Methodology for Generating Standardized Datasets with Characteristic Diagnostic Parameters of Rare Diseases in Form of HPO-Terms
Received: 21-Apr-2022 / Manuscript No. DPO-22-61378 / Editor assigned: 25-Apr-2022 / PreQC No. DPO-22-61378(PQ) / Reviewed: 09-May-2022 / QC No. DPO-22-61378 / Revised: 16-May-2022 / Manuscript No. DPO-22-61378(R) / Published Date: 23-May-2022 DOI: 10.4172/2476-2024.7.S11.002.
Abstract
Background: Finding a diagnosis for rare diseases is a challenge for patients and those treating them. Establishing a uniform methodology for specifying the symptoms of a patient seems useful. This, as well as a database with clinical parameters reported in patients already diagnosed with the corresponding disease or that has led to the diagnosis, would facilitate the global data exchange between specialists and subsequently diagnosis. This work aims to introduce a methodology for generating data sets with characteristic diagnostic parameters of rare diseases using exemplarily the three rare metabolic diseases late-onset Pompe disease, Gaucher disease Type I and Smith-Lemli-Opitz syndrome. For these data sets, a standardized word form is to be chosen that enables European or even worldwide exchange.
Methods and results: A systematic literature review of characteristic symptoms and diagnostic criteria was performed for each of the three disorders. These parameters were converted into vocabulary standardized by The Human Phenotype Ontology (HPO), so-called HPO terms. Subsequently, a retrospective analysis of the patient files of 23 late-onset Pompe disease patients, 21 Gaucher disease Type I patients and 25 Smith-Lemli-Opitz syndrome patients was carried out together with the University Children's Hospital Magdeburg and the Center of excellence for Rare Metabolic Diseases at the Charité Berlin. Features present in ≥ 40% of the cohort and collected simultaneously in a certain minimum number of patients were filtered out. The analysis resulted in data sets with 22 diagnostic parameters for late-onset Pompe disease, 16 features for Gaucher disease Type I and 17 parameters for Smith- Lemli-Opitz syndrome. After the statistical evaluation, the results were discussed comparatively with similar studies.
Conclusion: Using the introduced methodology data sets with characteristic diagnostic criteria for three rare diseases could be established. The developed datasets provide a good basis for expansion with further patient examples and for extending the methodology to other diseases to improve the diagnostic pathway and thus the health care of patients with rare diseases.
Keywords: Diagnosis of rare diseases; HPO-Terms; Generation of standardized datasets; Delay of diagnosis
Introduction
The European Union defines a disease as rare if no more than 5 out of 10,000 people are affected by this disease [1]. Of the approximately 30,000 known diseases, about 5,000 to 8,000 are counted as rare diseases [2]. In Germany, about 4 million people live with a rare disease [3]. Europe-wide about 30 million [2]. Due to their rarity, the knowledge of these diseases among physicians in normal care is often rather low and the number of corresponding specialists is limited. Moreover, in many cases, clinical symptoms are non-specific and seem to be unrelated and there are no standardized diagnostic criteria. Thus, diagnosis is often delayed, misleading and the result of multiple doctor visits. Additionally, treatment and care may also be challenging.
Publications and case reports exist on most diseases. However, the varying availability of corresponding studies and the frequent lack of guidelines for the individual diseases pose a challenge for those treating patients who are confronted with these diseases. At the same time, variable and diverse descriptions of the symptomatology of diseases in publications make it difficult to search for a specific disease based on the symptoms of those affected with an unclear, possibly rare disease.
With the establishment of self-help groups, the promotion of drug development, the creation of information portals such as ZIPSE (Central Information Portal on Rare Diseases) or se-atlas (Care Atlas for People with Rare Diseases) or the founding of the National Action Alliance for People with Rare Diseases (NAMSE), a lot is already being done in Germany to make things easier for all those involved. At the European level, European Reference Networks (ERNs), virtual networks involving health care providers across Europe, are established. They are intended to improve prevention and diagnosis and guarantee high-quality healthcare for patients with rare diseases throughout Europe. The Clinical Patient Management System (CPMS) designed for the ERNs is intended to facilitate cross-border diagnostic and therapeutic consultations and the safe exchange of patient data through the cooperation of specialists across European borders in compliance with data protection regulations.
