Research Article Open Access
Microfibrillar-Associated Protein 4 (MFAP4) Genes in Pelteobagrus fulvidraco play a Novel Role in Innate Immune Response
| Ming-Ming Han1,2, Le-Wang1, Li-Na-Peng1, Shahidd Mahboob3,4, Khalid A Al-Ghanim4, Jian-Guo Lu1* and Xiao-Wen Sun1* | |
| 1Heilongjiang River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Harbin, China | |
| 2Shanghai Ocean University, College of Fisheries and Life Science, Shanghai, China | |
| 3Department of Zoology, College of Science, King Saud University, Riyadh, Saudi Arabia | |
| 4Department of Zoology, GC University, Faisalabad, Pakistan | |
| Corresponding Authors : | Xiao-Wen Sun Heilongjiang River Fisheries Research Institute, Chinese Academy of Fishery Sciences 232 Hesong Road, Daoli District, Harbin 150070, China Tel: +86-451-84869349 Fax: +86-451-84604803 E-mail: sunxw2002@163.com |
| Jian-Guo Lu Heilongjiang River Fisheries Research Institute Chinese Academy of Fishery Sciences, 232 Hesong Road Daoli District, Harbin 15007, China Tel: +86- 451-84869349 Fax: +86-451-84604803 E-mail: jianguonk@gmail.com |
|
| Received: January 07, 2016; Accepted: February 05, 2016;Published: February 12, 2016 | |
| Citation: Han MM, Wang L, Peng LN, Mahboob S, Al-Ghanim KA, et al. (2016) Microfibrillar-Associated Protein 4 (MFAP4) Genes in Pelteobagrus fulvidraco play a Novel Role in Innate Immune Response. Biochem Physiol 5:198. doi:10.4172/2168-9652.1000198 | |
| Copyright: © 2016 Han MM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Visit for more related articles at Biochemistry & Physiology: Open Access
Abstract
Both mannose-binding lectins (MBLs) and ficolins recognize and bind carbohydrates in pathogens and activate complement leading to opsonization, leukocyte activation, and direct pathogen clean. While MBLs have been reported in many fish species, the function of ficolins had not been identified in Pelteobagrus fulvidraco, despite their importance in invertebrate and higher vertebrate innate immunity. Although microfibrillar-associated protein 4 (MFAP4) this protein has similar fibrinogen-like domain, has expression in fish, the role of it has not been dissected. We cloned three MFAP4 genes (1MFAP4, 2MFAP4 and 3MFAP4) from the yellow catfish, P. fulvidraco. The homology similarity of MFAP4 was 78% with Astyanax mexicanus of Cypriniformes. The yellow catfish MFAP4 transcripts expression analysis revealed difference of patterns of homeostatic expression among the genes in gill, blood, muscle, gonad, liver, brain, spleen, kidney, heart, intestine from male and female, respectively. Expression of the three MFAP4 transcripts showed significant expression changes in 4 h after infection with either Edwardsiella ictaluri or Flavobacterium columnare, which has modulation of expression continuing up to 24 h or 7 d following pathogen exposure. Several different tissues and gene-specific patterns were captured and transcript expression changes of >11.45-fold were observed over the course of the bacterial challenges. This information elucidates the functions of individual MFAP4 genes in regards to pathogen recognition, binding, and their larger immune context.
