Research Article Open Access
Microarray Analysis of Gene Expression in Rams Experimentally-Infected with the Virulent Strain of Brucella ovis
João Marcelo Azevedo de Paula Antunes1, Susan Dora Allendorf2, Camila Michele Appolinário2, Marina Gea Peres2, Acácia Ferreira Vicente2, Didier Quevedo Cagnini2, Larissa de Castro Demoner2, Paula Ripamonte Figueiredo3, José Buratini Júnior3, Ruth Cecilia Galindo4, Katherine M Kocan5, José de La Fuente4,5 and Jane Megid1*1Universidade Federal Rural do Semi-Árido - UFERSA, Mossoró, RN, Brazil
2Faculdade de Medicina Veterinária e Zootecnia, Botucatu, UNESP - Univ Estadual Paulista, Botucatu, São Paulo, CEP 18618-000, Brazil
3Departamento de Fisiologia, Instituto de Biociências, Univ Estadual Paulista, Botucatu, São Paulo, CEP 18618-000, Brazil
4SaBio. Instituto de Investigación en Recursos Cinegéticos, IREC (CSICUCLM- JCCM), 13005 Ciudad Real, Spain
5Department of Veterinary Pathobiology, Center for Veterinary Health Sciences, Oklahoma State University, Stillwater, OK, 74078, USA
- Corresponding Author:
- Jane Megid
Departamento de Higiene Veterinária e Saúde Pública (Prédio 1)
Faculdade de Medicina Veterinária e Zootecnia-FMVZ
Universidade Estadual Paulista “Júlio de Mesquita Filho”-UNESP, Botucatu
São Paulo, Brazil, CEP-18.618-970.
Tel: +55 14 3811 6270
Fax: +55 14 3811 6270
E-mail: jane@fmvz.unesp.br
Received date: August 20, 2015; Accepted date: October 09, 2015; Published date: October 15, 2015
Citation: de Paula Antunes JMA, Allendorf SD, Appolinário CM, Peres MG, Vicente AF, et al. (2015) Microarray Analysis of Gene Expression in Rams Experimentally- Infected with the Virulent Strain of Brucella ovis. J Biotechnol Biomater 5:203. doi:10.4172/2155-952X.1000203
Copyright: © 2015 de Paula Antunes JMA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Biotechnology & Biomaterials
Abstract
Microarray analysis of gene expression profiles of total mRNA was studied in rams experimentally-infected with the virulent strain of Brucella ovis (B. ovis). The tissues studied included reproductive organs (epididymus, testicles, ampolae, vesicular glands, bulbourethral glands) and a pool of lymph nodes (inguinal and scrotal). The microarray analysis of each tissue was done at three time points: acute infection (60 days post infection [dpi]), chronic infection phase I (120 dpi), and chronic infection phase II (240 dpi). The gene expression profiles associated to B. ovis infection were determined using the Affymetrix Bovine Genome Array and expression levels of infected and uninfected rams (control group, 0 dpi) were compared. Of the 23,000 genes analyzed in the microarrays, 139 during acute phase of infection, 930 during the chronic phase I and 744 genes from the chronic phase II were identified as Differentially Expressed Genes (DEGs). Thirty known genes and fourteen unknown genes were expressed during the three phases of infection in all tissues. The biological functions of these genes included immune cell trafficking, immunological disease, infectious disease, inflammatory disease, inflammatory response and cellular movement, and significant differences were observed at the three phases of infection and in all infected tissues. For the first analysis, 8 genes in common during the three time points and to all infected tissues were chosen for the microarray validation. Acute phase of infection supported the relevance of the ataxia telangiectasia-mutated ATMIN (ATM interactor) gene. RCHY1 (ring finger and CHY zinc finger domain containing 1), ANKFY1 (ankyrin repeat and FYVE domain containing 1) and EFEMP1 (EGF-containing fibulin-like extracellular matrix protein 1) were also highlighted by the analysis during chronic phase I of infection. ZRAB2 (Zinc finger, RAN-binding domain containing 2) was validated at chronic phase II. This study represents the first analysis of microarray gene expression in different tissues of rams infected with of B. ovis at various times during infection and many of the genes identified require further study. These results expand the knowledge of the pathogenesis of this B. ovis strain infection in rams and suggest new genes and pathways for further investigation.
Keywords
Brucella ovis; Brucellosis; Microarray; Sheep; Rams; Gene expression
Introduction
Brucella ovis is the causative agent of ovine brucellosis or contagious epididymitis, a disease mainly characterized by infertility in rams [1]. Brucella spp. replicate and persist within macrophages in host tissues, including male and female reproductive tissues [2]. Infection of these tissues with B. ovis results in a moderate inflammatory response, and virulence factors promote evasion of the immune response favoring persistent infection [3,4]. In rams at 30 dpi with B. ovis in the acute phase of infection, the host immune response was notable with an up-regulation of several cytokines in reproductive tissues. During the development of infection, cytokine gene expression levels decreased, providing evidence of immunosuppression and immune evasion which favoured persistence of chronic B. ovis infection [5,6]. Previous microarray studies of gene expression patterns were done up to 60 dpi in rams experimentally-infected with B. ovis using buffy coat cells [7], and the use of heterologous array hybridization (screening for gene expression in one species using a microarray developed for another species) was also done [7,8]. In this study, characterization of gene expression using total mRNA was done by microarray hybridization and real-time PCR (qRT-PCR) in multiple tissues collected over of eight months (240dpi) from rams experimentally-infected with B. ovis.