To facilitate this exchange, it seems useful to establish a uniform methodology for specifying the symptoms of a patient. Furthermore, a type of database with clinical parameters that are reported in patients already diagnosed with the corresponding disease or that have led to the diagnosis would facilitate the exchange of data and the subsequent diagnosis.
Within the scope of this work, a methodology for generating data sets with characteristic diagnostic parameters is to be introduced. For these data sets, a standardized word form is to be chosen that enables European or even worldwide exchange and can also be used for the CPMS.
The methodology was applied exemplarily for three rare metabolic diseases: Late-Onset Pompe Disease (LOPD), Gaucher disease Type I and Smith-Lemli-Opitz Syndrome (SLOS). Both LOPD and Gaucher Disease, Type 1 are lysosomal storage diseases caused by genetic enzyme deficiencies. SLOS is caused by a genetic defect in cholesterol biosynthesis.
For all three diseases, there are already published studies of the symptomatology and diagnostic parameters of differently sized patient cohorts. However, the literature research carried out shows how difficult it is to bring symptom descriptions from different studies to a common denominator, to summarize them or even to derive diagnoses for patients who have not yet been diagnosed, due to the different focal points and vocabulary used. Compressed and standardized data sets should contribute to facilitating the path to diagnosis and the exchange with specialists for those treating and affected by the disease.
Materials and Methods
First, a systematic literature analysis was performed with regards to characteristic symptoms and diagnostic criteria of the three diseases mentioned. For this purpose, the book Vademecum metabolicum [4]. Specialist portals such as Orphanet and OMIM (Online Mendelian Inheritance in Men) as well as numerous other publications were used. The result was a table with 67 diagnostic parameters for Pompe disease. In addition to a literature analysis for late-onset Pompe disease, the parameters for the infantile-onset form of the disease were also researched. For Gaucher's disease, a table of 63 symptoms and diagnostic criteria was obtained, again differentiating between Gaucher types I, II and III. The literature search for SLOS syndrome yielded 84 symptoms and diagnostic criteria. If a classification of the frequency of occurrence of the diagnostic parameter was found in the literature, it was adopted in the table created. There is a subdivision of frequency into very frequent, frequent, occasional and rare, whereby mainly very frequent, frequent and partly occasional occurring features were adopted in the table. However, since these frequency data are not available for all parameters, a complete ordering of the parameters according to their frequency as reported in the literature is not possible.
Conversion of the parameters into HPO terms
Subsequently, these terms of parameters, formulated freely or based on the sources, were converted into vocabulary standardized by The Human Phenotype Ontology (HPO), so-called HPO terms. HPO is an online database of standardized vocabulary of phenotypic abnormalities of human diseases. Firstly published in 2008, it “provides comprehensive bioinformatic resources for the analysis of human diseases and phenotypes, offering a computational bridge between genome biology and clinical medicine” [5]. Therefore, clinical descriptions of diseases listed in the Online Mendelian Inheritance in Man (OMIM) database [6], were extracted, analyzed, compared and finally a term in HPO was created [7]. Each HPO-term encodes a phenotypic abnormality. The terms are connected in the form of directed acyclic graphs. That means, there is a hierarchical ordering from a generalized abnormality with various subgroups to increasingly detailed characterizations. But a specialized term can be assigned to several higher-level features [7]. Each class of the HPO is provided with a list of synonyms and textual definitions created by clinical experts [8]. Currently, the HPO contains around 13,000 terms [9]. And it is constantly being expanded. HPO consists of several sub-ontologies, with phenotypic abnormality being the main sub-ontology. Additional subontologies describe inheritance patterns, clinical modifiers, clinical course and frequency [9,10]. Since also disease-phenotype annotations are provided, HPO-Terms are linked with specific diseases or genes [8]. Besides OMIM and Orphanet [11]. Nomenclatures of rare diseases, many other public-facing clinical databases are using HPO to annotate patient data for disease-gene discovery projects [5]. This allows deep phenotyping of rare diseases [8]. Besides that, many other tools and applications use HPO [5]. Meanwhile, HPO is no longer used only for rare diseases, but for many common diseases [5].