| Keywords |
| Pelteobagrus fulvidraco; MFAP4; Innate immune response |
| Introduction |
| Fibrinogen-related proteins (FREPs) are a family of glycoproteins that encode a fibrinogenlike (FBG) domain in the C-terminal end [1]. FREP members include fibrinogen/fibrin, ficolins, angiopoietin, tenascins, tachylectins, fibroleukin, FIBCD1, and microfibrillarassociated protein 4 [2]. It was found that large clusters of FREPs in invertebrate species would be induced tremendous coding diversity and immune responses by pathogen challenge [1,3]. Microfibrillarassociated protein 4 (MFAP4) is a multicellular glycoprotein that colocalizes with elastic fibers and is highly expressed in the spleen and gill [4]. MFAP4 is a collagen-binding molecule containing a C-terminal fibrinogen-like domain and an N-terminal located integrin-binding motif [4]. Structurally, MFAP4 possesses a characteristic FBG domain at the C-terminal end. In addition, MFAP4 has an integrin-binding motif with a single cysteine residue and an Arg-Gly-Asp (RGD) sequence located at the N-terminal region in mammals [4]. The RGD sequence motif is often associated with cell adhesion activity and is known to be a ligand motif for cell surface integrins [5]. |
| In previous research on the innate immune responses of blue catfish (Ictalurus furcatus), channel catfish (Ictalurus punctatus) and catfish (Silurus asotus) to a Gram-negative bacterium [6-9], significant upregulation of FREP family member microfibrillar-associated protein 4 (MFAP4) by microarray analysis were observed. However, little is known about MFAP4 function in fishes. |
| Yellow catfish widely distributed in the ocean of eastern China, and is an important food with economic value in nutrition, nourishing and ornamental. As the human population grows, the trends of large-scale fish culture show an increase in recent years, leading to the outbreak of fish diseases. The various diseases have become the main factors which restrict the development of yellow catfish breed. Yellow catfish P. fulvidraco MFAP4 genes and the distribution of mRNA expression have not been reported. The aims of this study were to: (1) clone the MFAP4 cDNA sequence of the P. fulvidraco; (2) compare its deduced amino acid sequence with those of other known MFAP4; (3) examine the mRNA expression of 1MFAP4, 2MFAP4 and 3MFAP4 in different tissues between male and female fish; (4) examine the three kind of MFAP4 genes changes in transcript expression after either Edwardsiella ictaluri or Flavobacterium columnare infection. Our results revealed the first functional characterization of MFAP4 genes in fish and suggest a novel role for MFAP4 in teleost immune responses. |
| Materials and Methods |
| RNA isolation |
| The P. fulvidraco were obtained from the Chinese Academy of Fishery Sciences Pelteobagrus sp. Breeding Engineering Center. All yellow catfish were 1 year old, 30.6 ± 2.7 g weight, 13.5 ± 3.4 cm length. Total RNA of the yellow catfish was extracted with Trizol reagent (Invitrogen, China) following the manufacturer’s instructions. The total RNA concentration was determined by measuring the absorbance at OD260. RNA integrity was checked by electrophoresis. The total RNA was reverse-transcribed into cDNA using the aM-MLV RTase cDNA Synthesis Kit (TaKaRa, Japan). |
