Research Article Open Access
Maximum α-Amylase Production by Molecular and Biochemical Characterized Soil Microorganism
Pushpendra Singh1*, Rajesh Sharma2 and Ravindra Singh11Department of Biological sciences, Mahatma Gandhi Chitrakoot Gramodaya Vishwavidhyalaya, Chitrakoot, Madhya Pradesh, India
2Department of Biotechnology, Veer Bahadur Singh Purvanchal University, Jaunpur, Uttar Pradesh, India
- Corresponding Author:
- Singh P
Department of Biological sciences
Mahatma Gandhi Chitrakoot Gramodaya Vishwavidhyalaya
Chitrakoot, Madhya Pradesh, India
Tel: 07309890484
E-mail: pushpendrasinghbiotech@gmail.com
Received date: June 13, 2017; Accepted date: June 17, 2017; Published date: July 24, 2017
Citation: Singh P, Sharma R, Singh R (2017) Maximum α-Amylase Production by Molecular and Biochemical Characterized Soil Microorganism. J Biotechnol Biomater 7:266. doi:10.4172/2155-952X.1000266
Copyright: © 2017 Singh P, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited
Visit for more related articles at Journal of Biotechnology & Biomaterials
Abstract
The present investigation was undertaken with an aim to produce α-amylase in cost effective way keeping its market potential into consideration. This result obtained from present study prove that Bacillus licheniformis is the maximum α-amylase producing strain isolated from soil under suitable conditions. This study will very helpful for upcoming researchers to uncover the various aspect of enzyme production by microorganism. The present investigation was undertaken with an aim to produce higher amount of amylase with pure compounds in submerged fermentation. To screen the isolates for promising α-amylase producing capability and one of the most promising isolate is further scale up. Amylase constitutes a class of industrial enzymes having approximately 20% of the enzyme market. α-amylase has found its application in a range of industries including food, brewing, distilling industry, textile, paper pharmaceutical and bioconversion of solid waste etc. Microorganism used in present investigation, isolated from rhizospheric regions of giant newar variety of Raphanus sativus grown in district Jaunpur, (Uttar Pradesh) India. A total of 30 natural isolates were scanned for α-amylase activity out of which 20 isolates were biochemically characterized. Based on ribotyping all the isolates were identified and one of the most promising amylase producer, i.e., Bacillus licheniformis was finally selected for further studies. The experiments were carried out in triplicates and mean value was taken. Nutritional source such as carbon and nitrogen sources were optimized for the production of α-amylase in free cell condition. Maximum α-amylase production was observed at 24 h. of incubation. Three different types of media such as Semi-synthetic, complex media I and complex media II tested and various parameters were optimized for enhance the alpha-amylase yield. It was found that identified soil microorganism Bacillus licheniformis is the best strain for α-amylase production and complex media-I gave maximum yield.
Keywords
α-amylase; Submerged fermentation; Starch hydrolysis; Ribotyping; Bio processing
Introduction
α-Amylases (E.C. 3.21.1.) are starch-degrading enzymes that catalyze the hydrolysis of internal α-1,4 glycosidic bonds in polysaccharides with the retention of α-anomeric configuration in the products. These enzymes account for about 30% of the world’s enzyme production and the most important enzymes and are of great significance for biotechnological applications. With the advancement in biotechnology, the amylase application has expanded in many fields such as clinical, medicinal and analytical chemistry, as well as their widespread application in starch saccharification and in the textile, food, brewing and distilling industries. Amylases of microorganisms have broad spectrum of industrial applications as they are more stable than when prepared with plant and animal amylases.
The major advantage of using microorganisms for the production of amylases is the economical bulk production capacity and the fact that microbes are easy to manipulate to obtain enzymes of desired characteristics. The further support of energy consumption studies, such as those on thermodynamics, pinch technology, and environment-friendly technologies, has hastened the large-scale production of the enzyme [1]. Today a large number of extracellular microbial amylases are available commercially and they have almost completely replaced chemical hydrolysis of starch. Bacillus sp. is a soil borne bacterium has been found to be the best candidate for commercial production of this enzyme. Thermophilic Bacillus sp. was found to produce thermostable amylase. Large scale industrial production of this enzyme is carried out mainly in submerged culture using different types of microorganisms. Further increase in incubation period did not show any significant increase in enzyme production rather it was decreased [2]. Some Bacillus strains produce enzyme in the exponential phase, whereas some others in the mid stationary phase. The designed model based upon the multi-variable analysis provides the strategy to produce amylase at a large scale from this strain. Further study is in progress to scale up enzyme production in a batch fermenter using Box-Behnken design (BBD) optimized conditions [3].