Experimental Procedures
Experimental infection
Twelve, 1 year-old Santa Inês rams, confirmed negative for brucellosis were selected for the study. In accordance with Marin et al. [7] serum samples tested negative by the AGID technique for ovine brucellosis. The rams were kept in an isolated pen and experimentallyinfected with the B. ovis PA strain (ATCC25840) [9] provided by Dr. Renato L. Santos (Universidade Federal de Minas Gerais, UFMG, Belo Horizonte, Minas Gerais, Brazil). Experiments were conducted with the approval of Ethical Committee of UNESP (Ethics committee protocol nr.: 69/2008). The B. bovis strain was cultured on blood agar base plates, and incubated for 48h at 37ºC and 10% CO2. Bacteria were then washed with 10mM phosphate buffered saline (PBS; pH 6.8), and the concentration was adjusted using a spectrophotometer (550nm). The rams were then infected with a total of 2.2 x 109 B. ovis CFU in 60 μl administered in two doses (30 μl administered conjunctivally and 30 μl intrapreputially). Uninfected (control) rams were inoculated in the same manner with sterile distilled water. Rams were then assigned four to groups of 3 rams each: (1) control (0 dpi), (2) acute phase (60 dpi) (3) chronic phase I (120 dpi) and (4) chronic phase II (240 dpi). From each group, samples of epididymus, testicles, ampolae, vesicular glands, bulbourethral glands, and a pool of lymph nodes (inguinal and scrotal) were collected at necropsy from each ram. Gene expression changes associated to B. ovis infection were determined using using microarray analysis and data from the infected and control groups were compared. All tissues were tested by PCR for B. ovis infection [10]. After 15 dpi, infection in rams was also confirmed by serology using the Agar Gel Immunodiffusion (AGID; Instituto de Pesquisas Veterinárias Desidério Finamor – IPVDF, Eldorado do Sul/RS, Brazil).
Microarray hybridization and analysis
Total RNA was isolated from tissue samples collected at 0, 60, 120, 240 dpi using Invisorb® Spin Tissue RNA Mini Kit (Invitek Gmbh, Germany) according to manufacturer’s instructions, and purified with the RNeasy Mini-kit (Qiagen, Mississauga, ON, Canada). Total RNA was quantified by absorbance at 260 nm. The quality of RNA preparations was confirmed by electrophoresis and the Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Only samples with a preserved rRNA ratio (28S/18S), no evidence of RNA degradation and values ≥ 8 using the Bioanalyzer were used for the microarray hybridization and qPCR studies. Expression measurements of 23,000 genes were performed with the Affymetrix GeneChip® Bovine Genome Array (Affymetrix, USA) used previously for microarray analysis of sheep tissues [11], and the RNA labeling and hybridization protocols were carried out according to manufacturer’s instructions. After array scanning, quality control was performed using the GCOS software (Affymetrix, USA) according to the manufacturer’s recommendations. The quality control also was verified using Expression Console (http:// www.affymetrix.com/browse/level_seven_software_products_only.jsp productId=131414&categoryId=35623#1_1). The unwanted variables (batch effects) were removed using ComBat methodology (http:// jlab.byu.edu/ComBat/Abstract.html) [12]. The IQR (Inter-Quartile- Range) filter available at R/Bioconductor [13] was then applied. Gene expression values were obtained using the three-step Robust Multiarray Average (RMA) pre-processing method, implemented in the Affy package from R/Bioconductor [13]. In order to select DEG’s the non-parametric RankProd method was used with a p-value ≤ 0.05 and adjusted for FDR (False Discovery Rate) [14,15], a method which uses hierarchical clustering (HCL) to group data with the Pearson correlation coefficient in order to calculate the distance and linkage between groups (available at http://acgt.cs.tau.ac.il/expander/index.html) [16]. In order to perform a functional annotation analysis of DEG’s, IPA (Ingenuity Pathway Analysis, http://www.ingenuity.com) tools were used, considering the default parameters: molecules per network=35; networks per analysis=25; and direct relationships between genes and the “Ingenuity Expert Information” data source, including the new option of “Ingenuity Expert Assist Findings” in which the information was manually reviewed and curated from full-text scientific publications. In this first microarray analysis of rams experimentally infected with B. ovis aimed to the comparison in each time points among the six infected reproductive tissues and the six controls tissue. Only repeated DEGs or commom genes to all infected tissues for each time points were recorded and used for functional analysis. The microarray data were deposited in the NCBI Gene Expression Omnibus (GEO) under the platform accession number GSE35615.
Validation of microarray expression using real-time qRTPCR
The total RNA extracted from tissues of experimentally-infected rams collected at 0, 60, 120 and 240 dpi was analyzed by qRT-PCR using gene-specific primers and the iScript One-Step RT-PCR Kit with SYBR Green and an iQ5 thermal cycler (Bio-Rad, Hercules, CA, USA) following manufacturer’s recommendations. The mRNA levels were normalized against Ovis aries glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and the ratios between infected (60, 120 and 240 dpi) and uninfected (0 dpi) values were calculated. Data were normalized to a calibrator sample using the ΔΔCt method with correction for amplification efficiency [17]. The validation of microarray data was done by comparisons between DEGs. Comparison (1) acute vs chronic I, (2) chronic I vs chronic II, (3) acute vs chronic II and (4) comparison 1 vs comparison 3. To validate the microarray data, DEGs between the three stages of infection (comparison 4) and with the most significant biological functions (IPA tools) were chosen for analysis by qRT-PCR. The correlation between microarray and qRT-PCR assays for the infected and control rams was calculated using Spearman’s correlation test.
Results
All rams from infected groups were seropositive after 15 dpi, confirming the infection. During the experiment, tissues samples (epididymus, testicles, ampolae, vesicular glands, bulbourethral glands, and a pool of lymph nodes (inguinal and scrotal) from each infected group (time points: 30 dpi, 60 dpi, 120 dpi, 240 dpi) were collected after euthanasia and tested B. ovis PCR positive. All DEGs found in each time points should be understood as the result of repeated gene or gene in commom to all infected tissues at each period of infection.