| MFAP4 cloning |
| Every organization total RNA used cDNA reverse treasure article number for RR047A transcription kit and used minus 20 degrees to put on cDNA template. According to Ietalurus Punetaus in Genbank madtoms MFAP4 (serial number FD303964, FD309065A and FD290690), we designed the PCR primers 1MFAP4 - F1, 1MFAP4 - R1, 2MFAP4 - F1, 2MFAP4 - R1, 3MFAP4 - F1 and 3MFAP4 - R1 for MFAP4 cloning (Table 1). |
| PCR was conducted for 5 min at 94ºC followed by 35 cycles at 94ºC for 30 s, 56ºC for 30 s, and 72ºC for 30 s. The final extension step was kept at 72ºC for 2 min. Each of the 20 μL reaction mixtures contained 1.5 mM of MgCl2, 0.2 mM of dNTP, 0.3 mM of relevant primers, 1 U of Taq DNA polymerase (TaKaRa, Japan), and 5 ng cDNA templates. Amplicons of expected sizes were purified using an Agarose Gel DNA Purification Kit (TaKaRa, Japan), and then subcloned into the pMD- 18T cloning vector (TaKaRa) connection at 16ºC for the night. Tiangen Top 10 into competent E. coli in LB Amp tablet on 37ºC, or 230 r/ min, Pick the bacteria , expand training and gas-bacilli PCR detection, expand training and gas-bacilli PCR detection. PCR products were 0.5% gel electrophoresis detection of target DNA in bacteria. Positive clones containing inserts of an expected size were sequenced using M13 primers and sequenced at BGI (Shanghai, China). The sequencing results on NCBI homology retrieval. |
| Sequence analysis of MFAP4 |
| Deduced amino acid sequences of MFAP4 cDNA were analyzed using the software Vector NTI Advance 11. MFAP4 sequences from different organisms were obtained using the NCBI BLAST search program. The cDNA sequence and deduced amino acid sequence from Yellow catfish were analyzed using the BLAST algorithm (www.ncbi. nlm.nih.gov/blast). Translation and protein analyses were performed using ExPASy tools (us.expasy.org/tools/). A multiple sequence alignment was created with ClustalW (www.ebi.ac.uk/clustalw/). Protein sequences retrieved from the public database were used for ORF and domain searches; alignment; and phylogenetic reconstruction. ORFs were predicted using Open Reading Frame Finder (www.ncbi. nlm.nih.gov/gorf/gorf.html), and signal peptides and fibrinogen-like domain (FBG) were identified by the NCBI conserved domain feature of blastp (www.ncbi.nlm.nih.gov/BLAST) and by the Simple Modular Architecture Research Tool (SMART) (smart.emblheidelberg.de). |
| Phylogenetic analysis |
| MFAP4 gene cloning CDS coding regions is derived using the Primer 5 software is deduced amino acid sequence, excerpts from GenBank (number for KT778802, KT778805 and KT318368 GenBank landed). Sequences of MFAP4: microfibrillar-associated protein 4: angiopoietin of Astyanax mexicanus, Danio rerio, Oreochromis niloticus, Haplochromis burtoni, Perca flavescens, Stegastes partitus, Maylandia zebra, Fundulus heteroclitus, Poecilia formosa, Xiphophorus maculatus and Oryzias latipes retrieved from databases were aligned using the ClustalW2 program (www.ebi.ac.uk/Tools/clustalw2/). The complete list of species, gene names and accession numbers are listed in Supplementary Figure 1. Phylogenetic trees were constructed using the neighbour-joining (NJ) method based on the deduced full-length amino acid sequences with 1,000 bootstrapping replications within the Molecular Evolutionary Genetics Analysis (MEGA 4.0) package [10]. |