Materials and Method
Experimental organisms
The root adhering samples were collected from radish (cv. Jaunpuri giant) grown field at 45 days of growth. The plants were sampled from radish grown area of district Jaunpur 82°44’E longitude and 25°46’N latitude, Uttar Pradesh (UP), India. The plants were carefully removed from the soil and the roots with adherent soil were put in plastic bags for isolation of bacteria. For the isolation of bacteria, the roots were shaken to remove excess soil. Ten gram of closely associated rhizospheric soil from each sample was added to 90 ml of sterile water and shaken for 30 min on a mechanical rotary shaker. Tenfold dilution were made and plated on nutrient agar (NA) medium. Bacterial cultures were maintained on the NA slants and stored at 4°C till further use. 30 isolates were picked randomly for preliminary screening having positive results for amylase activity by addition of iodine solution and observed for zone of clearance. Isolates having positive results will be further biochemically characterized and identified based on their ribotypes. On the basis of zone of clearance one of the most potential amylase producers will be finally selected for further scale up studies.
Biochemical characterization
All of 30 selected isolates were scanned for maximum amylase activity on starch agar plates. The composition of starch agar (in g/L) is soluble starch 20.0 g, Peptone 5.0 g, Beef Extract 3.0 g, Agar agar 20.0 g, Double Distilled water and pH at 7.0.
Starch hydrolysis
STARCH AGAR MEDIUM COMPOSITION (in g/L): Peptone 5.0 g, Beef extract 3.0 g, Soluble starch 2.0 g, Agar agar 25.0 g, Distilled water 1000 ml and pH 7.0, Iodine solution. Starch agar medium was prepared by adding the constituents separately in double distilled water. The medium was autoclaved and poured into Petri Plates under laminar flow. Using sterile technique, a single line streak inoculation of each isolates was done as a stab inoculation on agar. All the inoculated plates were incubated at 37°C for 24-48 h. The plates were flooded with iodine solution and allowed to remain in the medium for 30 s, excess starch is poured off. The cultures were observed for presence of blue black color surrounding the each isolates.
Molecular Characterization of Selected Isolates
Preparation of template DNA of isolates
Isolated bacterial samples were grown at 30°C in 25 ml LB Medium on a shaker until OD reached 0.3 to 0.5. Centrifuged the culture broth of bacteria at 10000 rpm, at 28°C for 5 min and collected the pellet by discarding supernatant. Pellet was rinsed with 1 ml TE (10 mM Tris-HCl pH-8.0 and 1 mM EDTA pH-8.0), repeated this step twice and supernatant were discarded. Resuspended the pellet in 0.5 ml SET buffer (75 mM NaCl, 25 mM EDTA pH-8.0, 20 mM Tris-HCl pH-8.0). 10 μL (10 mg/ml) was added to above suspension and incubated for 30-60 min at 37°C. 0.1 ml of the 10% SDS and 10 μL of protinase K was added to above suspension and incubated for 60 min at 55°C. 0.3 ml of 5 M NaCl and equal volume phenol:chloroform:isoamyl alcohol (25:24:1) (Tris/water saturated phenol) were added and incubated at room temperature for 15 min followed by gentle extraction. Centrifuged (Remi) at 10000 rpm and 4°C for 5 min and supernatant was discarded carefully. Washed the pellet with 70% ethanol and again centrifuged at 10000 rpm at 4°C for 5 min (this step was repeated twice). Dried the pellet and dissolved in 40 μL TE and stored at 4°C.
PCR amplification
DNA was amplified by mixing template DNA (50 ng) with the polymerase reaction buffer, dNTP mix (100 μM). Primers PA and PH (100 ng each) and 1.0 U Taq polymerase. The reaction master mix was prepared by using the following reagents- Deionized water 78.0 μL, 10X Taq Pol buffer 10.0 μL, Taq polymerase (1.0 U) 1.0 μL, dNTP’s (2 mM) 6.0 μL, Primer PA (100 ng) 1.5 μL, Primer PH (100 ng) 1.5 μL, Target DNA (50 ng) 1.0 μL.
Added the reagents in the required volumes and all the operations had been carried out on ice. In addition to the samples, negative control (i.e., without template DNA) had been maintained simultaneously to check for possible contamination. Capped all the PCR tubes and vortexed them to mix well. Pulsed the tubes for 2-3 s in a micro centrifuge to pool the contents at the bottom. Overlayed the reaction mixture with 50 μL of sterilized mineral oil and capped the tubes then the DNA amplification was done in thermal cycler (Biorad). Amplified DNA will be examined by horizontal electrophosesis in 0.8% agarose gel.