Acute phase of infection (60 dpc)
The differentially expressed genes (DEGs) found at this time of infection were noted only genes that were repeated in all infected tissues. After microarray analysis at 60 dpi of the 23,000 genes, 139 repeated DEGs were found in all infected tissues. From the 139 DEGs in this group, 119 (≥ 1.3 fold change) were upregulated (83 known and 36 unknown genes) and 20 were downregulated (16 known and 4 unknown genes). The IPA tool (Ingenuity Pathway Analysis, http:// www.ingenuity.com) was subsequently used to characterize biological functions and main canonicals pathways of the 139 DEGs (Table 1). After the clustering analysis, the ram tissues were grouped according to their disease condition (Figure 1, Supplementary material). The IPA software demonstrated an activation of different cellular functions after the infection, the top functions included: infectious diseases, immunological disease, inflammatory disease, protein degradation, protein synthesis, cancer, gene expression, cell cycle, nucleic acid metabolism, small molecule biochemistry, cell signaling, DNA replication, recombination, and repair, and cell-to-cell signaling and interaction. The IPA tools also identified DEGs significantly associated with biological functions: immunological disease, inflammatory diseases, infectious diseases, tumor morphology and reproductive system disease.
| Canonical Pathwaysa | p-valueb |
|---|---|
| IL-3 Signaling | 1.98E00 |
| Production of Nitric Oxide and Reactive Oxygen Species in Macrophages | 1.86E00 |
| p53 (tumor protein 53 ) Signaling | 1.82E00 |
| Role of NFAT (nuclear factor of activated T cells) in Regulation of the Immune Response | 1.8E00 |
| T Cell Receptor Signaling | 1.73E00 |
| CD28 Signaling in T Helper Cells | 1.52E00 |
| EGF (Epidermal growth factor )Signaling | 1.42E00 |
| IL-2 Signaling | 1.29E00 |
| GM-CSF (Granulocyte-macrophage colony-stimulating factor ) Signaling | 1.21E00 |
| CD40 Signaling | 1.19E00 |
| Macropinocytosis Signaling | 1.15E00 |
| Regulation of IL-2 Expression in Activated and Anergic T Lymphocytes | 1.04E00 |
| CTLA4 (Cytotoxic T-Lymphocyte Antigen 4) Signaling in Cytotoxic T Lymphocytes | 1.03E00 |
| SAPK/JNK (stress-activated protein kinase/c-Jun NH2-terminal kinase ) Signaling | 1.02E00 |
| IL-12 Signaling and Production in Macrophages | 8.96E-01 |
| IGF-1 (insulin-like growth factors ) Signaling | 8.78E-01 |
| TNFR2 (Tumor necrosis facrorrecptor 2) Signaling | 7.65E-01 |
| IL-9 Signaling | 7.32E-01 |
| MIF (Macrophage migration inhibition factor ) Regulation of Innate Immunity | 7.02E-01 |
| B Cell Receptor Signaling | 6.89E-01 |
| B Cell Activating Factor Signaling | 6.62E-01 |
| IL-17A Signaling in Fibroblasts | 6.62E-01 |
| Clathrin-mediated Endocytosis Signaling | 6.3E-01 |
| Endothelin-1 Signaling | 6.24E-01 |
| TNFR1(Tumor necrosis facrorrecptor 1) Signaling | 6.14E-01 |
| CXCR4 (C-X-C chemokine receptor type 4) Signaling | 6.13E-01 |
| CD27 Signaling in Lymphocytes | 6.03E-01 |
| Toll-like Receptor Signaling | 5.82E-01 |
| LPS/IL-1 Mediated Inhibition of RXR (Retinoid X receptor beta (RXR-beta ) Function | 5.72E-01 |
| Acute Phase Response Signaling | 5.53E-01 |
| IL-8 Signaling | 5.43E-01 |
| Leukocyte Extravasation Signaling | 5.39E-01 |
| Molecular Mechanisms of Cancer | 5.37E-01 |
| Role of JAK1 (Janus kinase 1)and JAK3 (Janus kinase 3) in γc Cytokine Signaling | 5.27E-01 |
| IL-4 Signaling | 5.19E-01 |
| IL-17A Signaling in Airway Cells | 5.11E-01 |
| JAK/Stat (Janus kinase/ Signal Transducer and Activator of Transcription) Signaling | 4.95E-01 |
| Chemokine Signaling | 4.95E-01 |
| IL-10 Signaling | 4.87E-01 |
| IL-15 Signaling | 4.87E-01 |
| Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses | 4.66E-01 |
| CCR5 (C-C chemokine receptor type 5 )Signaling in Macrophages | 4.53E-01 |
| IL-17 Signaling | 4.46E-01 |
| TGF-β (Transforming growth factor beta ) Signaling | 4.22E-01 |
| Natural Killer Cell Signaling | 3.64E-01 |
| IL-1 Signaling | 3.55E-01 |
| VEGF (Vascular endothelial growth factor ) Signaling | 3.5E-01 |
| Virus Entry via Endocytic Pathways | 3.46E-01 |
| IL-6 Signaling | 3.46E-01 |
| Fc Epsilon RI Signaling | 3.37E-01 |
| CCR3 C-C chemokine receptor type 3 )Signaling in Eosinophils | 3.21E-01 |
| Cdc42 (Cell division control protein 42) Signaling | 2.99E-01 |
| Dendritic Cell Maturation | 2.6E-01 |
| NF-κB ((nuclear factor kappa-light-chain-enhancer of activated B cells) Signaling | 2E-01 |
aFunctional annotation analysis of the differentially expressed genes (DEGs) with the IPA (Ingenuity Pathway Analysis, http://www.ingenuity.com) tools. The IPA tool defines
the most significant biological functions through association with the largest number of DEGs correlated to these functions. Canonical pathways are known classic routes
and well-established of biological functions evidenced by validated genes.
bP value determined from the average log2 ratio using Bioconductor (http://www.bioconductor.org). All values of these canonical pathways were significant.
Table 1: Canonical pathways associated with the DEGs from 60 dpc.
