Research Article Open Access
Metagenomic Study of the Liver Microbiota in Liver Cancer-Metagenomic and Metatranscriptomic Analyses of the Hepatocellular Carcinoma- Associated Microbial Communities and the Potential Role of Microbial Communities in Liver Cancer
Asmaa Ezzat1, Mohd Noor Mat Isa2, Irni Suhayu Sapian2, Zeinat Kamel3, Abd ElHady Abd ElWahab4,Ekram Hamed5 and Mahmoud ElHefnawi1,6*1Biomedical Informatics and Chemo-Informatics Group, Centre of Excellence for Advanced Sciences (CEAS), National Research centre, Ciro, Egypt
2Malaysia Genome Institute, Malaysia
3Microbiology Department, Faculty of Science, Cairo University, Egypt
4Cancer Biology Department, National Cancer Institute, Cairo, Egypt
5Radiotherapy Department, National Cancer Institute, Cairo, Egypt
6Informatics and Systems Department, Engineering Division, National Research Centre, Cairo, Egypt
- *Corresponding Author:
- Mahmoud ElHefnawi
Informatics and Systems Department
Engineering division, National Research Centre
Cairo, Egypt
Tel: (02) 01111177817
Fax: (+202) 33370931C E-mail: mahef111@gmail.com
Received date: June 21, 2014; Accepted date: October 09, 2014; Published date: October 15, 2014
Citation: Ezzat A, Mat Isa MN, Sapian IS, Kamel Z, ElHady Abd ElWahab A, et al. (2014) Metagenomic Study of the Liver Microbiota in Liver Cancer-Metagenomic and Metatranscriptomic Analyses of the Hepatocellular Carcinoma-Associated Microbial Communities and the Potential Role of Microbial Communities in Liver Cancer. J Gastroint Dig Syst 4:228. doi:10.4172/2161-069X.1000228
Copyright: © 2014 Ezzat A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Gastrointestinal & Digestive System
Abstract
Objective: Metagenomics is a new science that revolutionized microbiology for its ability to study the microbiota of a given environment without the need of culture. Human microbiota is the collection of microbes that inhabit different sites of the human body and recently its alterations were related to different human diseases especially cancers. Liver cancer incidence is continually increasing in Egypt with a high mortality rate. This study aimed to identify the abundant microbial communities that inhabit the liver of the hepatocellular carcinoma patient and may be associated with disease incidence or at least disease progression.
Methods: Fresh liver biopsy samples of two hepatocellular carcinoma Egyptian patients were obtained. DNA from one sample and RNA from other sample were extracted followed by Illumina sequencing. Taxonomic and functional analyses were performed using the MG-RAST server.
Results: Proteobacteria was the dominant phylum followed by Firmicutes and Actinobacteria in both DNA and RNA samples. Some other phyla as Chlorobi, Bacteria and Cyanobacteria had the same abundance in the two dataset. But it was noted that the bacterial diversity and presence of useful bacteria in sample 2 of grade 1 disease (RNA sample) were more than it in sample 1 of grade 2 disease (DNA sample). Also, some other phyla are found in the cDNA dataset annotations. On the other hand, infectious diseases pathways analysis showed the enrichment of infectious diseases pathways of Staphylococcus aureus infection, Vibrio cholera infection, pathogenic Escherichia coli infection, Hepatitis c, Tuberculosis, Epithelial cell signalling in Helicobacter pylori infection, Bacterial invasion of epithelial cells, and salmonella infection.
Conclusions: There is a potential link between some definite microbial communities and liver cancer that need some attention for improving early diagnosis and preventing disease progression. Further studies are required to support this
Keywords
Bacterial translocation; Gut microbiota; Hepatocellular carcinoma; Human microbiome; Liver cancer; Metagenomics; Metatranscriptomics
Introduction and Review of Relevant Literature
All surfaces of the human body, including the skin, mucosal surface and genital and gastrointestinal tracts are occupied by habitat-specific microorganisms whose cells outnumber those of the human host by ten times [1]. The human ‘metagenome’ is a composite of Homo sapiens genes and genes present in the genomes of the trillions of microbes that colonize our adult bodies. ‘Our’ microbial genomes (the microbiome) encode metabolic capacities that we have not had to evolve wholly on our own [2]. This means that the human genome is only one of the many genomes that affect Homo sapiens function. In reality, the organism we call Homo sapiens is controlled by a metagenome, a tremendous number of different genomes working in parallel [3]. Recent studies have shown that microbiota imbalances (dysbiosis) are associated with a wide variety of health problems. Inflammatory bowel diseases, obesity, atopic syndromes, various forms of colitis, and even autism, have been linked to disruptions in human-associated microbiota [4]. Metagenomics is an applied science that deals with throughput analysis of Environmental Genomic isolates [5]. It is a rapidly evolving field that emerged from rapid advances in DNA sequencing methods, with a focus on the use of culture-independent methods to study the structures, functions and dynamic operations of microbial communities [6]. Recent metagenomic assays have shown that human microbial communities can be used as biomarkers for host factors such as lifestyle and disease [7]. The complement of gene transcripts produced by the organisms in a particular environment under particular conditions (the transcriptome of the notional “meta-organism”) is the metatranscriptome. As well as representing a “giant microarray experiment” (or more accurately a giant RNA-Seq experiment), metatranscriptomics is also useful for elucidating the presence of highly-expressed genes which might have been missed by chance when sampling the metagenome itself [8]. The use of meta-transcriptomics facilitates detection of novel organisms with pathogenic potential to human and animals [9].
Liver diseases have become one of the most important causes of morbidity and mortality in the world. The gut microbiota is associated with a number of hepatic diseases from hepatocellular lipid accumulation to stellate cell activation, immune cell recruitment, and cancer development [10]. There is a strong relationship between liver and gut: the portal system receives blood from the gut, and intestinal blood content activates liver functions. The liver in turn, affects intestinal functions through bile secretion into the intestinal lumen [11]. Recent studies have proved that addition of a single agent to the gut microbiome can disrupt enterohepatic homeostasis and promote liver cancer [12].