Genomic DNA extraction and amplified ribosomal DNA restriction analysis (ARDRA)
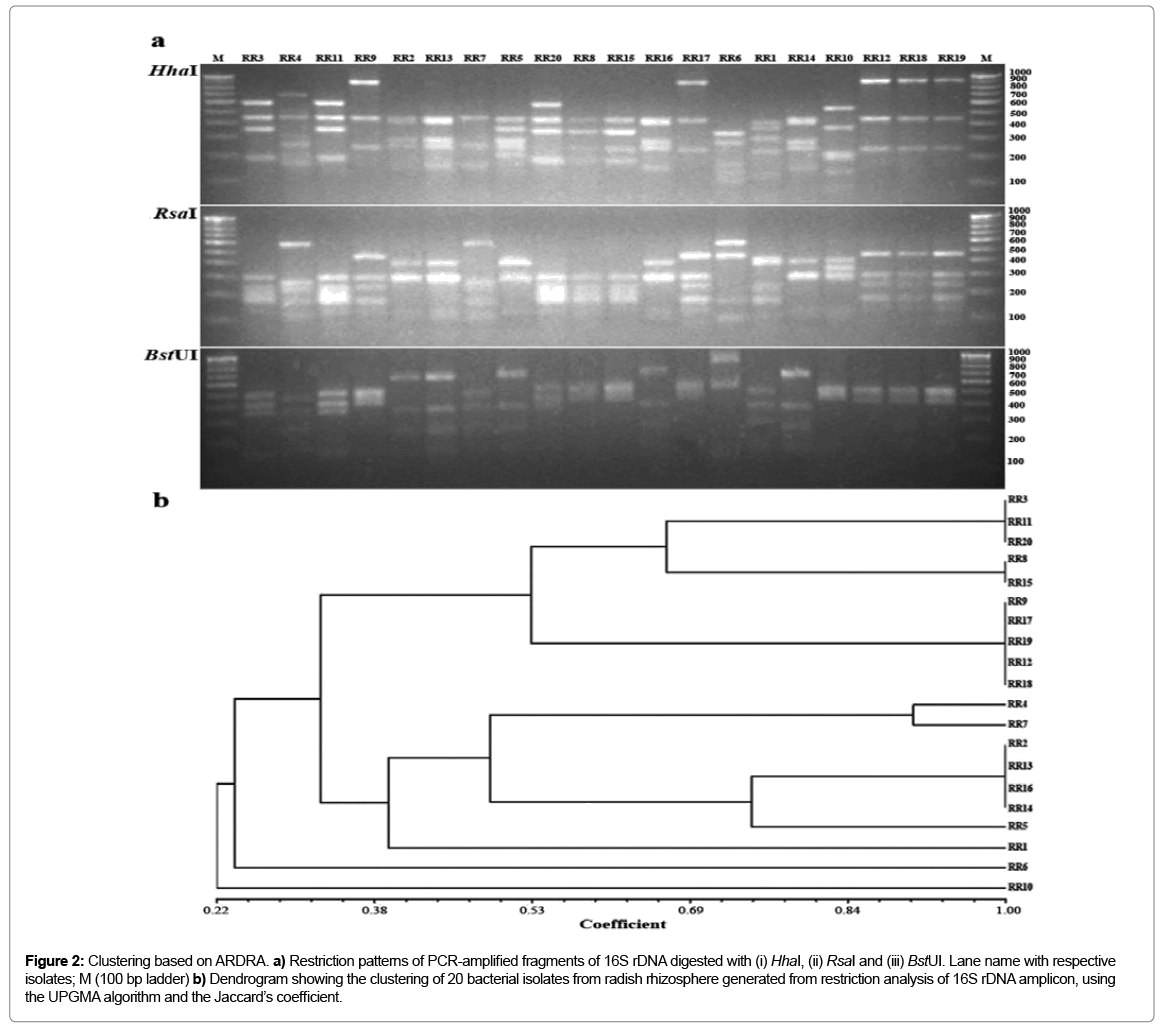
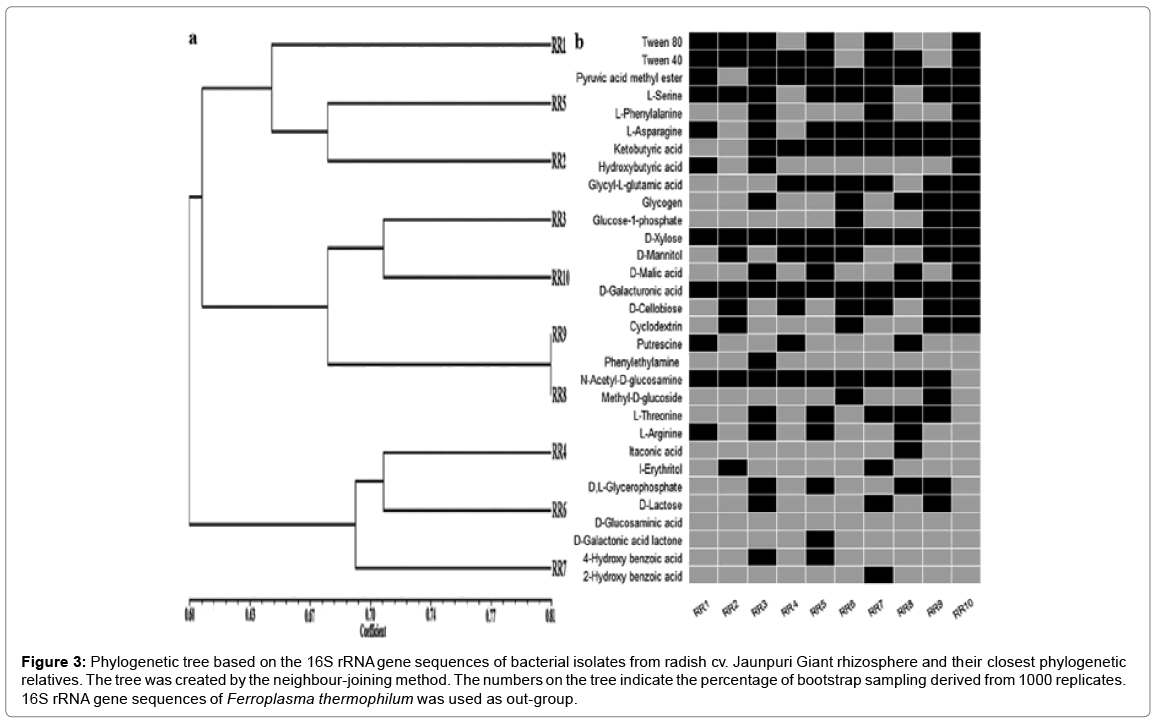
Total genomic DNA extraction from selected isolates was performed as per the method of Pospiech and Neumann [4]. The gene encoding 16S rDNA from selected isolates was amplified using universal primers pA (5’AGAGTTTGATCCTGGCTAG-3’) and pH (5’AGGAGGTGAT CCAGCCGCA-3’) as per method described by Edwards et al. The PCR products were monitored through gel electrophoresis (1% agarose, w/v), followed by ethidium bromide staining and UV transillumination. Approximately 1 μg of PCR-amplified 16S r-RNA gene fragments were restricted with endonucleases HhaI, RsaI and BstUI (Fermentas, USA) separately at 37°C for overnight and resolved by electrophoresis in 2.5% agarose. Banding pattern was visualized by ethidium bromide staining and documented in gel documentation and analysis system (Alphaimager, USA). Strong and clear bands were scored for similarity and clustering analysis using the software, NTSYS-PC2 package (Numerical taxonomy analysis program package, Exeter software, USA). Similarity among the strains was calculated by Jaccard’s coefficient and dendrogram constructed using UPGMA method [5]. 16S r-DNA gene sequencing and phylogenetic analysis Purified 16S r-RNA gene of representative isolates from each cluster was used as a template in cycle sequencing reactions with fluorescent dye-labeled terminators (Big Dye, Applied Biosystems). Both primers pA and pH were used for sequencing and run in ABI prism automated DNA sequencer (3130xl). Partial 16S r-DNA sequence was analyzed by alignment with the GenBank database using BLAST (NCBI BLASTR home page). Identification to the species level was determined based on 16S rRNA gene sequence similarity (>97%) with that of a prototype strain sequence. Multiple sequence alignment of approx. 1500 bp sequences was performed using CLUSTAL W, version 1.8 [6]. A phylogenetic tree was constructed using the neighbor-joining method [7]. Tree topologies were evaluated through bootstrap analysis of 1,000 data sets by MEGA 4.0 package [8]. The nucleotide sequences of partial 16S rDNA segment determined in this study have been deposited in NCBI GenBank data base under accession numbers HM367733–HM367742 [9,10].