Chronic phase I of infection (120 dpc)
After microarray analysis of the 23,000 genes in tissues collected at chronic phase I of infection, 930 genes were found to be DEGs at the same time in all infected tissues, 800 of these genes displayed a ≥ 1.3 fold change in expression. From the total of 930 DEGs, 800 genes were upregulated (613 known and 187 unknown genes), and 130 were downregulated (88 known and 42 unknown genes). Subsequently, the 930 DEGs were characterized using the IPA tool detection of possible biological functions and main canonicals pathways (Table 2). In table 2 new pathways associated with the DEGs were also observed providing evidence of innate and adaptive immune responses. The IPA tool demonstrated that the DEGs were mainly associated with genetic disorders such as cell death, immune cell trafficking, gene expression, inflammatory response, antigen presentation, inflammatory/ immunological/infectious diseases, and cancer.
| Canonical Pathwaysanuity Canonical Pathways | pp-valueb |
|---|---|
| Inhibition of Angiogenesis by TSP1 (tumor suppressor region 1) | 1.73E00 |
| Mechanisms of Viral Exit from Host Cells | 1.25E00 |
| PKCθ (protein kinase C theta ) Signaling in T Lymphocytes | 8.05E-01 |
| Caveolar-mediated Endocytosis Signaling | 6.7E-01 |
| FcγRIIB (Fc receptor or CD32) Signaling in B Lymphocytes | 6.03E-01 |
| Phospholipase C Signaling | 4.97E-01 |
| RhoA (RhoA kinase) Signaling | 4.79E-01 |
| Complement System | 4.79E-01 |
| NRF2(nuclear factor erythroid 2) mediated Oxidative Stress Response | 4.36E-01 |
| Lipid Antigen Presentation by CD1 | 4.28E-01 |
| Role of JAK2 (Janus Kinase 2) in Hormone-like Cytokine Signaling | 4.18E-01 |
| Coagulation System | 3.63E-01 |
| Crosstalk between Dendritic Cells and Natural Killer Cells | 3.55E-01 |
| Cytotoxic T Lymphocyte-mediated Apoptosis of Target Cells | 3.34E-01 |
| Retinoic acid Mediated Apoptosis Signaling | 3.25E-01 |
| Endoplasmic Reticulum Stress Pathway | 2.99E-01 |
| Role of JAK1 (Janus Kinase 1), JAK2 and TYK2 (tyrosine kinase 2) in Interferon Signaling | 2.81E-01 |
a Functional annotation analysis of the differentially expressed genes (DEGs) with the IPA (Ingenuity Pathway Analysis, http://www.ingenuity.com) tools. The IPA tool defines
the most significant biological functions through association with the largest number of DEGs correlated to these functions. Canonical pathways are known classic routes
and well-established of biological functions evidenced by validated genes.
bP value determined from the average log2 ratio using Bioconductor (http://www.bioconductor.org). All values of these canonical pathways were significant.
Table 2: Canonical pathways associated with the DEGs from 120 dpc.
Chronic phase II of infection (240 dpc)
From the 23,000 genes during the chronic phase II of infection at 240 dpi analyzed by microarrays, 744 were identified as DEGs. Of these DEGS, 658 genes displayed a ≥ 1.3 fold change, and 658 were upregulated genes (575 known and 83 unknown genes), while 83 were downregulated (63 known and 23 unknown genes). The IPA tool was used to detect biological significance in these DEGs. The pathways, IL-8 signaling, clathrin-mediated endocytosis signaling, leukocyte extravasation signaling, antigen presentation, Fcγ receptor, dendritic cell maturation, coagulation system, endocytic pathways, LPS/IL-1, JAK1, JAK2 and TYK2 related to interferon signaling, NF-κB activation, IL-22 signaling and interferon signaling pathways were suggested by these results. An association between DEGs and top function networks was also performed using the IPA, which demonstrated many functions including cell morphology, gene expression, cellular movement, cellular assembly and organization, immunological disease, cell-tocell signaling and interaction, cancer, infectious disease, reproductive system disease, inflammatory response, cell death, DNA replication, recombination, and repair.
Comparison of DEGs among time points (acute, chronic I and chronic II) in B. ovis infected rams
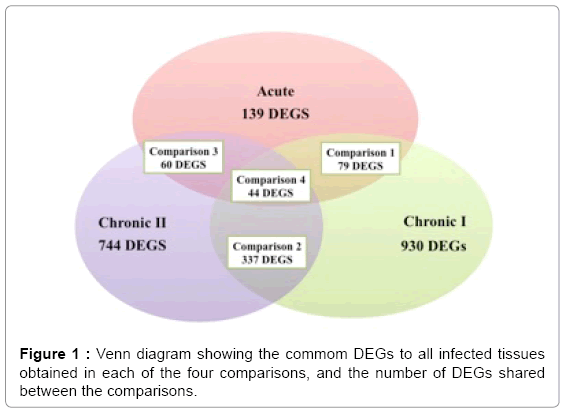
The Venn diagram (Figure 1) illustrates the number of DEGs obtained in each of the four time points, as well as the number of DEGs shared between groups. From the 44 common DEGs (31 known genes) obtained in the comparison among the three groups (comparison 4), only 8 had significant biological functions (Table 3). In Table 4, qPCR validated five DEGS: ATMIN (ATM interactor) gene, RCHY1 (ring finger and CHY zinc finger domain containing 1), ANKFY1 (ankyrin repeat and FYVE domain containing 1), EFEMP1 (EGFcontaining fibulin-like extracellular matrix protein 1) and Zinc finger, RAN-binding domain containing 2 (ZRANB2). The IPA tool allowed for comparison of the results of different tests. This feature enabled identification of biological functions and/or enriched canonical pathways common to different analyses. When the results of the three IPA analysis were compared, the biological functions: immune cell trafficking, immunological disease, infectious disease, inflammatory disease, inflammatory response and cellular movement were significant for the three conditions tested (acute, chronic I and II), and a greater significance was observed for phase chronic II.