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors worldwide [13]. HCC in Egypt affects between 5 and 7 cases per 100,000 annually with a mortality rate of 6 per 100,000 reflecting a high disease fatality [14]. Major etiologic factors associated with HCC include infection with hepatitis C (HCV) and hepatitis B (HBV) viruses, excess alcohol intake and aflatoxin B1 exposure [15]. Lehman et al. calculated age- and sex-specific and age- and sex-standardized HCC incidence rates for the eight districts in Gharbiah. They also compared rates from Gharbiah with the USA (US Surveillance Epidemiology and End Results [SEER] database). The findings of this study document the heterogeneous distribution of HCC at regional and international levels [16]. Increasing evidence suggests that derangement of gut flora is of substantial clinical relevance to patients with cirrhosis. Recent studies suggest that, gut flora contributes to the pro-inflammatory state of cirrhosis even in the absence of overt infection. Furthermore, manipulation of gut flora to augment the intestinal content of lactic acid-type bacteria at the expense of other gut flora species with more pathogenic potential may favourably influence liver function in cirrhotic patients [17]. Several recent reports have shown that HCC can occur in patients with longstanding Crohn’s disease (CD) in the absence of other underlying high-risk liver diseases [18]. Preclinical and clinical studies over the last decade have suggested a role for bacterial translocation (BT) in the pathogenesis of nonalcoholic steatohepatitis (NASH) [19]. Bacterial translocation (BT) is a phenomenon in which live bacteria or its products cross the intestinal barrier [20] to the mesenteric lymph nodes, systemic circulation, or extraintestinal organs [21]. Non-alcoholic fatty liver disease (NAFLD) comprises a spectrum of diseases ranging from simple steatosis to non-alcoholic steatohepatitis (NASH), fibrosis, and cirrhosis [22]. The liver is continually exposed to gut derived factors including bacteria and bacterial components. To combat this influx, the liver contains a large number of resident immune cells. Together, these immune cell populations in conjunction with other nonparenchymal cells orchestrate a controlled and organized response to these potentially highly inflammatory factors. However, when normal liver physiology is disrupted and inflammatory cells are activated, gut-derived factors likely augment or exacerbate certain liver diseases leading to enhanced tissue damage and propagation of inflammation [10]. Approximately 80% of hepatocellular carcinomas (HCCs) are preceded by chronic liver disease, hepatic fibrosis and cirrhosis. Chronic liver disease is also accompanied by the translocation of intestinal bacteria and pathogen-associated molecular patterns (PAMPs) —molecular motifs only found in microbes, including intestinal bacteria — which induce inflammatory immune responses [23].
In this study, we thought to investigate and characterize the abundant microbial species from liver biopsies of liver cancer patients. This could point out to a potential role of microbial species in liver cancer occurrence or development. This is performed by sequencing of one DNA sample and one RNA sample after performing microbial enrichment from two liver cancer patients liver biopsies. Then, bioinformatics analysis is conducted to identify the abundant bacterial species as well as enriched functions and infections associated with these sequences. Several bacterial species and enriched infection pathways are identified which could influence the oncogenic transformation of the liver.
Methods
Subjects
Liver biopsy samples were obtained from two donors where Liver biopsy was a part of their requested diagnosis procedures for hepatocellular carcinoma, at the National Cancer Institute in Cairo under supervision of Prof.Dr.Abdel Hady Abdel Wahab and Prof. Dr.Ikram Hamid. The Liver biopsies were obtained under informed consent and the approval of the medical research ethics committees at the National Research Centre and the National Cancer Institute. Liver samples were frozen immediately in liquid nitrogen till used.
Total DNA extraction and purification (Metagenomic study on subject 1)
Total DNA was extracted from biopsy sample of subject 1 using the QIAamp DNA Mini Kit According to the manufacturer protocol followed by ethanol purification.
Total RNA extraction and microbial mRNA enrichment (Metatranscriptomic study on subject 2)
Total RNA was extracted from the second biopsy sample of subject 2 using Thermo Scientific GeneJET RNA Purification Kit According to the manufacturer protocol of RNA purification from mammalian tissues. Ambion® DNA-free™ kit was used to remove contaminating DNA from RNA preparations, and to subsequently remove the DNase and divalent cations from the sample. Microbial RNA enrichment was done using Ambion® MICROBEnrich™ Kit. Because rRNA represents more than 90% of the bacterial RNA, Enrichment of bacterial mRNA from purified total RNA was performed using Ambion® MICROBExpress™ Kit.
DNA and RNA samples preparation and sequencing
Sample preparation and sequencing were done in collaboration with Malaysia Genome Institute
DNA sample preparation was done using Nextera XT DNA sample preparation kit according to user guide and RNA sample preparation was done using the TruSeq® RNA Sample Preparation v2 kit according to Low Sample (LS) protocol
Cluster generation and sequencing
Cluster generation reagents were prepared using TruSeq® PE Cluster Kit v3 and Sequencing was performed on Illumina HiSeq 2000 machine according to user guide.
Bioinformatics analyses
The resulted data files were uploaded to the MG-RAST (Meta Genome Rapid Annotation using Subsystem Technology, v3.3.5.4) server at (http://metagenomics.anl.gov). At the manage inbox section, compressed files were unpacked. Also, join paired –ends option was selected to join the paired end reads of each sample to produce longer reads for analysis. The DNA dataset and the cDNA dataset were submitted as eight MG-RAST jobs for the DNA dataset under the accession numbers 4523587.3 (DNA1), 4523588.3 (DNA2), 4523589.3 (DNA3), 4523590.3 (DNA4), 4523591.3 (DNA5), 4523592.3 (DNA6), 4523593.3 (DNA7) and 4523594.3 (DNA8) and as two MG-RAST jobs for the cDNA dataset under the accession numbers 4523596.3 (cDNA1) and 4523597.3 (cDNA2).
Taxonomic analysis
Taxonomic classification of Best similarity hits was reported for the DNA and the cDNA dataset separately by sequence comparison to M5NR protein database (Non-redundant multi-source protein annotation database) and Refseq protein database (NCBI reference sequences database) using a maximum e-value of 1e-5, a minimum identity of 60%, and a minimum alignment length of 15.
Functional analysis
Infectious diseases pathways analysis: The DNA and the cDNA datasets were compared to KO (KEGG orthology) using a maximum e-value of 1e-5, a minimum identity of 60 %, and a minimum alignment length of 15.
Results and Discussion
After demultiplexing, the DNA Dataset was generated as 16 files which is 8 for forward reads (R1) and 8 for reverse reads (R2) (Total size~5 GB), fragment size=350 bp. Whereas, the cDNA dataset was generated as 4 files (2 R1 and 2 R2 reads) (Total size~1.5 GB), and the fragment size=250 bp.
Subject 1 diagnosis was Grade 2 Hepatocellular carcinoma on top of liver cirrhosis and diagnosis of subject 2 was Grade 1 Hepatocellular carcinoma. Bacterial infections are very frequent in advanced cirrhosis and become the first cause of death of these patients [24]. Spontaneous bacterial peritonitis (SBP) is defined as an infection of initially sterile ascitic fluid (AF) without a detectable, surgically treatable source of infection. It is a frequent and severe complication of cirrhotic ascites, first described in the middle of the 1960s [25]. Also, Patients with cirrhosis have altered and impaired immunity, which favours bacterial translocation [26].