Medium Selection for α-Amylase Production
Three media namely synthetic, complex were selected for amylase production. One of the media giving best result will be used throughout entire investigation.
Semi synthetic medium composition
Tryptone 2.0 g, MgSO4.7H2O 0.05 g, KH2PO4 1.0 g, Na2HPO4 2.5 g, NaCl 1.0 g, (NH)4SO4 2.0 g, CaCl2.2H2O 0.05 g, Distilled water 1000 ml and maintain pH=7 [9].
Complex medium (No. I) composition
Soluble Starch 4.0 g, (NH4)2SO4 5.0 g, Tryptone 6.0 g, FeCl3 0.01 g, MgCl2.6H2O 0.01 g, CaCl2.2H2O 0.01 g, K2PO4 4.0 g, KH2PO4 7.5 g, Distilled water 1000 ml and maintain pH=7 [10].
Complex medium (No. II) composition
Trypticase 10.0 g, Peptone 5.0 g, (NH4)2SO4 3.0 g, K2HPO4 2.0 g, Cys- HCl 0.5 g, MgSO4 0.2 g Distilled water 1000 ml and maintain pH=7 [11].
All the three media were prepared by adding the constituents separately in double distilled water and autoclaved for amylase production.
Amylase quantification
Estimation of amylase activity was done as per described by Miller [12] using DNS reagent (3,5-Dinitrosalicylic acid). 1 ml of 1% starch was incubated with different dilutions of the enzymes extract and 1ml of citrate phosphate buffer (pH 6.0). The reaction mixture was incubated at 37°C for 30 min. The reaction was stopped by adding 2 ml of DNS and kept in boiling water bath for 10 min. The absorbance was read at 540 nm using spectrophotometer (Systronic, Model. 2305) against glucose standard. One unit of enzyme activity is defined as the amount of enzyme, which releases 1 μmol of reducing sugar as glucose per minute under the assay condition (U/ml/min).The experiments were carried out in triplicates and mean was taken.
Optimization of medium for α-amylase production by free cells
Based on productivity performance of finally selected isolate (Bacillus licheniformis) in different media one of the most suitable medium was selected for further optimization. The experiments were carried out in triplicates and mean value was taken.
Results and Discussion
Isolation and biochemical characterization
Out of 30, 20 isolates were exhibiting clear zone on starch agar after addition of iodine were selected for further investigation.
On the basis of performance of the isolates in terms of zone of clearance Isolates number RR9 which was identified as Bacillus licheniformis was selected for further scale up studies.
On the basis of 16 Sr-DNA sequences analysis the selected isolate was identified as Bacillus licheniformis.
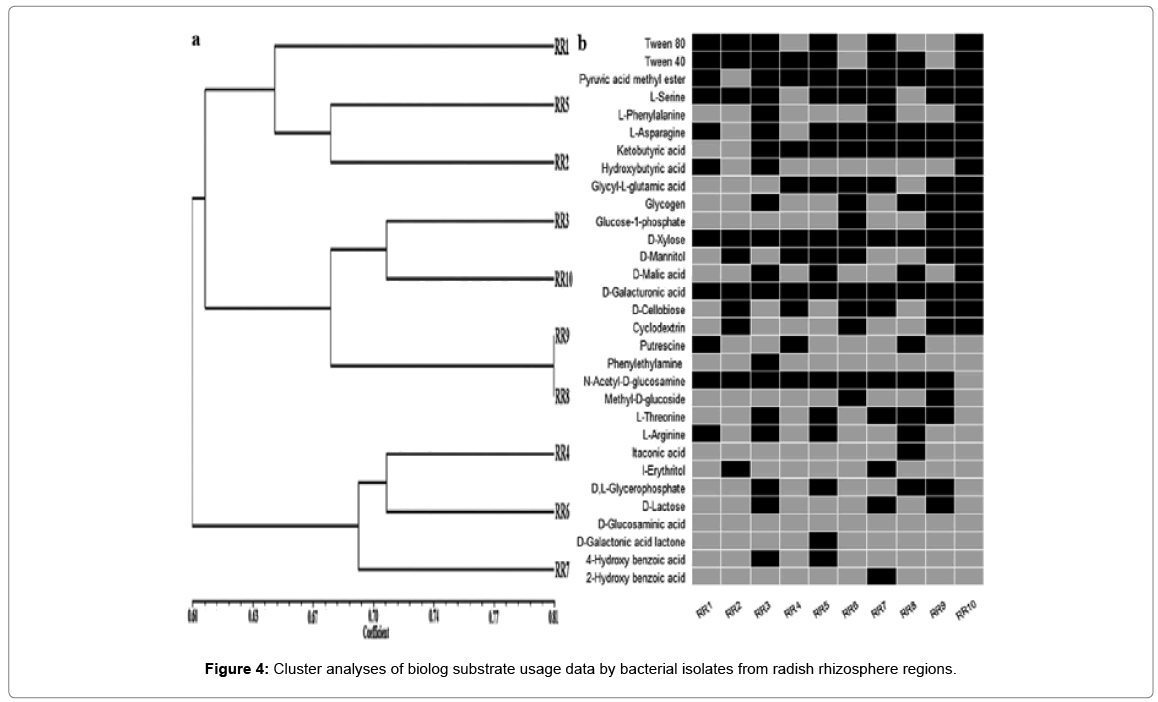
a. Dendrogram showing clustering of 10 representative bacterial isolates based on simple matching (SSM) coefficient and unweighted pairgroup method with average (UPGMA) clustering.