In our current study, all symptoms and clinical findings were converted into HPO-Terms by one researcher.
Analysis of patient data
In the third step, medical records of patients with a confirmed diagnosis of the respective diseases were analyzed according to these criteria. In cooperation with the University Children's Hospital Magdeburg and the Center of excellence for Rare Metabolic Diseases at the Charité Berlin, three hereditary metabolic diseases were selected for the development of the data sets: LOPD, Gaucher disease Type I and SLOS. For these diseases, both centers have their own experience and larger patient collectives. A retrospective analysis of the medical records of 23 patients with LOPD, 21 patients with Gaucher disease Type I and 25 patients with SLOS was performed. In the tables created for the literature analysis with parameters transformed to HPO-Terms, it was marked which of the parameters occur in which patient. A subdivision was made into the characteristic present, characteristic not present or no specification. In this context, the table was expanded to include characteristics that were not included in the table during the literature research but did occur in patients. The symptoms, clinical presentation and results of laboratory tests were translated from German to English and assigned to corresponding HPO-Terms by one researcher.
In addition, the early and first symptoms, including the time of onset, were extracted from the patient records for all LOPD and Gaucher Type I patients.
Statistical analysis
The percentage frequencies and the corresponding confidence intervals for the occurrence of a characteristic in the patient group were calculated. There were two variants of the calculation: in the first variant, the percentages were calculated for the entire patient cohort for each characteristic, including the patients with missing information. In the second variant, the patients without information for this characteristic were excluded. Accordingly, the percentage frequencies of occurrence here are partly related to smaller numbers of patients. Concerning the latter variant, characteristics were filtered out that were detected and documented in ≥ 40% of the patients assessed according to this characteristic and collected simultaneously in a certain minimum number of patients.
Furthermore, for the three most frequent early or first symptoms of a disease, the time periods between the occurrence of the early symptoms and the time of diagnosis were calculated and the arithmetic means were given. For each of these early symptoms, an age range with frequent occurrence was determined on the basis of graphs. Correlations between individual early symptoms of a disease were also graphically traced.
Evaluation of datasets
Subsequently, the datasets were compared with the results of the literature search and discussed. Comparative tables have been prepared. In the event of significant discrepancies from the literature, the results of the study may need to be questioned.
Results
The evaluation of the frequency of occurrence of the diagnostic parameters in the examined patients following the literature analysis was carried out separately for each of the three diseases. Using the method described above, data sets for all three diseases could be developed. For LOPD, 22 diagnostic parameters were filtered out. The data set for Gaucher disease Type I includes 13 features, while for SLOS 17 parameters were found.
The average age of SLOS patients at the time of diagnosis is 12.8 months. The average age of diagnosis in the studied Gaucher disease type I patients is 32.7 years and in LOPD patients 39.8 years. The average time between the onset of early symptoms and the time of diagnosis is 7.7 years for Gaucher type I disease and 14.0 years for LOPD. The most frequent early symptoms of Gaucher disease type 1 and LOPD are shown in Tables 1 and 2.
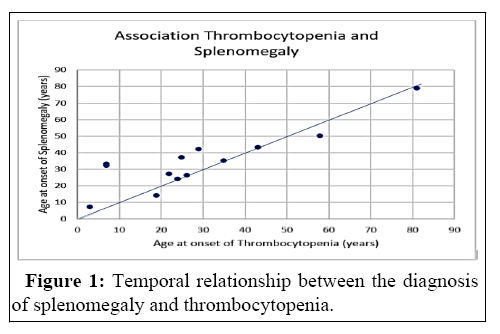
Table 1 shows that the most common and at the same time earliest documented symptom in Gaucher type 1 patients is thrombocytopenia. Thrombocytopenia is a result of Hypersplenism, a complication of Splenomegaly. Figure 1 shows that, on average, splenomegaly is only diagnosed after thrombocytopenia.