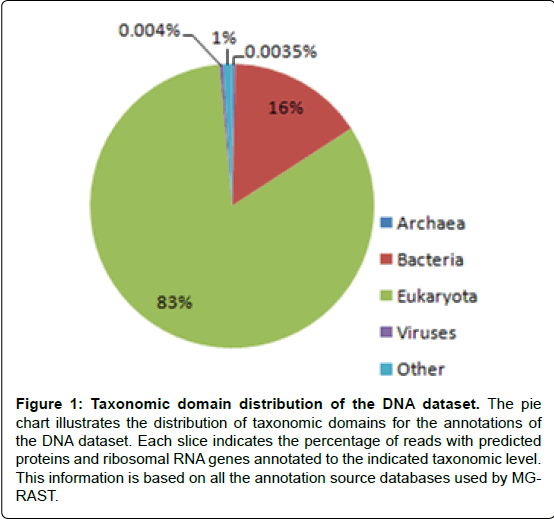
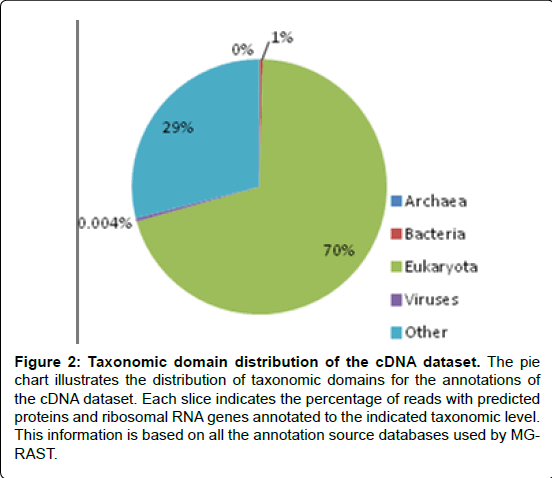
In the current study, Analysis results (Figures 1 and 2) showed that bacteria accounted for 16% of DNA sequences that passed the QC and 1% of cDNA sequences that passed the QC. The noticeable difference between the percentage of bacterial sequences between the DNA and the cDNA sequences probably resulted from the more quick degradation of bacterial mRNA than eukaryotic mRNA [27].
Figure 1: Taxonomic domain distribution of the DNA dataset. The pie chart illustrates the distribution of taxonomic domains for the annotations of the DNA dataset. Each slice indicates the percentage of reads with predicted proteins and ribosomal RNA genes annotated to the indicated taxonomic level. This information is based on all the annotation source databases used by MGRAST.