| Tissue expression of MFAP4 genes |
| MFAP4 gene in accordance with full design relative fluorescence quantitative PCR primers for 1MFAP4-F5, 1MFAP4-R5, 2MFAP4 - F5, 2MFAP4 - R5, 3MFAP4 - F4 and 3MFAP4 - R4 is shown in Table 1.It can enter the yellow catfish beta actin genes for internal reference. Primers respectively has actins-F and actins-R. The length of amplified fragment is 200 pb. RT-PCR reverses transcription using the reverse transcription kit ABI. The PCR reaction was performed in a 25 lL volume with a SYBR Premix Ex TaqTM Kit (TaKaRa, Japan). Experiment according to the instructions, the reaction fluorescence quantitative instrument (ABI 7500) on the conduct for analysis using 2 μM of each specific primer, and 1 L of cDNA using the following procedure: initial denaturation at 95ºC for 2 min followed by 40 cycles of amplification (95ºC for 10 s and 55ºC for 30 s). Experimental analysis need to â�?³â�?³CT. Relative quantitative calculation method: relative quantitative is equal to 2- â�?³â�?³CT multiplied by 100%. To determine MFAP4 gene expression in various healthy male and female catfish tissues, samples of 10 tissues including gonads, brain, liver, kidney, spleen, intestine, blood, gills, muscle and heart from control channel catfish were isolated, pooled, and flash-frozen in liquid nitrogen. Tissues were homogenized under liquid nitrogen using a mortar and pestle, and stored at −80ºC until RNA extraction. |
| E. ictaluri and F. columnare challenges |
| In the first challenge, the P. fulvidraco, with an average body weight of 30.6 g and average body length of 13.5 cm, kept at 26ºC in a flowthrough system utilizing heated. Sterile water was used for the challenge. Fish were treated in each of two groups: (1) control group (phosphate buffered saline, 100 μL PBS injected); (2) E. ictaluri challenged group (injection). To inoculate bacteria for the challenge, a single colony of E. ictaluri was isolated and cultured in BHI broth at 27ºC overnight. The bacterial culture was diluted with PBS (pH 7.4), then 1 × 105 CFU of bacteria in 100 μL PBS were injected intraperitoneally into the channel yellow catfish. |
| In the second challenge, health P. fulvidraco with a mean weight 30.6 g were used. The fish were divided into two groups: (1) control group (without bacteria); (2) F. columnare challenge group (immersed with the dose of 3 ml of 107 CFU ml−1 of F. Columnare for 1 h). To inoculate bacteria for the challenge, a single colony of F. columnare was isolated and cultured in BHI broth at 28ºC overnight. The spleen and gill tissues from 60 fish (three pools of 15 fish each) at 4 h, 24 h, 3 d and 7 d after E. ictaluri treatment and from 60 fish (three pools of 15 fish each) at 4 h, 24 h and 48 h after F. columnare treatment were isolated for RNA extraction, respectively. Corresponding uninfected control samples were taken at each time interval. |