b.Carbon source usage of representative isolates via BIOLOG ECO assay plates. Black boxes represent statistically significant substrate usage at any point during experiments. Experiments were conducted in triplicate.
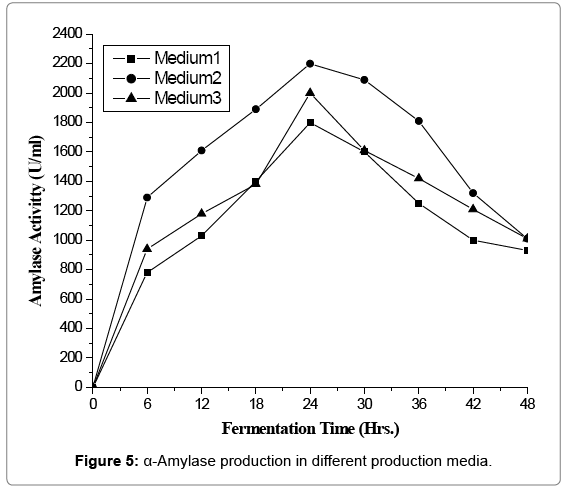
α-Amylase production in different production medium
Cost of fermentation media has profound effect on overall cost of a fermentation process and account for 38 to 73% of the total production cost [13]. All microorganisms require water, source of energy, nitrogen trace element. On a small scale it is relatively simple to devise a medium, supporting satisfactory growth and optimum yield of product. In the present investigation three media namely semi-synthetic, complex medium-I and complex medium-II. The performance of natural isolate B. licheniformis was evaluated in terms of amylase productivity (Tables 1 and 2 and Figures 1-5).
| S. No. | Isolates | Zone of Clearance (in cm) |
|---|---|---|
| 1 | RR3 | 2.6 |
| 2 | RR4 | 0.8 |
| 3 | RR11 | 1.9 |
| 4 | RR9 | 2.8 |
| 5 | RR2 | 0.6 |
| 6 | RR13 | 1.3 |
| 7 | RR7 | 1.9 |
| 8 | RR5 | 1.8 |
| 9 | RR20 | 2.1 |
| 10 | RR8 | 1.7 |
| 11 | RR15 | 2.2 |
| 12 | RR16 | 2.1 |
| 13 | RR17 | 1.5 |
| 14 | RR6 | 1.7 |
| 15 | RR1 | 2.3 |
| 16 | RR14 | 1.8 |
| 17 | RR10 | 1.6 |
| 18 | RR12 | 1.4 |
| 19 | RR18 | 2.1 |
| 20 | RR19 | 1.6 |
Table 1: Zone of starch hydrolysis by isolates.