Figure 2: Taxonomic domain distribution of the cDNA dataset. The pie chart illustrates the distribution of taxonomic domains for the annotations of the cDNA dataset. Each slice indicates the percentage of reads with predicted proteins and ribosomal RNA genes annotated to the indicated taxonomic level. This information is based on all the annotation source databases used by MGRAST.
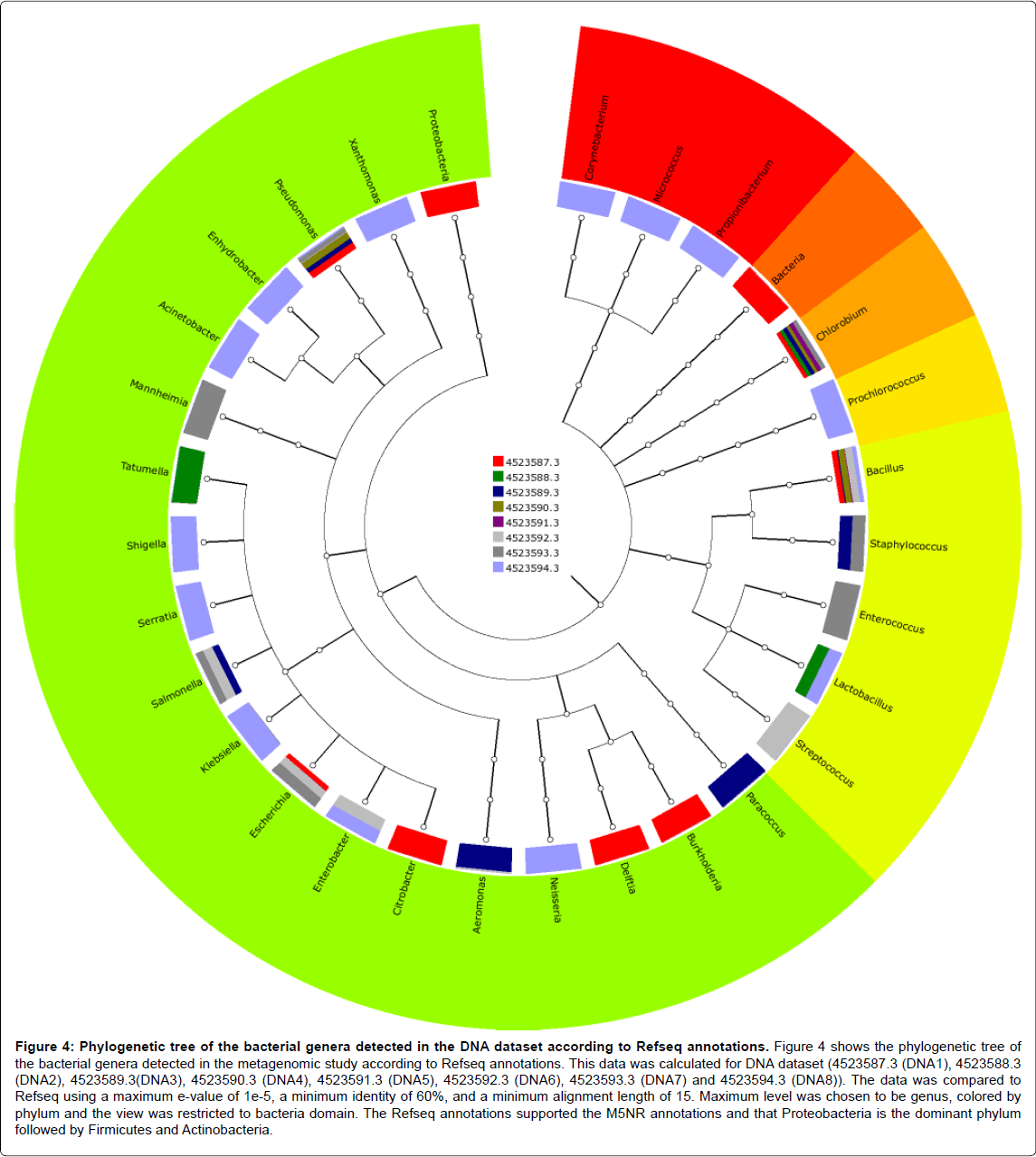
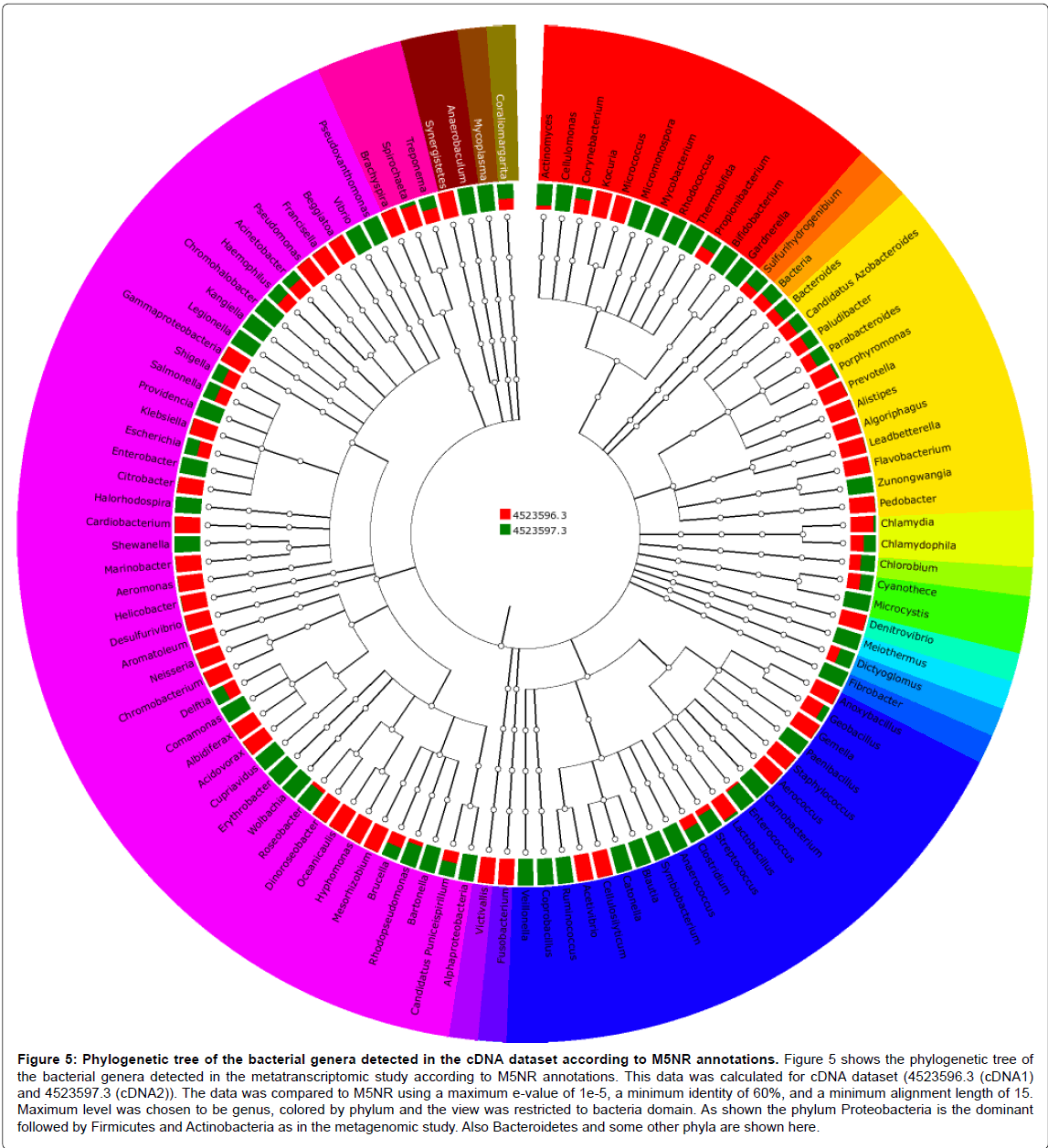
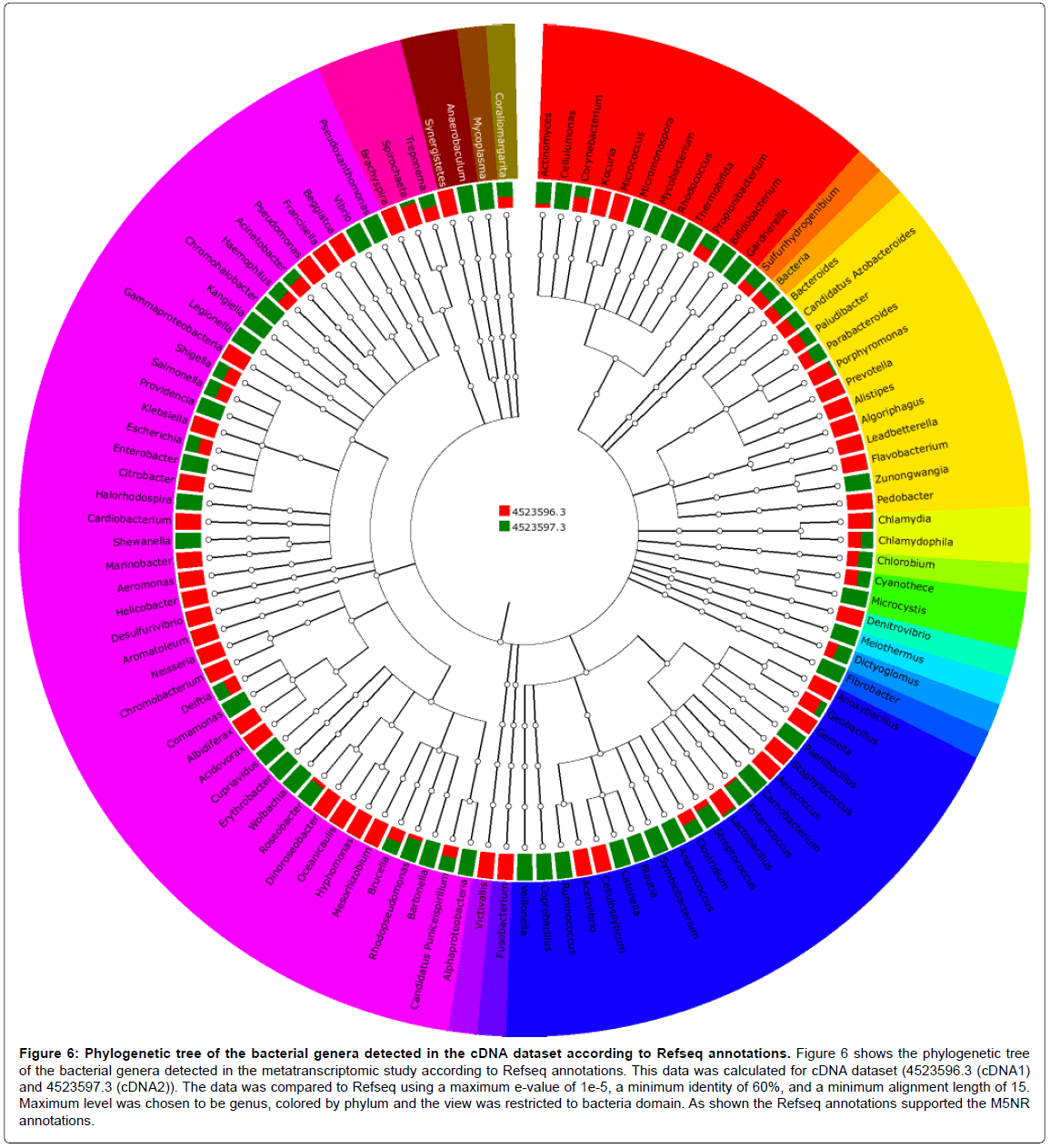
Annotation of taxonomic affiliation of DNA and cDNA sequences with M5NR and Refseq databases of MG-RAST showed that the dominant bacterial phylum in the two samples is Proteobacteria followed by Firmicutes and Actinobacteria as shown in Figures 3-6. Some other phyla as Chlorobi, Bacteria and Cyanobacteria had the same abundance in the two dataset. Also, Bacteroides and some other phyla are found in the cDNA dataset annotations. The Proteobacteria are a major group (phylum) of bacteria. They include a wide variety of pathogens, such as Escherichia, Salmonella, Vibrio, Helicobacter, and many other notable genera [28].
