MicroRNA and Gene Expression Changes in Parkinson’s Disease Patients Blood Leukocytes: A Short Communication
Received: 02-Jun-2022 / Manuscript No. JADP-22-65683 / Editor assigned: 07-Jun-2022 / PreQC No. JADP-22-65683 (PQ) / Reviewed: 21-Jun-2022 / QC No. JADP-22-65683 / Revised: 28-Jun-2022 / Manuscript No. JADP-22-65683 (R) / Published Date: 05-Jul-2022 DOI: 10.4172/2161-0460.1000544
Abstract
Parkinson’s disease has increasing prevalence worldwide. Several genes were identified as involved in the disease so far (e.g., Park, SNCA, MAPT, PINK1 as well as several molecular pathways (e.g., alternative spicing, metal ion channels, inflammation) but there are still mainly unknown genes involved in the disease. I previously applied SOLID small and long RNA-Seq, junction and exon arrays on PD patient’s and matching control samples. Furthermore, model MPTP (1-methyl-4-phenyl-1, 2, 3, 6-tetrahydropyridine) mice studies found similar patterns in functional groups of genes (using Kolmogorov-Smirnov- KS statistical method. Subsequently, Gene Ontology (GO) functional analysis of Biological Processes (BP) and molecular functions. Additionally, PD RNA expression datasets can be compared to those from other neurodegenerative diseases including Alzheimer’s disease (AD) and Motor neuron disease. Also, stem cells from patients can be studied in parallel for RNA expression pattern changes through RNA-Seq or Cerebro Spinal Fluid (CSF).
Keywords:
Keywords
Blood; Mental disorders; Deep brain stimulation; Leucocytes; Parkinson’s disease; Microarrays; RNA-Seq
Abbrevations
DBS: Deep Brain Stimulation; BG: Basal Ganglia; STN: Sub Thalamic Nucleus; PD: Parkinson’s Disease.
Description
Parkinson’s disease (PD) is a devastative motor disease with no current early detection or effective treatment methods. It is a prevalent, multifaceted neurodegenerative disease caused by mostly unknown factors. However, there are many postulated pathophysiological hypotheses to explain it. The DA population of SNpc is affected by PD. By the time of disease diagnosis, the majority of brain dopaminergic cells have already died. RNA expression studies of patient’s blood leukocytes prior to and following Deep Brain Stimulation (DBS) treatment can reveal important molecular changes in patient’s gene expression patterns including of coding and non-coding RNAs. The data can be generated using microarrays or RNA-Seq (including single cell RNA-Seq). Here, we comment on our recent studies related to this subject highlighting the importance of future similar studies [1-7].
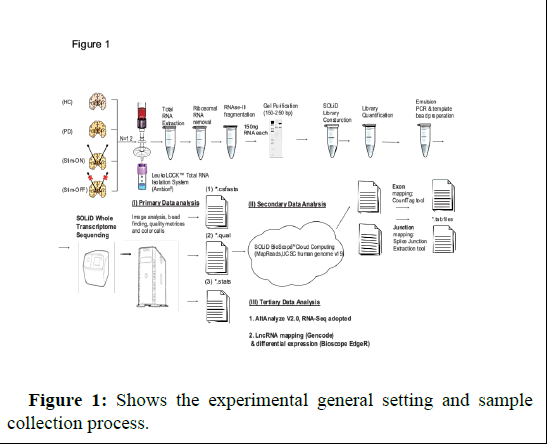
In my recent paper published in American journal of neurodegenerative diseases journal (AJND) this year [8], I reported significance changes found in miRNA genes in PD patient’s leukocytes proper to and following DBS detected by analysis of microRNA arrays expression data from 3 patients and 3 age and gender matching control volunteers. I isolated leukocytes from patients and control blood samples using LeukoLockTM filter and produce the RNA from each filter following -80 degrees storage. The study received approval by Hadassah ethics committee (approval number 6-07.09.07 code 2507). Each recruited patient signed informed consent. The age and gender matched healthy control volunteers were recruited to the study in Givat Ram campus (signed on informed consent as well). In a previous study I profiles using SOLID RNA-seq on blood leukocyte microRNAs in another set of PD patients prior to and following DBS and controls and quantified long non coding RNAs (lncRNAs) in patients and controls blood leucocytes samples [4]. My results may reveal significance genes altered in the disease and potential targets for future genomic intervention using CRISPR gene editing technique. Analysis of slow and rapid progressive PD patients’ microarray data also identified significance changes in specific genes and pathways [9]. Future larger studies will enable a better understanding of the disease underlying molecular pathways. I measured the RNA for quality using BioAnalyzer and received RIN values for each sample to assure samples integrity and quality (Figure 1).