Table 2 shows that, on average, the first early symptom in LOPD patients is the elevation of transaminases, followed by muscle weakness and elevated creatine kinase.
| Thrombocytopenia | Splenomegaly | Hepatomegaly | |
|---|---|---|---|
| number of patients with early symptom* | 13/16 | Dec-19 | Jul-20 |
| Time interval between 1st documentation and diagnosis | 7,3 | 1,6 | -0,4 |
| Age with frequent first onset | 19-35 years (n=8) | 22-43 years (n=11) | 22-51 years (n=11) |
Table 2: Early symptoms of late-onset Pompe disease.
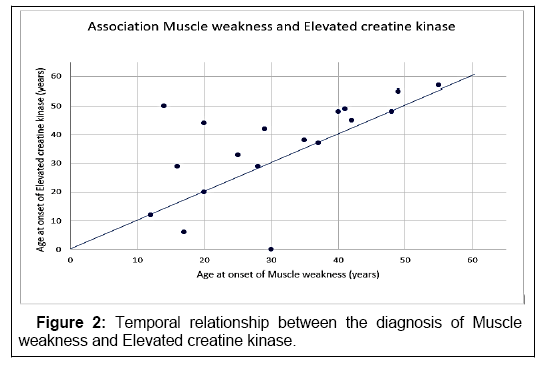
The temporal relationship between the diagnosis of muscle weakness and elevated creatine kinase is shown in Figure 2. It highlights that muscle weakness, associated with the elevation of creatine kinase, is diagnosed on average after a rise in creatine kinase.
Discussion
The introduced methodology was successfully used to create datasets with the most characteristic symptoms and diagnostic criteria, given in form of HPO-Terms exemplarily for three rare metabolic diseases.
A methodological challenge was the retrospective nature of the analysis of the patient records. Both reference centers involved indeed care for their patients according to a detailed care scheme specific to each disease so that certain diagnoses and examinations are always carried out. However, some patients came from other centers or other countries, so in some cases only the diagnosis and no or incomplete findings at diagnosis were available. For this reason, and also due to different types of documentation, no information was available for some parameters that were established in the literature analysis. In some cases, it was also not possible to determine whether the characteristic had not been investigated or whether it had been investigated but was not present and therefore not documented. Accordingly, the evaluation of some characteristics was only possible on the basis of a smaller group of patients than the original number, since missing data had to be subtracted. If, as planned, the data sets are expanded by other physicians or within the framework of studies with further patient examples, this will increase their informative value. At the same time, biases that arise due to differences in the focus of different physicians in the study will be compensated for. It should also be considered to what extent an additional temporal classification of the occurrence of the characteristics can be documented. This study also tried to find out which initial symptoms occur and when. The limiting factor was that concrete dates of the first occurrence were not always apparent or it was unclear whether the first date mentioned corresponded to the date of the first occurrence. Possibly, interviews with the patients in addition to the analysis of the files could provide more concrete information. It would then be possible to distinguish between initial symptoms and late symptoms. This precision of the information could further facilitate the diagnostic pathway.
One could argue that only one researcher extracting and converting the patient data into HPO-Terms makes the results error-prone. It would have been better, to have two researchers are doing these tasks and maybe a third one, deciding in case of varying results. But since also a comparison with similar studies in the literature was done major errors can be excluded. However, more patients should be added to the data sets by other investigators to further objectify and complement the results.