Figure 3: Phylogenetic tree of the bacterial genera detected in the DNA dataset according to M5NR annotations. Figure 3 shows the phylogenetic tree of the bacterial genera detected in the metagenomic study according to M5NR annotations. Genera that belong to the same phylum were colored with the same color. This data was calculated for DNA dataset (4523587.3 (DNA1), 4523588.3 (DNA2), 4523589.3(DNA3), 4523590.3 (DNA4), 4523591.3 (DNA5), 4523592.3 (DNA6), 4523593.3 (DNA7) and 4523594.3 (DNA8)). The data was compared to M5NR using a maximum e-value of 1e-5, a minimum identity of 60%, and a minimum alignment length of 15. Maximum level was chosen to be genus, colored by phylum and the view was restricted to bacteria domain. It is noted from the figure that Proteobacteria is the dominant phylum followed by Firmicutes and Actinobacteria.
Figure 4: Phylogenetic tree of the bacterial genera detected in the DNA dataset according to Refseq annotations. Figure 4 shows the phylogenetic tree of the bacterial genera detected in the metagenomic study according to Refseq annotations. This data was calculated for DNA dataset (4523587.3 (DNA1), 4523588.3 (DNA2), 4523589.3(DNA3), 4523590.3 (DNA4), 4523591.3 (DNA5), 4523592.3 (DNA6), 4523593.3 (DNA7) and 4523594.3 (DNA8)). The data was compared to Refseq using a maximum e-value of 1e-5, a minimum identity of 60%, and a minimum alignment length of 15. Maximum level was chosen to be genus, colored by phylum and the view was restricted to bacteria domain. The Refseq annotations supported the M5NR annotations and that Proteobacteria is the dominant phylum followed by Firmicutes and Actinobacteria.
Figure 5: Phylogenetic tree of the bacterial genera detected in the cDNA dataset according to M5NR annotations. Figure 5 shows the phylogenetic tree of the bacterial genera detected in the metatranscriptomic study according to M5NR annotations. This data was calculated for cDNA dataset (4523596.3 (cDNA1) and 4523597.3 (cDNA2)). The data was compared to M5NR using a maximum e-value of 1e-5, a minimum identity of 60%, and a minimum alignment length of 15. Maximum level was chosen to be genus, colored by phylum and the view was restricted to bacteria domain. As shown the phylum Proteobacteria is the dominant followed by Firmicutes and Actinobacteria as in the metagenomic study. Also Bacteroidetes and some other phyla are shown here.
Figure 6: Phylogenetic tree of the bacterial genera detected in the cDNA dataset according to Refseq annotations. Figure 6 shows the phylogenetic tree of the bacterial genera detected in the metatranscriptomic study according to Refseq annotations. This data was calculated for cDNA dataset (4523596.3 (cDNA1) and 4523597.3 (cDNA2)). The data was compared to Refseq using a maximum e-value of 1e-5, a minimum identity of 60%, and a minimum alignment length of 15. Maximum level was chosen to be genus, colored by phylum and the view was restricted to bacteria domain. As shown the Refseq annotations supported the M5NR annotations.
According to both M5NR and Refseq annotations in the current study, some specific genera were specified in both DNA and cDNA datasets as shown in the taxonomic Tables 1-4 (Included as supplementary data)[see Additional file.doc]. These genera included: Chlorobium, Acinetobacter, Propionibacterium, Pseudomonas, Micrococcus, Nesseria, Escherichia, Corynebacterium, Salmonella, Shigella, Staphylococcus, Enterococcus, Lactobacillus, and Delftia.
Klebsiella, Streptococcus and Enterobacter genera were found in the M5NR and Refseq annotations of the DNA dataset and M5NR annotations of the cDNA dataset. Also Citrobacter, Xanthomonas and Aeromonas were found in M5NR and Refseq annotations of the DNA dataset and Refseq annotations of the cDNA dataset Whereas, Geobacillus and Comamonas were found in M5NR annotations of the DNA dataset and M5NR and Refseq annotations of the cDNA dataset.
In the same time there was an abundance of Bacillus, Enhydrobacter, Paracoccus, Burkholderia, Serratia, Tatumella and Mannheimia in both M5NR and Refseq of the DNA dataset only.
Also, M5NR and Refseq annotations of the cDNA dataset only supported the abundance of Cyanothece, Beggiatoa, Brucella, Clostridium, Treponema and Bacteroides and the presence of Vibrio, Kangiella, Legionella and Helicobacter and some other genera.
On the other hand, in the functional analysis, the datasets were annotated with KO of MG-RAST. Cancers, Endocrine and metabolic diseases, Immune diseases and Infectious diseases had the highest hit numbers in both DNA and cDNA datasets.
Samaras et al. searched Pubmed, Cochrane, and Scopus without time limits for relevant articles to evaluate the evidence regarding the association of such infections with the development of malignancy, excluding the overwhelming evidence of the association of Helicobacter pylori and cancer. They found that there is evidence that some bacterial and parasitic infections are associated with cancer development. The level of evidence of this association varies from high to low; in any case, a long time interval is mandatory for the development of cancer. A high level of evidence exists for the association of Salmonella typhi with gallbladder and hepatobiliary carcinoma; Opisthorchis viverrini and Clonorchis sinensis with cholangiocarcinoma and Schistosoma hematobium with bladder cancer [29].