| Gene description | Genbank accession number | qPCR annealing conditions |
|---|---|---|
| Arylsulfatase family, member K (ARSK) F- tgatgcaagtggactggaag R- atctaccgggtgcatttctg |
NM_001035400 | 55°C, 30s |
| Ring finger (RCHY1) F - atgccttgcaatgaatctcc R- gcgcatacacaatggacatc |
NM_001083754 | 55°C, 30s |
| Ankyrin (ANKFY1) F- gactttgctgctgtggatga R- ccaaaatgtgcagtggtgac |
XM_606825 | 55°C, 30s |
| EGF-containing fibulin-like extracellular matrix protein 1 (EFEMP1) F- gtgcaatcctgggtttcagt R- ccaggttcgttgacacattg |
NM_001081717 | 55°C, 30s |
| ATM interactor (ATMIN) F- tacaccacaccgcagttcat R- ttccgctggtctgagtttct |
XM_001249565 | 55°C, 30s |
| Zinc finger, RAN-binding domain containing 2 (ZRANB2) F- aatgtgaattgggccagaag R- ccaactgccttccctctgta |
NM_001105346 | 550C, 30s |
| Histocompatibility complex, class II, DQ alpha, type 1(BOLA-DQA1) F- agcctctgtggaggtgaaga R- gctgccagacagtctccttc |
NM_001013601 | 55°C, 30s |
| MHC class II antigen (BLA-DQB) F- cagatcaaggttcggtggtt R- aagcctccaacaccactcag |
NM_001034668 | 55°C, 30s |
| Ovis ariesglyceraldehyde-3-phosphate dehydrogenase (GAPDH) F- gggtcatcatctctgcacct R- ggtcataagtccctccacga |
NM_001190390 | 56°C, 30s |
Table 3: A: acute phase; C1: chronic phase 1; C2: chronic phase II.
| Genes | Relative fold-change of genes analysed by qRT-PCR. | Relative fold-change of genes analysed by microarray | Spearman’s correlation test (r-value) |
| ZRANB2 (C2) Zinc finger, RAN-binding domain containing 2 | 3.880 (±0.07; p=0.03) | 6.33(±0.29) | r=0.77 |
| RCHY1 (C1) Ring finger | 1.38(±0.21;p=0.04) | 1.32 (±0.11) | r=0.85 |
| ANKFY (C1)Ankyrin | 1.46(±0.54;p=0.03) | 1.35(±0.11) | r=0.9 |
| ATMIN (A) ATM interactor | 6.35(±0.04;p=0.01) | 6.25 (±0.51) | r=0.8 |
| EFEMP1 (C1) EGF-containing fibulin-like extracellular matrix protein | 1.76(±0.04;p=0.02) | 1.88(±0.17) | r=0.8 |
Table 4: Genes validated through qRT-PCR.
Discussion
Acute phase of infection (60 dpc)
An association was demonstrated between significant DEGs and the canonical pathways at 60 dpi using the IPA tool which was indicative of an innate immune response in all infected tissues (Table 1). The clinical presentation of ovine brucellosis, characterized by epididymitis and orchitis, was highlighted by general upregulation of genes in reproductive organs and lymph nodes (Figure 1 Supplementary material) [1]. Previous studies demonstrated the upregulation of pro-inflammatory genes as IL-1, IL-2 in infected rams [7]. Recent developments have shown that the inflammatory response induced by moderate brucelas likely represent a response directed toward escape and supression of the host immune response [6]. The upregulation of DEGs involved with the innate immune response in reproductive tissues and lymph nodes may have been directed toward elimination of the bacteria. The upregulation of genes during the acute phase of infection suggests a general antibacterial host mechanism in response to the intracellular pathogen [7]. The presence of genes associated with the IL-6 pathway, a Th2 cytokine, provided evidence of macrophage and humoral immune response activation which was described previously [18]. Activation of IL-12 suggested a response to bacterial infection and enhanced cell-mediated cytotoxicity [19]. As reported by Galindo et al. [7], B. ovis infection in rams at 60 dpi induces an activation of inflammatory and innate immune response pathways which contribute to development of a chronic B. ovis infection. By use of the IPA tool, evidence of an anti-inflammatory response against B. ovis was evident because of demonstration of macrophage migration inhibition factor (MIF) and IL-10 pathway. As reported [20], the presence of IL-10 contributes to the control of the intracellular growth of Brucella spp.
Previous studies have reported that expression of the BOLADQA and BOLA-DQB genes may contribute to the progression of mastitis [21]. In this study expression of upregulated genes during acute infection, including the BOLA-DQA1 and BOLA-DQB, and also suggestting that these genes were involved in the progression of B. ovis infection. BOLA genes, especially the major histocompatibility complex (MHC), genes have been associated with the genetic control of immune responses, during which they promote resistance or susceptibility to clinical mastitis [21]. In this study, the major histocompatibility complex class IIDQ alpha 2 (BOLA-DQA2) from the group of Bovine Leukocyte Antigen (BOLA) class II genes in the group of downregulated DEGs are involved in the immune response and were reported previously to be associated to mastitis resistance in dairy cattle [22]. Downregulation of BOLA-DQA2 during the acute phase infection may represent a mechanism that B. ovis for evasion in infected rams of the immune response that was not reported previously reported. An understanding of the role of BOLA genes in ovine brucellosis may contribute to a better knowledge of different phases, including susceptible, chronic, and resistant disease. During the acute phase of infection, the pathogenicity of B. ovis in the infected tissues activates the immune response with participation of gene expression, pathways and networks directed primarily toward elimination of the bacteria.