| Results |
| Phylogenetic analysis of gene MFAP4 |
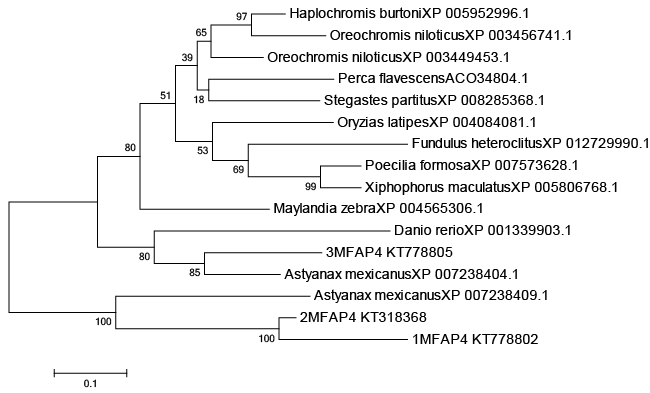
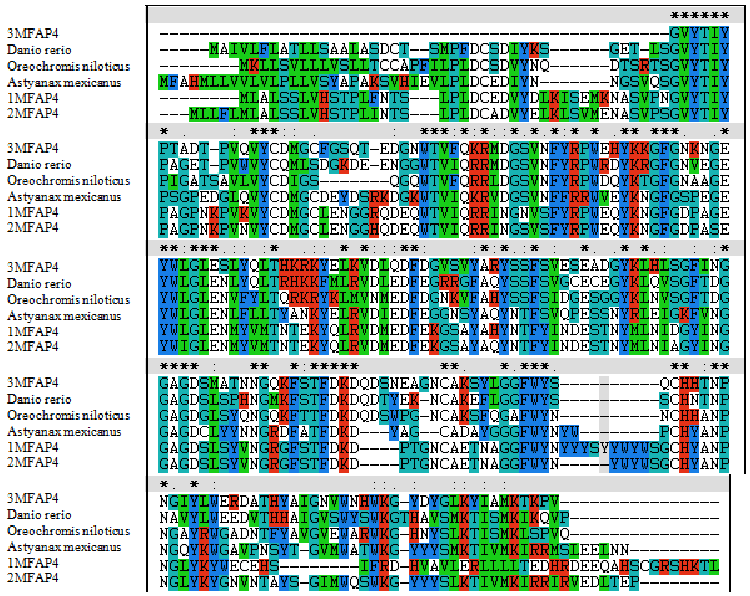
| A phylogenetic tree was constructed based on the amino acid sequences of selected MFAP4 using the neighbour-joining (NJ) method (Figures 1 and 2). Data were analyzed using Poisson’s correction, and gaps were removed by complete deletion. The results showed that the molecular MFAP4 and the corresponding species exhibited similar evolutionary relationships. The homology similarity of MFAP4 (1MFAP4, 2MFAP4 and 3MFAP4) was 78% compared with Astyanax mexicanus of Cypriniformes (Figure 1). The homology similarity of 1MFAP4 was 92% compared with 2MFAP4. The homology similarity of 1MFAP4 was 69% compared with 3MFAP4. |
| Tissue expression of MFAP4 genes |
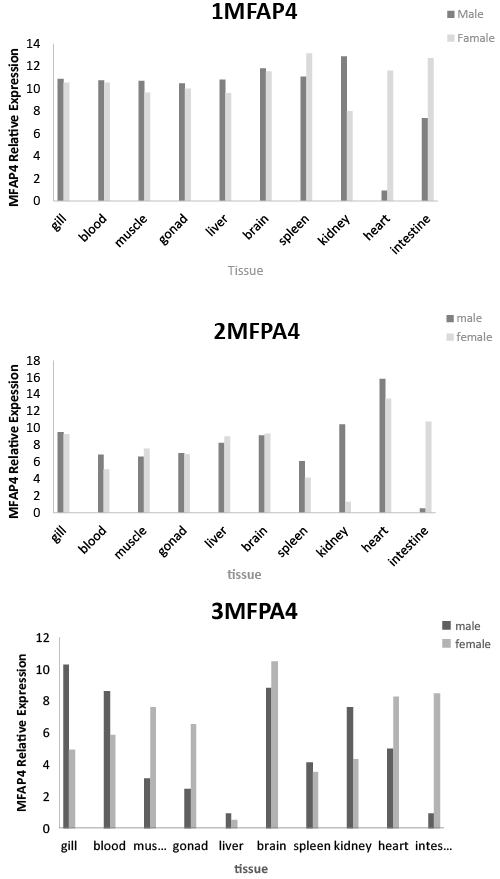
| Quantitative real-time PCR was performed to determine the tissue distribution of MFAP4 gene expression in both male and female P. fulvidraco. MFAP4 genes were widely expressed in all 11 tissues tested (gonads, brain, liver, kidney, spleen, intestine, blood, gills, muscle and heart). 1MFAP4, 2MFAP4 and 3MFAP4 gene expression levels in male kidney were higher than those in female. 1MFAP4, 2MFAP4 and 3MFAP4 gene expression levels in female spleen and intestine were higher than those in males. 1MFAP4 and 3MFAP4 gene expression levels in female heart were higher than those in males. 3MFAP4 gene expression level in male gill and blood were higher than those in females. 3MFAP4 gene expression level in female muscle and gonads were higher than those in males. MFAP4 gene expression level in the brain and gill of both male and female P. fulvidraco were highest (Figure 3). |
| Expression of MFAP4 after infection of E. ictaluri |
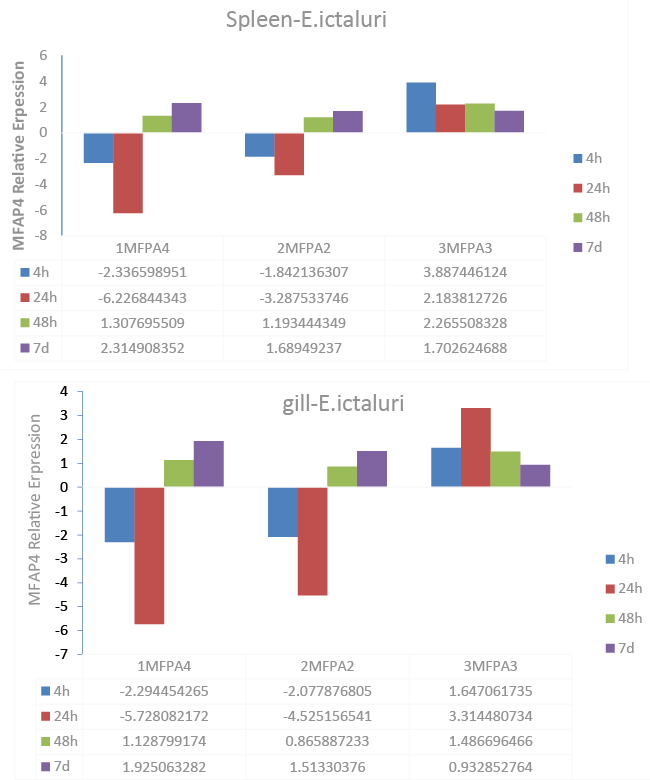
| In order to investigate whether MFAP4 genes are involved in responses to disease infection with the catfish pathogen and Gram negative intracellular bacterium E. ictaluri, qRT-PCR analysis was performed to determine the expression patterns of MFAP4 genes in infected spleen and gill tissues (Figure 4). In spleen, 1MFAP4 and 2MFAP4 genes exhibited highly similar expression patterns, as in the cases of expression in uninfected tissues. Transcript expression of these genes was suppressed initially after infection at 4 h and 24 h after infection, but up-regulated at 7 d. Strongest suppression (>5-fold) was observed at 24 h. The 3MFAP4 gene was significantly up-regulated, especially at 4 h after infection (Figure 4). Similar transcript expression patterns of the three catfish MFAP4 genes were observed in gill tissue. 1MFAP4 and 2MFAP4 expression was significantly up-regulated at 7 d after infection. Significant suppression of gene expression was observed in 1MFAP4 and 2MFAP4 at 24 h in gill (Figure 4). MFAP4 expression was expressed as fold change over control samples taken at the same time interval as normalized to change in expression in the 18S control. The bars indicate mean expression of 3 tested pools (8 fish each) ± SE. |