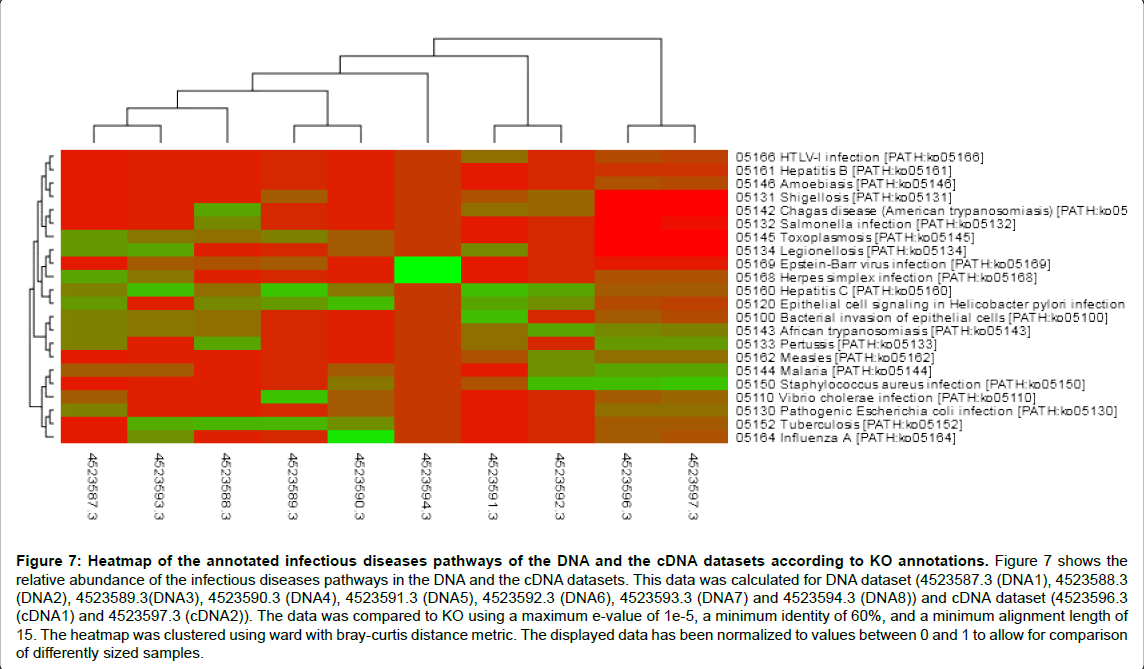
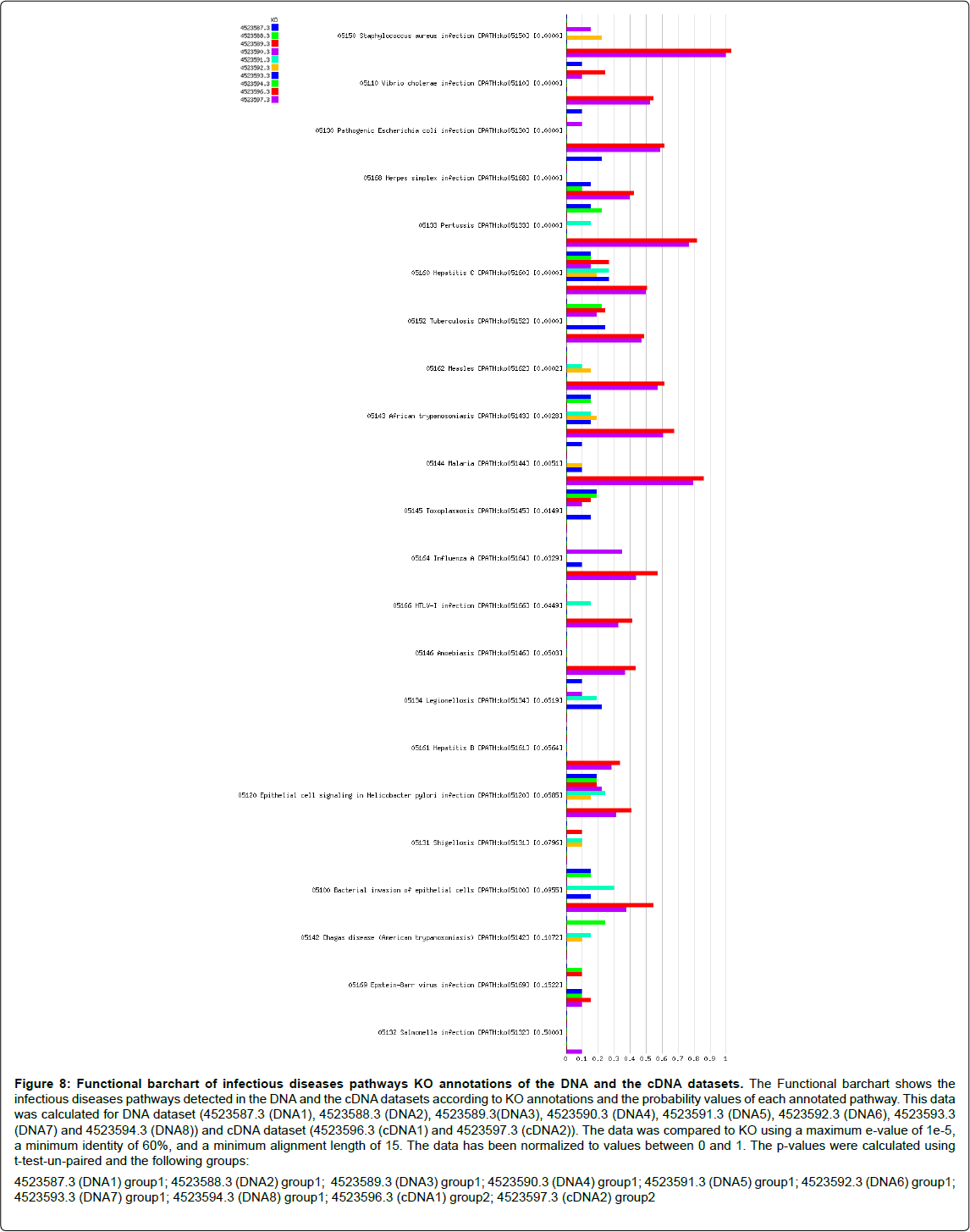
Here, in infectious diseases pathways analysis, level 3 KO annotations showed that among the determined infections pathways enriched in both DNA and cDNA datasets were Staphylococcus aureus infection, Vibrio cholera infection, pathogenic Escherichia coli infection, Pertussis, Hepatitis c and Tuberculosis with a probability value equal 0.0000 for all. Also, African trypanosomiasis (p value=0.0028), Malaria (p value=0.0051), Epithelial cell signaling in Helicobacter pylori infection (p value=0.0585), Bacterial invasion of epithelial cells (p value=0.0955), and salmonella infection (p value=0.5000) were detected as shown in Figures 7 and 8. Tables 5 and 6 (Included as supplementary data) show this in more detail [see Additional file.doc]. Also, Toxoplasmosis, Legionellosis, Shigellosis and Chagas disease were detected in DNA dataset whereas; Amoebiasis and Hepatitis B were detected in cDNA dataset. Infectious diseases pathways analysis showed the expression of anti-inflammation proteins against inflammation caused by some infectious organisms which reveal the potential role of these organisms in disease incidence or by sure in disease progression. Despite of abundance of Shigella and Legionella species in the taxonomic classification of cDNA dataset, Shigellosis and Legionellosis were not determined as infectious diseases in functional analysis although, they were determined as infectious diseases in DNA dataset of sample 1 of grade 2 disease. This probably means that those species may lead to disease progression with time. These infections may be also associated with cirrhosis. Bacterial infection is a frequent and severe complication of cirrhosis [30]. Also it was noted that sample one have more aggressive bacterial species from the same genus than sample two and this confirm its correlation with disease development.
Figure 7: Heatmap of the annotated infectious diseases pathways of the DNA and the cDNA datasets according to KO annotations. Figure 7 shows the relative abundance of the infectious diseases pathways in the DNA and the cDNA datasets. This data was calculated for DNA dataset (4523587.3 (DNA1), 4523588.3 (DNA2), 4523589.3(DNA3), 4523590.3 (DNA4), 4523591.3 (DNA5), 4523592.3 (DNA6), 4523593.3 (DNA7) and 4523594.3 (DNA8)) and cDNA dataset (4523596.3 (cDNA1) and 4523597.3 (cDNA2)). The data was compared to KO using a maximum e-value of 1e-5, a minimum identity of 60%, and a minimum alignment length of 15. The heatmap was clustered using ward with bray-curtis distance metric. The displayed data has been normalized to values between 0 and 1 to allow for comparison of differently sized samples.