Chronic phase of infection I (120 dpc)
Many DEGs were observed in the infected tissues at the chronic phase I, with an increase in downregulated genes. These results are in accordance with Galindo et al. [7] and Antunes et al. [5], who reported that B. ovis induces downregulation of gene expression in order to supress the inflammatory host defenses infected rams. The inflammatory response induced by B. ovis represents an attempt to evade the immune response and suppress the host immune response [5,6]. In Table 2, the IPA tools through the canonical pathways emphasized an immune response associated with chronic of B. ovis infection. While this intracellular bacteria stimulates a cellular response [23], an effective response to Brucella spp. requires both humoral and cellular immune responses [24]. The canonical pathway NFAT remained significant until 120 dpi, and this family of transcription factors plays an important role in the immune response [25]. At 120 dpi the JAK1 pathway was significantly altered, and this pathway is an essential signaling protein used by cytokines type I and type II in response to tumors [26]. TYK2 was the first member of the JAK family described, and has been implicated in IFN-α, IL-6, IL-10 and IL-12 signaling [27]. The genes found in association with the canonical pathway of FcγRIIB receptor provided evidence of protective immune functions. The Fc receptor is a protein found on the surface of natural killer cells, macrophages, neutrophils, and mast cells that contributes to the activity of cytotoxic cells trough the antibody-dependent cell-mediated cytotoxicity [28]. IGF-1 canonical pathway was significantly upregulated in this study, which has been related to macrophages activation, thus contributing to the pathogenesis of B. ovis in rams [7,29]. Upregulation of genes associated with the CTLA4 canonical pathway suggested a tentative response directed toward suppression of the immune response to B. ovis because this signaling is transmitted as an inhibitory signal to T cells [30]. The upregulated genes associated to SAPK/JNK canonical pathway wich was significant at both 60 and 120 dpi demonstrated a cellular response direct toward elimination of B. ovis infection. This signaling is responsible for the nuclear production of NF-kB, TNF-α and IL-2 during T lymphocyte activation, which, in turn, regulates cell survival, apoptosis, and proliferation [31]. The canonical pathways VEGF remained activated at chronic phase I of infection. VEGF genes are associated with inflammatory reaction induced by intracellular bacterial infection [32]. The JAK-STAT canonical pathway appears to be relevant during acute phase of infection and chronic phase I. This pathway is a major signaling alternative to the second messenger system which is activated by a signal from interferon, interleukin, growth factors, or other chemical messengers that act in basic cell functions, such as cell growth, differentiation and death. Failure of JAK-STAT pathway can result in immune deficiency syndromes and cancer [33-36]. During the chronic phase I of infection, upregulation of BOLA genes (BOLA-DQA1 and BOLA-DQB) was observed in infected tissues, and suggested a role of these genes during ovine brucellosis. The BOLADQA2 gene (MHC class I heavy chain) remained downregulated after the acute phase of infection. At the infected tissues, the upregulation of genes linked in canonical pathways associated with immune responses may represent an antibacterial mechanism of the host for elimination of B. ovis infection.
Chronic phase of infection II (240 dpc)
At this stage of infection a strong cellular activation was noticed in the infected tissues, demonstrating the chronic establishment of ovine brucellosis. Chronic B. ovis infection is mainly recognized by the biological functions inflammation and coagulation. According to Bergmann and Hammershmidt [37], the interplay between blood coagulation and inflammation is essential for a host defense against intracellular infectious agents. However, this interpolation favors dissemination of bacteria within the host and contributes to evasion of the immune response. At 240 dpi activation of inflammatory and LPS/IL-1 inhibition pathways were observed. Changes of gene expression during B. ovis late infection may be transient in order to extend the intracellular life of the pathogen, and may result due to upregulation of genes involved in bacterial phagocytosis and downregulating of genes resulting from a protective host defense mechanism [7]. Of the 86 downregulated genes observed, the analysis emphasizes downregulation of cytokine IL-2, BOLA and aquaporin. Galindo et al. [7] reported a Th1 profile and a suppression of Th2-type response at 60 dpi. During chronic infection II a notable upregulation of pro-inflammatory and phagocytosis genes as BOLA-DQA1, BOLADQB and annexin was observed, demonstrating a polarization of Th1 response. A Th2 response was also observed in the DEGs studied. The aquaporin genes are involved in the transport of small solutes and water channels, and alteration of these genes has been associated to several human diseases [35]. Dowregulation of the aquaporin genes appears to be a finding not reported previously as a mechanism of B. ovis during the onset of the disease. The the gene profile results in the late phase of infection suggested that B. ovis employs mechanisms that facilitate the persistence on the host.
Comparison of DEGs among infection phases (acute, chronic I and chronic II) and validation of the microarray analysis
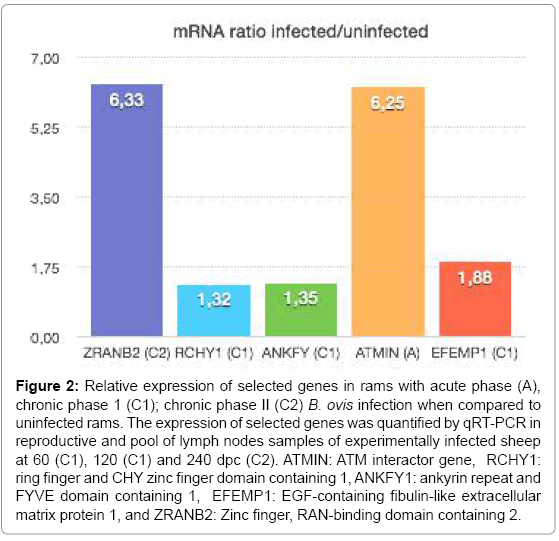