| Expression of MFAP4 after infection of F. columnare |
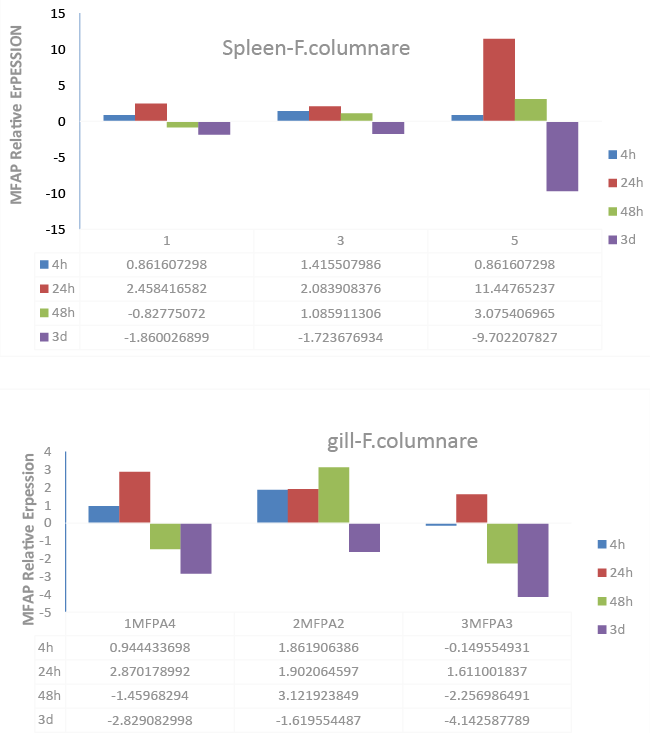
| The transcript expression levels of the three MFAP4 genes after bacterial infection with F. columnare in spleen and gill tissues (Figure 5) was also examined using qRT-PCR. All MFAP4 genes were significantly up-regulated at 24 h after columnaris infection in spleen tissue. 3MFAP4 showed the most sensitive response to infection, increasing expression 11.45-fold at 24 h, before suppressing expression -9.70-fold at 3d (Figure 5, Spleen-F. columnare). In gill, 1MFAP4 and 2MFAP4 were significantly up-regulated at 4 h after infection, while 3MFAP4 showed the largest expression changes over the course of the infection (Figure 5). |
| Discussion |
| A remarkable array of innate immune strategies has been revealed in recent years by large-scale sequencing in invertebrate and primitive vertebrate species [1]. Tracing the maintenance, loss and/or adaptation of these strategies through vertebrate evolution can highlight innate immune elements that may be overlooked in single species studies. Study of the FREP family in yellow catfish has highlighted a littlestudied MFAP4 protein. Gene MFAP4 has a putative role domain and a conserved fibrinogen-like in the first line of immune defense. In this study, we have identified three yellow catfish P. fulvidraco MFAP4 genes. The copies of MFAP4 genes likely exist in the catfish genome, and they may exist in clusters of tandem duplicates. MFAP4 genes were similar to FREPs reported in invertebrate species. Transcript expression of P. fulvidraco three MFAP4 genes was modulated through exposure to two common bacterial pathogens F. columnare and E. ictalur. Analysis of transcript expression in healthy and infected tissues indicated subfunctionalization of the three P. fulvidraco MFAP4 genes in tissue specificity, pathogen response and potential coordinated expression between groups of P. fulvidraco MFAP4 genes. |
| MFAP4 is extracellular matrix proteins that interact with fibrillin to influence microfibril function. The microfibril-associated proteins include MFAP4 are related with fibrillin through a 60 amino acid matrixbinding domain, but their sequences differ outside of this region. The lectin pathway of complement system is characterized by two groups of soluble pattern recognition molecules, MBL and ficolins. These molecules recognize and bind carbohydrates in pathogens and activate supplement leading to opsonization, direct pathogen killing, and leukocyte activation. MBL is expressed almost exclusively in the liver of healthy catfish [11] and has been described as a pattern recognition receptor and an activity factor in the complement system [12-15]. MBL in rainbow trout is expressed exclusively in liver and spleen, respectively [16]. Ficolins and fibrillin serve as pathogen recognition molecules, activating the complement cascade that is pivotal many innate immune strategies [9,17]. Fibrillin