Figure 8: Functional barchart of infectious diseases pathways KO annotations of the DNA and the cDNA datasets. The Functional barchart shows the infectious diseases pathways detected in the DNA and the cDNA datasets according to KO annotations and the probability values of each annotated pathway. This data was calculated for DNA dataset (4523587.3 (DNA1), 4523588.3 (DNA2), 4523589.3(DNA3), 4523590.3 (DNA4), 4523591.3 (DNA5), 4523592.3 (DNA6), 4523593.3 (DNA7) and 4523594.3 (DNA8)) and cDNA dataset (4523596.3 (cDNA1) and 4523597.3 (cDNA2)). The data was compared to KO using a maximum e-value of 1e-5, a minimum identity of 60%, and a minimum alignment length of 15. The data has been normalized to values between 0 and 1. The p-values were calculated using t-test-un-paired and the following groups: 4523587.3 (DNA1) group1; 4523588.3 (DNA2) group1; 4523589.3 (DNA3) group1; 4523590.3 (DNA4) group1; 4523591.3 (DNA5) group1; 4523592.3 (DNA6) group1; 4523593.3 (DNA7) group1; 4523594.3 (DNA8) group1; 4523596.3 (cDNA1) group2; 4523597.3 (cDNA2) group2
However, the more abundance of Lactobacillus species in the cDNA dataset (generated from sample 2 of Grade 1 disease) than in the DNA dataset and the abundance of Bifidobacterium species in the cDNA dataset may be associated with the positive role of some normal intestinal microbiota in playing a defensive role against pathogens.
This was in agreement with the results obtained by Singh et al. that investigated the status of bacterial colonization and inflammation in the normal, cirrhotic primary biliary cirrhosis (PBC), and nonalcoholic steatohepatitis (NASH) human liver tissues. Comparatively normal livers showed increased bacterial colonization than PBC and NASH. They analyzed mRNA levels of Toll-like receptors (TLR) 2 and TLR4, and protein levels of TLR4. Phosphorylated IKKα (pIKKα) protein estimation served as a marker for nuclear factor-kappa B (NF-κB) activation. In spite of the increased bacterial colonization in normal liver tissues, lower levels of TLR2/4 mRNA and TLR4 and pIKKα proteins were found compared to PBC and NASH indicating the maintenance of suppressed inflammation and immune tolerance in normal livers. They mentioned that this is the first clinical evidence showing suppressed inflammation despite bacterial colonization in normal human livers thus maintaining liver immune homeostasis [31].
The results of the current study supported the analysis results of fecal microbiome in patients with hepatitis B and alcoholic liver cirrhosis that demonstrated an increase in pathogenic Enterobacteriaceae and Streptococcaceae, while beneficial Bifidobacteria and Lachnospiraceae were decreased. In addition, liver cirrhosis induced by carbon tetrachloride in rats was also associated with high levels of Enterobacteraceae. Another study reported a decrease in Gram-positive anaerobic Clostridium groups and an increase in the aerobic/anaerobic bacterial ratio in mice with fibrosis. For animals with cirrhosis, treatments with antibiotics (e.g. norfloxacin) or probiotics (e.g. Bifidobacterium) reduces Enterobacter, while it increases Bifidobacterium and Lactobacillus, resulting in decreased systemic endotoxin levels and improved liver function [32]. Yilmaz et al. presented an immunocompetent case of liver abscess caused by Streptococcus anginosus originated most probably from oral flora [33]. Spencer et al. performed a 2-month inpatient study of 15 female subjects who were placed on well-controlled diets in which choline levels were manipulated. Variations between subjects in levels of Gammaproteobacteria and Erysipelotrichi were directly associated with changes in liver fat in each subject during choline depletion. The Gammaproteobacteria genera identified in their study, including Klebsiella spp, Enterobacter spp, and Escherichia spp, are known gramnegative bacteria with lipopolysaccharide- containing membranes [34].
Although most infectious microorganisms that reach the liver via the blood are rapidly cleared, it is evident from the success of certain pathogens — such as the hepatitis B and hepatitis C viruses and malariacausing Plasmodium spp. in establishing hepatic infections that the liver provides a favourable environment for escaping immune responses [35]. Also, Dapito et al. tested the hypothesis that the intestinal microbiota and Toll-like receptors (TLRs) promote hepatocellular carcinoma (HCC), a long-term consequence of chronic liver injury, inflammation, and fibrosis. Hepatocarcinogenesis in chronically injured livers depended on the intestinal microbiota and TLR4 activation in non-bone-marrowderived resident liver cells. TLR4 and the intestinal microbiota were not required for HCC initiation but for HCC promotion, mediating increased proliferation, expression of the hepatomitogen epiregulin, and prevention of apoptosis. Gut sterilization restricted to late stages of hepatocarcinogenesis reduced HCC, suggesting that the intestinal microbiota and TLR4 represent therapeutic targets for HCC prevention in advanced liver disease [36]. DeLeon et al. examined whether gallium maltolate (GaM), a novel formulation of Ga, had antibacterial activity in a thermally injured acute infection mouse model. Dose-response studies indicated that a GaM dose as low as 25 mg/kg of body weight delivered subcutaneously was sufficient to provide 100% survival in a lethal P. aeruginosa-infected thermally injured mouse model. Mice treated with 100 mg/kg GaM had undetectable levels of Pseudomonas aeruginosa in their wounds, livers, and spleens, while the wounds of untreated mice were colonized with over 108 P. aeruginosa CFU/g of tissue and their livers and spleens were colonized with over 105 P. aeruginosa CFU/g of tissue. GaM also significantly reduced the colonization of Staphylococcus aureus and Acinetobacter baumannii in the wounds of thermally injured mice. Furthermore, GaM was also therapeutically effective in preventing preestablished P. aeruginosa infections at the site of the injury from spreading systemically [37,38].
Conclusions
It can be concluded that there is a potential link between the microbial communities and liver cancer that needs further verification and studies. We conclude also that certain bacterial phyla such as proteobacteria were the most abundant type of bacteria in both samples. This is in agreement with previous studies highlighting the role of H. pylori in liver cancer and stomach and gut cancers and pathogenic E. coli in gut diseases.
Also, the present study indicates to a contributing probability of some infections in pathogenesis and progression of liver cancer. Also, these data indicate to the key role of bacterial translocation of some intestinal microflora or its products in disruption of liver homeostasis.
This study is a preliminary study with our available resources in order to shed a light on the question of the relation between the gut microbiota and liver cancer. More studies to confirm the conclusions of the paper are needed in the future.
Acknowledgements
We acknowledge useful discussions with Dr Rania Siam, head of biology department at AUC. We also acknowledge the help of both National Research Centre and Malaysia Genome Institute in performing the work.
References
- Nam YD, Jung MJ, Roh SW, Kim MS, Bae JW (2011) Comparative analysis of Korean human gut microbiota by barcoded pyrosequencing. PLoS One 6: e22109.