Of the 8 genes that were chosen from the microarray result, five correlated with the data generated from qRT-PCR (Table 4 and Figure 2). Differences in the reproductive tissues infected/uninfected ratio obtained by both methods may have resulted from variations in their sensitivity and from animal-to-animal variations, which were more evident in the qRT-PCR analysis with a larger number of animals [7]. The analysis during the acute phase of infection supported the relevance of the ataxia telangiectasia-mutated ATMIN (ATM interactor) gene. The ATM gene acts as a tumor suppressor, activated by DNA damage and interacts with a broad network of proteins as the tumor suppressor (p53), DNA repair factors (RAD50, RAD51, GADD45) and other signaling molecules (NF-kB). In addition to regulating DNA repair and the cell cycle, ATM can also trigger apoptosis in cells exposed to radiation [36]. ATM gene may be activated to cause apoptosis and release the bacteria from infected cells, thus contributing to immune response against R-B. ovis. The genes, RCHY1 (ring finger and CHY zinc finger domain containing 1), ANKFY1 (ankyrin repeat and FYVE domain containing 1) and EFEMP1 (EGF-containing fibulin-like extracellular matrix protein 1), were also highlighted by the analysis during chronic phase I of infection. The RCHY1 gene is expressed at higher levels in liver, testis and heart promoting p53 degradation. Higher levels of expression of RCHY1 have been associated to lung tumorigenesis [37]. Ankyrin proteins have been associated with a number of human diseases, including the cell cycle inhibitor p16, which is associated with cancer [38]. EFEMP1 gene has been shown to promote tumor growth in human adenocarcinoma [39]. The genes RCHY1, ANKFY1, and EFEMP1 were shown to be involved in cancer, which could be relevant in ovine brucellosis because of the role they play in the initiation of apoptosis, inhibition of cell cycle and progression of infection. ZRAB2 was validated at chronic phase II; this gene was implicated in regulation of mRNA processing [40]. This study represents the first analysis of microarray gene expression in different tissues of rams infected with of B. ovis at various times during infection [60 dpi (acute phase), 120 dpi (chronic phase I), and 240 dpi (chronic phase II)], and many of the genes identified require further study. Future studies are needed focusing on the common genes altered at the three times of infection using the PCR array tool. The microarray results of rams demonstrated a list of commom DEGS due the comparison in each time point among the six infected reproductive tissues and the six controls tissues, giving a pattern of gene expression in reproductive tissues and pool of lymph nodes experimentally infected with B. ovis. The microarray used for this study was designed for gene expression in cattle, and validation of these common DEGs will be necessary to avoid the underestimation of gene expression in rams involved in the pathogenesis of B. ovis..These results expand knowledge of the the pathogenesis of this B. ovis strain infection in rams and suggest new genes and pathways for futher investigation.
Figure 2: Relative expression of selected genes in rams with acute phase (A), chronic phase 1 (C1); chronic phase II (C2) B. ovis infection when compared to uninfected rams. The expression of selected genes was quantified by qRT-PCR in reproductive and pool of lymph nodes samples of experimentally infected sheep at 60 (C1), 120 (C1) and 240 dpc (C2). ATMIN: ATM interactor gene, RCHY1: ring finger and CHY zinc finger domain containing 1, ANKFY1: ankyrin repeat and FYVE domain containing 1, EFEMP1: EGF-containing fibulin-like extracellular matrix protein 1, and ZRANB2: Zinc finger, RAN-binding domain containing 2.
Conflict of Interest Statement
All authors disclose any financial and personal relationships with other people or organizations that could inappropriately influence their work.
Acknowledgements
This work was supported by Fundação de Amparo a Pesquisa do Estado de São Paulo-FAPESP (2008/03962-0), and by Conselho Nacional de Desenvolvimento Científico e Tecnológico-CNPQ (559023/2010-3). J.M.A.P. Antunes was granted with a full tuition FAPESP scholarship (2008/03837-0) in Brazil, and a full tuition of Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-CAPES scholarship (Programa Institucional de Doutorado Sanduíche no Exterior, Processo BEX 0987/11-5) in Spain. We are also grateful for the technical assistance during the experiment by Mr. Adilson Aparecido Romero and Mr. Roberto Martins.
References
- Megid J, Mathias LA, Robles CA(2010) Clinical Manifestations of Brucellosis in Domestic Animals and Humans. The Open Vet Sci J 4:119-126.
- Moreno E, Gorvel JP(2004) Invasion, intracellular trafficking and replication of Brucella organisms in professional and nonprofessional phagocytes. In: Lopes-Goni, I., Moriyon, I. (Eds.), Brucella Molecular and Cellular Biology. Horizon Bioscience, Wymondham, UK.
- Seleem MN, Boyle SM, Sriranganathan N (2008) Brucella: a pathogen without classic virulence genes. Vet Microbiol 129: 1-14.
- Xavier MN, Silva TM, Costa EA, Paixão TA, Moustacas VS, et al. (2010) Development and evaluation of a species-specific PCR assay for the detection of Brucellaovis infection in rams. Vet Microbiol 145: 158-164.
- Antunes JM, Allendorf SD, Appolinário CM, Cagnini DQ, Figueiredo PR, et al. (2013) Rough virulent strain of Brucellaovis induces pro- and anti-inflammatory cytokines in reproductive tissues in experimentally infected rams. Vet Microbiol 161: 339-343.
- Antunes JMAP, Megid J(2013)Brucellaovis: invasion, traffic, virulence factors and immune response. Semina. 34:1301-1312.
- Galindo RC, Muñoz PM, De Miguel MJ, Marin CM, Blasco JM, et al. (2009) Differential expression of inflammatory and immune response genes in rams experimentally infected with a rough virulent strain of Brucellaovis. Vet ImmunolImmunopathol 127:295-303.
- Pariset L, Chillemi G, Bongiorni S, Romano Spica V, Valentini A (2009) Microarrays and high-throughput transcriptomic analysis in species with incomplete availability of genomic sequences. N Biotechnol 25: 272-279.
- Marín CM, Jiménez de Bagués MP, Blasco JM, Gamazo C, Moriyón I, et al. (1989) Comparison of three serological tests for Brucellaovis infection of rams using different antigenic extracts. Vet Rec 125: 504-508.
- Blasco JM(1990)Brucellaovis. In: Nielsen K, Duncan JR. (Edr.), Animal Brucellosis. CRC Press Inc., Boca Raton, FL.
- Manterola L, Tejero-Garcés A, Ficapal A, Shopayeva G, Blasco JM, et al. (2003) Evaluation of a PCR test for the diagnosis of Brucellaovis infection in semen samples from rams. Vet Microbiol 92: 65-72.