The standardized English-language vocabulary developed by the Human Phenotype Ontology includes more than 13,000 terms to describe signs, symptoms, or phenotypic manifestations that characterize specific diseases [9]. Despite this comprehensive registry, converting the terms used by clinicians for documentation into HPO terms proved challenging in some cases. Different vocabulary was used for the same symptoms, or there were minimal differences or variations in the expression or presence of symptoms, making it difficult to assign them to a specific term. One possibility here is the categorization and hierarchical ordering of HPO terms from a generalized abnormality with various subgroups to increasingly detailed characterizations. Thus, in some cases, assignment to a somewhat more general HPO term was necessary. Furthermore, since HPO provides mainly phenotypic characteristics, this also explains why some very common traits were not standardized by an HPO term. In the future, due to the continuous expansion and updating of the HPO database, a more precise characterization of the symptomatology will be possible. However, this shows that the use of a standardized vocabulary is advantageous for the documentation and exchange of the symptomatology of certain diseases.
One could also ask why The Human Phenotype Ontology was used and not ontology. First, has to be argued, the European Common Data Set for Rare Disease Registration recommends recording the phenotype of undiagnosed patients, which is what the datasets will be used for, with HPO [12]. Berger, et al. evaluated in their study How to design a registry for undiagnosed patients in the framework of rare disease diagnosis: suggestions on software, data set and coding system [13] three terminologies concerning usefulness and feasibility for a registry for undiagnosed patients. The three terminologies that were compared are HPO, SNOMED CT and LOINC [14, 15]. Using ten patient files, they analyzed the matches of symptoms, extracted from patient records, with medical terms given by the different coding systems [13]. The results showed that with HPO at 73%, SNOMED CT at 98% and LOINC only at 31% the tested medical terms could be correctly coded and matched the clinical terms given by the ontologies [13].
Accepting also more generic coding terms, HPO reached 89%, SNOMED CT reached 99%, and LOINC reached 39% of matches [13]. Despite the high percentage of matches of SNOMED CT, the authors conclude that HPO is their ontology of choice [13]. Reasons for this decision given by Berger, et al. are the acceptable matching rate of 73% or 89%, the wide use of HPO for deep phenotyping in the field of rare diseases, and its continuous expansion [10]. Which will improve the matching rate over time? Furthermore, it can be argued, that phenotype profiles of undiagnosed diseases can be compared with disease profiles in the HPO database to identify genetic diseases [13]. Unlike SNOMED CT, HPO is also available free of charge and no national license is needed [13].
However, to ensure continuous evaluation and improvement the developed datasets will also be included in the CrescNet. CrescNet is a competence network, developed by the University of Leipzig that enables the detection of impaired growth and weight development in children at an early stage [16]. It is based on the voluntary association of pediatricians. This network increases the chance that children will receive help in advance if a maldevelopment is predicted. In CrescNet diseases can be defined by HPO-Terms. After adding the generated datasets of the three chosen diseases, clinicians participating in CrescNet can introduce their patients, suffering from these diseases with their clinical presentation. This kind of prospective try will give feedback on the composition of the datasets and show necessary additions and improvements after practical application.
In addition, the applicability of the developed parameters was tested with the help of the Phenomizer [17]. Phenomizer is a freely available web application intended to support the differential diagnostic process in human genetics. The user, usually the attending physician, enters the symptoms of the patient coded as HPO-Terms. The algorithm yields a list of differential diagnoses providing the significance of the proposed candidate diagnoses. The given p-value indicates whether the entered clinical features are very suggestive of a particular diagnosis or whether no diagnosis in the database significantly matches the terms [17]. The first suggestion of the Phenomizer after entering 12 of the 13 found features for Gaucher disease (one feature is not yet available in HPO) is Gaucher disease Type III with a p-value of 0.0968, giving already very clear hint to the right diagnosis. If only seven of the 13 features are entered the first mentioned differential diagnosis is Gaucher disease without specifying a type with a corresponding p-value of 0.0160. After entering all founded SLOSfeatures apart from the parameters Elevated 7-dehydrocholesterol and Elevated 8-dehydrocholesterol, which a practitioner would only have measured if there was already a clear suspicion of SLOS, Phenomizer mentioned SLOS as a possible differential diagnosis but not as part of the first suggestions. Entering fewer parameters exemplarily three or seven, which are very characteristic, the Phenomizer suggests SLOS as the first option. LOPD seems to not be listed in the Phenomizer, since no combination brought LOPD as differential diagnosis, even using the HPO-Terms listed in the HPO-database annotaded with Pompe. In this way, it was possible to show for SLOS and Gaucher disease that the data sets developed also guarantee delimitation to possible differential diagnoses.