latent transforming growth factor competed with the mannose binding to ManLAM [18]. MFAP4 can interact with fibrillin in dermal tissue, suggested that there is skin homeostasis during skin bleaching effect [19]. The most important is that, MFAP-4 plays essential roles not only in microfibril development but also in the maintenance of ECM proteins, including collagen I [19]. Some researchers used bind ficolin to up-regulate MFAP4 in catfish following infection [6-8,20]. MFAP4 and MBLs contact with fibrillin, of innate immune strategies system has certain help by an as yet undisclosed mechanism(s). |
| In conclusion, MFAP4 is a member of the functionally important FREP family, little is known about its role in vertebrate immunity. Yellow catfish P. fulvidraco, apparently lacking ficolin genes, have variable numbers of MFAP4 genes which may mediate similar roles during the innate immune response. We have cloned three MFAP4 genes (1MFAP4, 2MFAP4 and 3MFAP4) in P. fulvidraco. The deduced amino acid sequence showed considerable conservation when compared with those from other species. The mRNA encoding MFAP4 was predominantly produced in the liver, spleen and gill. Rapid and sensitive responses of P. fulvidraco MFAP4 genes to infection with two bacterial pathogens F. columnare and E. ictalur, coordination model between the expressions of gene subset. MFAP-4 plays an essential role in the maintenance of ECM proteins. Yellow catfish may be as a model organism to further investigation the ECM protein action mechanism, associated with aortic aneurysms, cirrhotic sickness and pulmonary hypertension. These informations will help us elucidate the functions of individual MFAP4 genes in regards to pathogen recognition, binding, and their larger immune context. |
| Acknowledgement |
| The research was funded and supported by grants from National Science Foundation of China (31302177), and Heilongjiang River Fisheries Research Institute Foundation (HSY201301). |
References
|
Tables and Figures at a glance
| Table 1 |
Figures at a glance
 |
 |
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 | Figure 5 |
Relevant Topics
- Analytical Biochemistry
- Applied Biochemistry
- Carbohydrate Biochemistry
- Cellular Biochemistry
- Clinical_Biochemistry
- Comparative Biochemistry
- Environmental Biochemistry
- Forensic Biochemistry
- Lipid Biochemistry
- Medical_Biochemistry
- Metabolomics
- Nutritional Biochemistry
- Pesticide Biochemistry
- Process Biochemistry
- Protein_Biochemistry
- Single-Cell Biochemistry
- Soil_Biochemistry
Recommended Journals
- Biosensor Journals
- Cellular Biology Journal
- Journal of Biochemistry and Microbial Toxicology
- Journal of Biochemistry and Cell Biology
- Journal of Biological and Medical Sciences
- Journal of Cell Biology & Immunology
- Journal of Cellular and Molecular Pharmacology
- Journal of Chemical Biology & Therapeutics
- Journal of Phytochemicistry And Biochemistry
Article Tools
Article Usage
- Total views: 9460
- [From(publication date):
March-2016 - Sep 18, 2024] - Breakdown by view type
- HTML page views : 8774
- PDF downloads : 686