- Stanton JB, Knowles DP, Call DR, Mathison BA, Baszler TV (2009) Limited transcriptional response of ovine microglia to prion accumulation. BiochemBiophys Res Commun 386: 345-350.
- Johnson WE, Li C, Rabinovic A (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8: 118-127.
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, et al. (2003) Summaries of AffymetrixGeneChip probe level data. Nucleic Acids Res 31:e15.
- Benjamini Y, Hochberg Y(1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57: 289-300.
- Sharan R, Maron-Katz A, Shamir R (2003) CLICK and EXPANDER: a system for clustering and visualizing gene expression data. Bioinformatics 19: 1787-1799.
- Shamir R, Maron-Katz A, Tanay A, Linhart C, Steinfeld I, et al. (2005) EXPANDER--an integrative program suite for microarray data analysis. BMC Bioinformatics 6: 232.
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45.
- Salas-Téllez E, Núñez del Arco A, Tenorio V, Díaz-Aparicio E, de la Garza M, et al. (2005) Subcellular fractions of Brucellaovis distinctively induce the production of interleukin-2, interleukin-4, and interferon-gamma in mice. Can J Vet Res 69: 53-57.
- De Rose R, Scheerlinck JP, Casey G, Wood PR, Tennent JM, et al. (2000) Ovine interleukin-12: analysis of biologic function and species comparison. J Interferon Cytokine Res 20: 557-564.
- Golding B, Scott DE, Scharf O, Huang LY, Zaitseva M, et al. (2001) Immunity and protection against Brucellaabortus. Microbes Infect 3: 43-48.
- Takeshima S, Matsumoto Y, Chen J, Yoshida T, Mukoyama H, et al. (2008) Evidence for cattle major histocompatibility complex (BoLA) class II DQA1 gene heterozygote advantage against clinical mastitis caused by Streptococci and Escherichia species. Tissue Antigens 72: 525-531.
- Hou Q, Huang J, Ju Z, Li Q, Li L, et al. (2012) Identification of splice variants, targeted microRNAs and functional single nucleotide polymorphisms of the BOLA-DQA2 gene in dairy cattle. DNA Cell Biol 31: 739-744.
- Vemulapalli R, HE Y, Boyle SM, Sriranganathan N, Schurig GG(2000)Brucellaabortus strain RB51 as a vector for heterologous protein expression and induction of specific Th1 type immune responses. Infect Immun 68: 3290-3296.
- Ficht TA, Adams G(2009) Vaccines for Biodefense and Emerging and Neglected Diseases. In: Barret, ADT, Stanberry LR (Eds.), Brucellosis. CRC Press, Boca Raton, USA, FL.
- Crabtree GR, Olson EN (2002) NFAT signaling: choreographing the social lives of cells. Cell 109 Suppl: S67-79.
- Jatiani SS, Baker SJ, Silverman LR, Reddy EP (2010) Jak/STAT pathways in cytokine signaling and myeloproliferative disorders: approaches for targeted therapies. Genes Cancer 1: 979-993.
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD (1998) How cells respond to interferons. Annu Rev Biochem 67: 227-264.
- Anderson R (2003) Manipulation of cell surface macromolecules by flaviviruses. Adv Virus Res 59: 229-274.
- Mora AL, Torres-González E, Rojas M, Corredor C, Ritzenthaler J, et al. (2006) Activation of alveolar macrophages via the alternative pathway in herpesvirus-induced lung fibrosis. Am J Respir Cell MolBiol 35: 466-473.
- Fernández-Mestre M, Sánchez K, Balbás O, Gendzekhzadze K, Ogando V, et al. (2009) Influence of CTLA-4 gene polymorphism in autoimmune and infectious diseases. Hum Immunol 70: 532-535.
- Nishina H, Wada T, Katada T (2004) Physiological roles of SAPK/JNK signaling pathway. J Biochem 136: 123-126.
- Sato F, Imaizumi T, Sashinami H, Yoshida H, Kusumi T, et al. (2007) Upregulation of vascular endothelial growth factor by heat-killed Listeria monocytogenes in macrophages. BiochemBiophys Res Commun 354: 608-612.
- Aaronson DS, Horvath CM (2002) A road map for those who don't know JAK-STAT. Science 296: 1653-1655.
- Bergmann S, Hammerschmidt S (2007) Fibrinolysis and host response in bacterial infections. ThrombHaemost 98: 512-520.
- Agre P, Kozono D (2003) Aquaporin water channels: molecular mechanisms for human diseases. FEBS Lett 555: 72-78.
- McKinnon PJ (2004) ATM and ataxia telangiectasia. EMBO Rep 5: 772-776.
- Beitel LK, Elhaji YA, Lumbroso R, Wing SS, Panet-Raymond V, et al. (2002) Cloning and characterization of an androgen receptor N-terminal-interacting protein with ubiquitin-protein ligase activity. J MolEndocrinol 29: 41-60.
- Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY (2004) Theankyrin repeat as molecular architecture for protein recognition. Protein Sci 13: 1435-1448.
- Camaj P, Seeliger H, Ischenko I, Krebs S, Blum H, et al. (2009) EFEMP1 binds the EGF receptor and activates MAPK and Akt pathways in pancreatic carcinoma cells. BiolChem 390: 1293-1302.
Relevant Topics
- Agricultural biotechnology
- Animal biotechnology
- Applied Biotechnology
- Biocatalysis
- Biofabrication
- Biomaterial implants
- Biomaterial-Based Drug Delivery Systems
- Bioprinting of Tissue Constructs
- Biotechnology applications
- Cardiovascular biomaterials
- CRISPR-Cas9 in Biotechnology
- Nano biotechnology
- Smart Biomaterials
- White/industrial biotechnology
Recommended Journals
Article Tools
Article Usage
- Total views: 12101
- [From(publication date):
December-2015 - Mar 31, 2025] - Breakdown by view type
- HTML page views : 11191
- PDF downloads : 910