Individual parameters of the data sets are not discussed in this methodologically oriented paper. The statistical analyses carried out with regard to the first and early symptoms show that the diagnoses for LOPD and Gaucher's disease Type 1 are made with an average delay of 14.0 and 7.7 years. Due to the early onset of symptoms, which are at least partly easily visible from the outside, the diagnosis of SLOS is usually made in the first year of life.
In LOPD patients, the increase of transaminases is on average the earliest documented initial symptom. The analysis of the age of onset shows that there are two age intervals with frequent first diagnoses: 0-25 years and 41-50 years. Muscle weakness is found on average before creatine kinase elevation. An elevation of the enzyme is indicative of muscle disease, in the case of LOPD explained by glycogen accumulation in muscle cells. The fact that muscle weakness, as an organic correlate, is diagnosed several years’ earlier on average shows that in diagnostics, despite characteristic organ symptoms, the corresponding laboratory values are often determined late.
The most common first symptom of Gaucher type 1 disease is thrombocytopenia. Splenomegaly, the symptom that follows, can be explained by the infiltration of Gaucher cells into the organ. As a complication of splenomegaly, hypersplenism can occur, which can manifest itself, among other things, in the form of thrombocytopenia. Accordingly, it would theoretically have been expected that splenomegaly would be diagnosed before thrombocytopenia. Thus, this remarkable observation is to be confirmed by standardized data collection. It should be noted that the retrospective nature of the study, as already mentioned, is a limiting factor for the analyses carried out.
These statistical calculations prove exemplarily for two rare diseases that there is a delay of diagnosis although early symptoms are already recognized. This emphasizes again the importance of standardized documentation and of opportunities to discuss patient cases with specialists as the CPMS is.
Conclusion
A literature search, the conversion of diagnostic parameters into HPO terms, the retrospective analysis of patient records as well as statistical analysis and comparative evaluation of the findings resulted in the successful application of a methodology for the generation of standardized data sets for three rare diseases. It was found that a retrospective analysis based on patient records makes standardizationdifficult due to missing data and different documentation. It can therefore be concluded that documentation of patient data using a template as well as standardized vocabulary would have facilitated this. In turn, the developed data sets can be used to create templates.
To obtain as complete and accurate a picture as possible, it is recommended that this study is supplemented with additional patient cases or cohorts to further expand and improve the data sets for CPMS. This and an application of the methodology to other diseases can improve the diagnosis and treatment of patients with rare diseases.
Declarations
Ethics approval
The ethical committee of the Otto-von-Guericke University in Magdeburg approved the study.
Consent for publication
Not applicable.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare that they have no competing interests.
Funding
This work was generated within the European Reference Network for Rare Hereditary Metabolic Disorders (MetabERN), co-funded by the European Union within the framework of the Third Health Programme ERN-2016-Framework Partnership Agreement 2017-2021, Project ID No. 739543 and was built upon the INEA CEFfunded Action 2017-DE-IA-0109, which aimed to set up the MetabERN Operational Helpdesk.
Authors' contributions
ACLK analyzed and interpreted the patient data and was a major contributor in writing the manuscript. AZ as well as KM concepted and designed the study, recruited patients and revised the manuscript. CVL, AB, CMB and MS advised on the conception of the study and revised the manuscript. KP contributed to patient recruitment and revised the manuscript. All authors read and approved the manuscript.
Acknowledgements
We would like to thank Anke Lux (Medical faculty-University Hospital Magdeburg, Institute of Biometry and Medical Informatics) for her assistance with statistical analysis. Moreover, we thank Laura Paneghetti, PhD for comments that greatly improved the manuscript.
References
- Regulation (EC) No 141/2000 of the European Parliament and of the Council. On Orphan Medicinal Products. December 1999.