| S. No. | Isolates | Gram’s Stain | H2S production | Nitrate reduction | Indole production | Urease activity | MR | VP | Carbohydrate fermentation | Citrate utilization | Starch hydrolysis | Catalase activity | Casein hydrolysis | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glucose | Lactose | Sucrose | |||||||||||||
| 1 | RR3 | +ve | +ve | +ve | -ve | +ve | +ve | -ve | A | A | A | -ve | +ve | -ve | +ve |
| 2 | RR4 | -ve | -ve | +ve | -ve | -ve | -ve | +ve | A | A | AG | -ve | +ve | -ve | +ve |
| 3 | RR11 | +ve | -ve | +ve | -ve | +ve | +ve | -ve | AG | -ve | A | -ve | +ve | -ve | +ve |
| 4 | RR9 | +ve | -ve | +ve | -ve | +ve | -ve | -ve | A | -ve | A | -ve | +ve | +ve | +ve |
| 5 | RR2 | -ve | +ve | -ve | -ve | -ve | -ve | -ve | A | -ve | A | +ve | +ve | +ve | +ve |
| 6 | RR13 | +ve | +ve | +ve | -ve | -ve | -ve | -ve | A | -ve | AG | +ve | +ve | +ve | +ve |
| 7 | RR7 | -ve | -ve | +ve | +ve | +ve | -ve | -ve | AG | AG | AG | -ve | +ve | +ve | +ve |
| 8 | RR5 | +ve | -ve | +ve | -ve | +ve | -ve | -ve | AG | AG | AG | -ve | +ve | -ve | +ve |
| 9 | RR20 | +ve | +ve | +ve | -ve | -ve | -ve | -ve | A | -ve | A | -ve | +ve | -ve | +ve |
| 10 | RR8 | +ve | -ve | +ve | -ve | -ve | -ve | -ve | AG | A | -ve | +ve | +ve | -ve | +ve |
| 11 | RR15 | -ve | -ve | +ve | +ve | -ve | -ve | -ve | AG | AG | AG | +ve | +ve | +ve | +ve |
| 12 | RR16 | -ve | -ve | +ve | -ve | -ve | +ve | -ve | AG | -ve | AG | +ve | +ve | -ve | +ve |
| 13 | RR17 | -ve | +ve | -ve | -ve | +ve | -ve | -ve | AG | -ve | -ve | +ve | +ve | +ve | +ve |
| 14 | RR6 | +ve | +ve | +ve | -ve | -ve | -ve | -ve | AG | -ve | AG | +ve | +ve | -ve | +ve |
| 15 | RR1 | -ve | +ve | -ve | -ve | +ve | -ve | +ve | A | A | AG | +ve | +ve | +ve | +ve |
| 16 | RR14 | -ve | -ve | +ve | -ve | -ve | -ve | -ve | A | AG | AG | +ve | +ve | +ve | +ve |
| 17 | RR10 | +ve | -ve | +ve | +ve | -ve | -ve | -ve | A | AG | AG | +ve | +ve | +ve | +ve |
| 18 | RR12 | +ve | -ve | +ve | -ve | -ve | -ve | -ve | AG | -ve | -ve | -ve | +ve | -ve | +ve |
| 19 | RR18 | +ve | -ve | +ve | +ve | -ve | -ve | -ve | AG | -ve | AG | -ve | +ve | +ve | +ve |
| 20 | RR19 | -ve | -ve | +ve | -ve | -ve | -ve | -ve | AG | AG | -ve | +ve | +ve | +ve | +ve |
Table 2: Result of biochemical tests.
Figure 2: Clustering based on ARDRA. a) Restriction patterns of PCR-amplified fragments of 16S rDNA digested with (i) HhaI, (ii) RsaI and (iii) BstUI. Lane name with respective isolates; M (100 bp ladder) b) Dendrogram showing the clustering of 20 bacterial isolates from radish rhizosphere generated from restriction analysis of 16S rDNA amplicon, using the UPGMA algorithm and the Jaccard’s coefficient.
Figure 3: Phylogenetic tree based on the 16S rRNA gene sequences of bacterial isolates from radish cv. Jaunpuri Giant rhizosphere and their closest phylogenetic relatives. The tree was created by the neighbour-joining method. The numbers on the tree indicate the percentage of bootstrap sampling derived from 1000 replicates. 16S rRNA gene sequences of Ferroplasma thermophilum was used as out-group.
Minimum amylase activity 1800 (U/ml) was observed in synthetic medium. It might be due to lack of nutritional component required by the process organism. Maximum productivity was found to be 2200 (U/ml) with complex medium-I while complex medium-II gave moderate yield of enzyme activity of 2000 (U/ml). Higher yield in complex medium may be due to salts of alkaline earth metal [14,15]. Based on productivity complex medium-I was finally selected for further optimization of various nutritional and cultural requirements for amylase production.
