-849.
- Urich T, Lanzén A, Qi J, Huson DH, Schleper C, et al. (2008) Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One 3: e2527.
- Hu ZZ, Huang H, Wu CH, Jung M, Dritschilo A, et al. (2011) Omics-based molecular target and biomarker identification. Methods MolBiol 719: 547-571.
Relevant Topics
- Agricultural biotechnology
- Animal biotechnology
- Applied Biotechnology
- Biocatalysis
- Biofabrication
- Biomaterial implants
- Biomaterial-Based Drug Delivery Systems
- Bioprinting of Tissue Constructs
- Biotechnology applications
- Cardiovascular biomaterials
- CRISPR-Cas9 in Biotechnology
- Nano biotechnology
- Smart Biomaterials
- White/industrial biotechnology
Recommended Journals
Article Tools
Article Usage
- Total views: 14095
- [From(publication date):
March-2016 - Mar 31, 2025] - Breakdown by view type
- HTML page views : 13010
- PDF downloads : 1085