Conclusion
RNA-Seq studies of PD patients’ blood leukocytes highlight the importance of disease studies in the hope for future identification of further genes and pathways involved in the disease. Future studies may enlarge the patient cohort, and apply single cell RNA-Seq and advanced computational/bioinformatic analyses to analyse the data.
Acknowledgement
I thank Prof. Hagai Bergman (the Hebrew university of Jerusalem Israel), Prof. Hermona Soreq (Hebrew university of Jerusalem Israel) and Dr. Zvi Israel (Hadassah faculty of medicine Israel) for supervision during my Ph.D.
Author Contributions
Lilach Soreq wrote and prepared the figure.
Conflict of Interest
None declared
References
- Pankratz N, Nichols WC, Uniacke SK, Halter C, Rudolph A et al., (2002) Genome screen to identify susceptibility genes for Parkinson disease in a sample without parkin mutations. Am J Hum Genet 71: 124-135.
[Crossref] [Google Scholar] [PubMed]
- Soreq L, Ben-Shaul Y, Israel Z, Bergman H, Soreq H (2012) Meta-analysis of genetic and environmental Parkinson's disease models reveals a common role of mitochondrial protection pathways. Neurobiol Dis 45: 1018-1030.
- Soreq L, Bergman H, Israel Z, Soreq H (2012) Exon arrays reveal alternative splicing aberrations in Parkinson's disease leukocytes. Neurodegener Dis 10: 203-206.
[Crossref] [Google Scholar] [PubMed]
- Pankratz N, Nichols WC, Uniacke SK, Halter C, Rudolph A et al., (2014) Long non-coding RNA and alternative splicing modulations in Parkinson's leukocytes identified by RNA sequencing. PLoS Comput Biol 10: e1003517.
[Crossref ][Google Scholar] [PubMed]
- Soreq L, Salomonis N, Guffanti A, Bergman H, Israel Z (2015) Whole transcriptome RNA sequencing data from blood leukocytes derived from Parkinson's disease patients prior to and following deep brain stimulation treatment. Genom Data 3: 57-60.
[Crossref ][Google Scholar] [PubMed]
- Soreq L, Salomonis N, Guffanti A, Bergman H, Israel Z., et al., (2015) Analyzing alternative splicing data of splice junction arrays from Parkinson patients' leukocytes before and after deep brain stimulation as compared with control donors. Genom Data 5: 340-343.
[Crossref] [Google Scholar] [PubMed]
- Benmoyal-Segal L, Soreq L, Ben-Shaul Y, Ben-Ari S, Ben-Moshe T, et al., (2012) Adaptive alternative splicing correlates with less environmental risk of parkinsonism. Neurodegener Dis 9: 87-98.
- Lilach S, Hagai B, Zvi I, Hermona S, Wael M (2021) MicroRNA expression changes in Parkinson's disease (PD) patients' leukocytes prior to and following deep brain stimulation (DBS). Am J Neurodegener Dis 10: 28-33.
[Google Scholar] [PubMed]
- Pinho R, Guedes LC, Soreq L, Lobo PP, Mestre T et al., (2016) Gene Expression Differences in Peripheral Blood of Parkinson's Disease Patients with Distinct Progression Profiles. PLoS One 11: e0157852.
[Crossref] [Google Scholar] [PubMed]
Citation: Soreq L (2022) MicroRNA and Gene Expression Changes in Parkinson’s Disease Patients Blood Leukocytes: A Short Communication. J Alzheimers Dis Parkinsonism. 12:544. DOI: 10.4172/2161-0460.1000544
Copyright: © 2022 Soreq L. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 1377
- [From(publication date): 0-2022 - Apr 02, 2025]
- Breakdown by view type
- HTML page views: 1026
- PDF downloads: 351