Conclusion
The present work describes the α-amylase production by bacteria isolated from rhizospheric soil of radish field. Isolates were characterized biochemically and identified by 16S ribotyping. One of the most promising isolate RR9 that is Bacillus licheniformis was used throughout the entire investigation. Bacillus species produce the amylase because of its heterotrophic nature as it can grow on variety of substrates ranging from pure compounds to solid wastes. It was observed from the experiments the natural isolates namely RR9 was used throughout the entire investigation. Three different types of media such as Semi-synthetic, complex media-I and complex media II tested and it was found that complex media-I gave maximum yield therefore this complex media-I used throughout entire studies and various parameters were optimized for enhance the alpha-amylase yield. Maximum production of α-amylase was observed at 24 h. of fermentation. To avoid any lag and optimum production of the enzyme inoculum was developed in the same production medium.
Significant Statement
This result obtained from present study prove that Bacillus licheniformis is the maximum α-amylase producing strain isolated from soil under suitable conditions. This study will very helpful for upcoming researchers to uncover the various aspect of enzyme production by microorganism. In present study submerged fermentation was found to be most suitable as compared with solid state fermentation and it is required less fermentation time. Economic consideration is the first and foremost criteria for any bioprocessing operations. In past few decades much attentions was paid to convert waste in useful material including production of food, feed and energy alongwith biochemicals and biocatalyst due to its cost effectiveness using as a substrate or sole source of carbon. Amylase and proteases are one of the world most best seller enzyme of microbial origin across the globe in its crude form.
Acknowledgement
The Authors is very thankful to Department of Biological sciences, Mahatma Gandhi Chitrakoot Gramodaya Vishwavidhyalaya, Chitrakoot satna (M.P.) and Department of Biotechnology, V.B.S. Purvanchal University, Jaunpur (U.P.).
References
- Gopinath SC, Anbu P, Arshad MK, Lakshmipriya T, Voon CH, et al. (2017) Biotechnological processes in microbial amylase production. Biomed Res Int 2017: 1272193.
- Singh P, Singh R, Sharma R, Gupta P (2012) Factor affecting a-amylase production on submerged fermentation by Bacillus sp. Int J Pharma Life Sci 3: 2243-2246.
- Khusro A, Barathikannan K, Aarti C, Agastian P (2017) Optimization of thermo alkali stable amylase production and biomass yield from Bacillus sp. Under submerged cultivation. Fermentation 3: 1-19.
- Pospiech A, Neumann B (1995) A versatile quick-prep of genomic DNA from gram-positive bacteria. Trends Genet 11: 217-218.
- Nei M, Li WH (1979) Mathematical-Model for studying genetic variation in terms of restriction endonucleases. Proceedings microbial diversity of radish rhizosphere of National Academy of Sciences, USA. 76: 5269-5273.
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673-4680.
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406-425.
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596-1599.
- Natasa B, Jordi R, Joseph Lopez S, Zoran V (2011) Optimization of the growth and α-amylase production of Bacillus subtilis IP 5832 in shake flask and laboratory fermenter batch cultures. J Serb Chem Soc 76: 965-972.
- Vengadaramana A, Balakumar S, Arasaratnam V (2012) Production and optimization of a-amylase by Bacillus licheniformis ATCC 6346 in lab bench-scale fermenter. J Microbiol Biotechnol Res 2: 190-211.
- Sethi S, Shyamsundar A, Sakhsam G, Lal B (2013) Amylase activity of a starch degrading bacteria isolated from soil. Arch Appl Sci Res 5: 15-24.
- Miller GC (1959) Use of dinitrosalicylic acid reagent for determination of reducing suger. Anal Chem 31: 426-428.
- Ratledge C (1977) Fermentation substrate. Ann Rep Ferment Proc 1: 49-71.
- Gupta R, Gigras P, Mohapatra H, Goswami VK, Chauhan B. (2003) Microbial α-amylases: A biotechnological perspective. Process Biochem 38: 1599-1616.
- Vidyalakshmi R, Paranthaman R, Indhumathi J (2009) Amylase production on submerged fermentation by Bacillus spp. World J Chem 4: 89-91.
Relevant Topics
- Agricultural biotechnology
- Animal biotechnology
- Applied Biotechnology
- Biocatalysis
- Biofabrication
- Biomaterial implants
- Biomaterial-Based Drug Delivery Systems
- Bioprinting of Tissue Constructs
- Biotechnology applications
- Cardiovascular biomaterials
- CRISPR-Cas9 in Biotechnology
- Nano biotechnology
- Smart Biomaterials
- White/industrial biotechnology
Recommended Journals
Article Tools
Article Usage
- Total views: 4522
- [From(publication date):
September-2017 - Jul 03, 2025] - Breakdown by view type
- HTML page views : 3606
- PDF downloads : 916