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, et al. (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027-1031.
- Proal AD, Albert PJ, Marshall T (2009) Autoimmune disease in the era of the metagenome. Autoimmun Rev 8: 677-681.
- Peris-Bondia F, Latorre A, Artacho A, Moya A, D'Auria G (2011) The active human gut microbiota differs from the total microbiota. PLoS One 6: e22448.
- Patake RS, Patake GR (2011) A Mini Review on Metagenomics and its Implications in Ecological and Environmental Biotechnology. Universal Journal of Environmental Research and Technology 1: 1-6.
- Turnbaugh PJ, Gordon JI (2009) The core gut microbiome, energy balance and obesity. J Physiol 587: 4153-4158.
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, et al. (2011) Metagenomic biomarker discovery and explanation. Genome Biol 12: R60.
- http://www.jic.ac.uk/staff/john-walshaw/metaomics.html.
- Wittekindt NE, Padhi A, Schuster SC, Qi J, Zhao F, et al. (2010) Nodeomics: pathogen detection in vertebrate lymph nodes using meta-transcriptomics. PLoS One 5: e13432.
- Son G, Kremer M, Hines IN (2010) Contribution of gut bacteria to liver pathobiology. Gastroenterol Res Pract 2010.
- Cesaro C, Tiso A, Del Prete A, Cariello R, Tuccillo C, et al. (2011) Gut microbiota and probiotics in chronic liver diseases. Dig Liver Dis 43: 431-438.
- Fox JG, Feng Y, Theve EJ, Raczynski AR, Fiala JL, et al. (2009) Gut microbes define liver cancer risk in mice exposed to chemical and viral transgenic hepatocarcinogens. Gut 59: 88-97.
- Blum HE (2005) Hepatocellular carcinoma: therapy and prevention. World J Gastroenterol 11: 7391-7400.
- The Egyptian Society for Liver Cancer ESfL (2012) The Egyptian Society for Liver Cancer third annual conference reveals Hepatocellular Carcinoma: Current and Future Prospects .
- Elgendy SM, Hessien M, Elsherbiny MM, Abd El-Salam IM, El-Attar IA, et al. (2005) A panel of molecular markers in hepatitis C virus-related hepatocellular carcinoma. J Egypt NatlCancInst 17: 270-278.
- Lehman EM, Soliman AS, Ismail K, Hablas A, Seifeldin IA, et al. (2008) Patterns of hepatocellular carcinoma incidence in Egypt from a population-based cancer registry. Hepatol Res 38: 465-473.
- Almeida J, Galhenage S, Yu J, Kurtovic J, Riordan SM (2006) Gut flora and bacterial translocation in chronic liver disease. World J Gastroenterol 12: 1493-1502.
- Zheng SM, Cui DJ, Ouyang Q (2011) Gut-liver axis plays a role in hepatocarcinogenesis of patients with Crohn's disease. World J Gastroenterol 17: 3171-3172.
- Ilan Y (2012) Leaky gut and the liver: a role for bacterial translocation in nonalcoholic steatohepatitis. World J Gastroenterol 18: 2609-2618.
- Balzan S, de Almeida Quadros C, de Cleva R, Zilberstein B, Cecconello I (2007) Bacterial translocation: overview of mechanisms and clinical impact. J GastroenterolHepatol 22: 464-471.
- Fouts DE, Torralba M, Nelson KE, Brenner DA, Schnabl B (2012) Bacterial translocation and changes in the intestinal microbiome in mouse models of liver disease. J Hepatol 56: 1283-1292.
- Hakansson A, Molin G (2011) Gut microbiota and inflammation. Nutrients 3: 637-682.
- Alderton GK (2012) Inflammation: the gut takes a toll on liver cancer. Nat Rev Cancer 12: 379.
- Fernández J, Gustot T (2012) Management of bacterial infections in cirrhosis. J Hepatol 56 Suppl 1: S1-12.
- Lata J, Stiburek O, Kopacova M (2009) Spontaneous bacterial peritonitis: a severe complication of liver cirrhosis. World J Gastroenterol 15: 5505-5510.
- Pleguezuelo M, Benitez JM, Jurado J, Montero JL, De la Mata M (2013) Diagnosis and management of bacterial infections in decompensated cirrhosis. World J Hepatol 5: 16-25.
- Yu K, Zhang T (2012) Metagenomic and metatranscriptomic analysis of microbial community structure and gene expression of activated sludge. PLoS One 7: e38183.
- http://en.wikipedia.org/wiki/Proteobacteria
- Samaras V, Rafailidis PI, Mourtzoukou EG, Peppas G, Falagas ME (2010) Chronic bacterial and parasitic infections and cancer: a review. J Infect DevCtries 4: 267-281.
- Yang YY, Lin HC (2005) Bacterial infections in patients with cirrhosis. J Chin Med Assoc 68: 447-451.
- Singh R, Bullard J, Kalra M, Assefa S, Kaul AK, et al. (2011) Status of bacterial colonization, Toll-like receptor expression and nuclear factor-kappa B activation in normal and diseased human livers. ClinImmunol 138: 41-49.
- Seki E, Schnabl B (2012) Role of innate immunity and the microbiota in liver fibrosis: crosstalk between the liver and gut. J Physiol 590: 447-458.
- Yalmaz H, Yalmaz EM, Karadag A, Esen S, Sunbul M, et al. (2012) Liver abscess associated with an oral flora bacterium Streptococcus anginosus. Microbiology and Infectious Diseases 2: 33-35.
- Spencer MD, Hamp TJ, Reid RW, Fischer LM, Zeisel SH, et al. (2011) Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology 140: 976-986.
- Protzer U, Maini MK, Knolle PA (2012) Living in the liver: hepatic infections. Nat Rev Immunol 12: 201-213.
- Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, et al. (2012) Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 21: 504-516.
- DeLeon K, Balldin F, Watters C, Hamood A, Griswold J, et al. (2009) Gallium maltolate treatment eradicates Pseudomonas aeruginosa infection in thermally injured mice. Antimicrob Agents Chemother 53: 1331-1337.
- http://metagenomics.anl.gov
Relevant Topics
- Constipation
- Digestive Enzymes
- Endoscopy
- Epigastric Pain
- Gall Bladder
- Gastric Cancer
- Gastrointestinal Bleeding
- Gastrointestinal Hormones
- Gastrointestinal Infections
- Gastrointestinal Inflammation
- Gastrointestinal Pathology
- Gastrointestinal Pharmacology
- Gastrointestinal Radiology
- Gastrointestinal Surgery
- Gastrointestinal Tuberculosis
- GIST Sarcoma
- Intestinal Blockage
- Pancreas
- Salivary Glands
- Stomach Bloating
- Stomach Cramps
- Stomach Disorders
- Stomach Ulcer
Recommended Journals
Article Tools
Article Usage
- Total views: 16648
- [From(publication date):
October-2014 - Nov 21, 2024] - Breakdown by view type
- HTML page views : 12034
- PDF downloads : 4614