- Council recommendation. For action in the field of rare diseases2009.
- Frank M, Eidt-Koch D, Aumann I, Reimann A, Wagner TOF, et al. (2014) Measures to improve the health situation of people with rare diseases in Germany: A comparison with the National Action Plan. Federal Health Sheet 57:1216-1223.
[Crossref] [Google Scholar] [Pubmed]
- Zschocke J, Hoffmann GF. (2012) Vademecum metabolicum: Diagnosis and therapy of hereditary metabolic diseases. (4th Edn). Thieme, Germany.
- Köhler S, Vasilevsky NA, Engelstad M, Foster E, McMurry J, et al. (2017) The Human Phenotype Ontology in 2017. Nucleic Acids Res 45:D865-876.
[Crossref] [Google Scholar] [Pubmed]
- Amberger J, Bocchini C, Scott A, Hamosh A. (2008) McKusick’s online Mendelian inheritance in man (OMIM(R)). Nucleic Acids Research 37:D793-6.
[Crossref] [Google Scholar] [Pubmed]
- Robinson PN, Köhler S, Bauer S, Seelow D, Horn D,et al. (2008) The human phenotype ontology: A tool for annotating and analyzing human hereditary disease. Am J Hum Genet 83:610-615.
[Crossref] [Google Scholar] [Pubmed]
- Köhler S, Doelken SC, Mungall CJ, Bauer S, Firth HV, et al. (2014) The human phenotype ontology project: Linking molecular biology and disease through phenotype data. Nucleic Acids Res 42:D966-974.
[Crossref] [Google Scholar] [Pubmed]
- Human Phenotype Ontology. 2021.
- Köhler S, Carmody L, Vasilevsky N, Jacobsen JOB, Danis D, et al. (2019) Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources. Nucleic Acids Research 47:D1018-27.
[Crossref] [Google Scholar] [Pubmed]
- Rath A, Olry A, Dhombres F, Brandt MM, Urbero B, et al. (2012) Representation of rare diseases in health information systems: The orphanet approach to serve a wide range of end users. Human Mutation 33:803-808.
[Crossref] [Google Scholar] [Pubmed]
- Set of Common Data Elements for Rare Diseases Registration. European Platform on Rare Disease Registration. February 17, 2022.
- Berger A, Rustemeier A-K, Göbel J, Kadioglu D, Britz V, Schubert K, et al. (2021) How to design a registry for undiagnosed patients in the framework of rare disease diagnosis: suggestions on software, data set and coding system. Orphanet J Rare Dis 16:198.
[Crossref] [Google Scholar] [Pubmed]
- Shahpori R, Doig C. (2010) Systematized nomenclature of medicine-clinical terms direction and its implications on critical care. J Crit Care 25:364. -364.e9.
[Crossref] [Google Scholar] [Pubmed]
- Forrey AW, McDonald CJ, DeMoor G, Huff SM, Leavelle D, et al. (1996) Logical Observation Identifier Names and Codes (LOINC) database: A public use set of codes and names for electronic reporting of clinical laboratory test results. Clin Chem 42:81-90.
[Crossref] [Google Scholar] [Pubmed]
- CrescNet.org n.d. 2022.
- Köhler S, Schulz MH, Krawitz P, Bauer S, Dölken S, et al. (2009) Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am J Hum Genet 85:457-464.
[Crossref] [Google Scholar] [Pubmed]
Citation: Liebers-Kyungay A, Mohnike K, Lingen C, Bressan A, Bellettato C, et al. (2022) Methodology for Generating Standardized Datasets with Characteristic Diagnostic Parameters of Rare Diseases in Form of HPO-Terms. Diagn Pathol Open 7:002. DOI: 10.4172/2476-2024.7.S11.002.
Copyright: © 2022 Liebers-Kyungay AC, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Open Access Journals
Article Tools
Article Usage
- Total views: 2309
- [From(publication date): 0-2022 - Mar 31, 2025]
- Breakdown by view type
- HTML page views: 1906
- PDF downloads: 403