Marker-Assisted Pyramiding Resistance Genes Against Angular Leaf Spot and Common Bacterial Blight Disease into Preferred Common Bean Cultivar "REDWOLAITA"
Received: 21-Nov-2018 / Accepted Date: 24-Jan-2019 / Published Date: 05-Feb-2019 DOI: 10.4172/2329-8863.1000416
Abstract
Angular leaf spot (ALS) caused by Pseudocercospora griseola and common bacterial blight (CBB) caused by Xanthomonas campestris pv phaseoli X. campestris pv. phaseoli var. fuscans are the most economically important diseases of common bean production in Ethiopia. This research was aimed at pyramiding the Phg-2 R gene for angular leaf spot resistance and two CBB major resistance quantitative trait loci (RQTLs) into the background of the most popular and susceptible common bean cultivar “REDWOLAITA” (RW). Marker-assisted Parallel Back Crossing (MAPBC) breeding scheme with three separate parallel backcrossing streams were adopted. This technique accelerated tracking three independent resistance loci linked to g796, SAP6 and SU91 genetic marker from two different donor parents into the RW recurrent parent. The donor parental line VAX-6 with known RQTLs for CBB and tagged by the SAP6 and SU91 genetic markers on linkage groups 10 and 8, respectively was used. The other donor parent MEX-54 with known Phg-2 R gene tagged by the g796 genetic marker at the linkage group 8 was used in the gene pyramiding program. After the BC4 generation, progenies that combined SAP6 and g796 genetic markers were created and selected from the BC4 inter-crossing of progenies. Then, further inter-crossing was made between selected progenies that combined the SAP6 and g796 genetic markers with selected progenies with the SU91 genetic marker. Finally, from this study we developed Monogenic Near Isogenic Lines (MNILs) with R genes tagged by the SAP6, g796, and SU91 molecular markers and Polygenic Near Isogenic Lines (PNILs) with different gene combination. The developed lines include MNILSAP6, MNILSU91 and MNILg796, PNILsSAP6/g796, PNILsSU91/g796, PNILsSAP6/SU, PNILsSAP6/g796/SAP6, with more than 97% genome recovered from the RW genetic background. Marker-assisted backcrossing facilitated selection of progenies that combined good agronomic traits with resistance loci were constructed from the RW common bean cultivar. The developed lines showed high level of disease resistance to the strains of CBB and ALS under the screening conditions. The developed lines will be evaluated under multiple environment under natural epidemics, before varietal release for wider production. We also recommend the developed NILs with RW back ground and known sources of R genes and good agronomic performance could be used as alternative donor parent for the future gene pyramiding program.
Keywords: Gene pyramiding; Parallel backcrossing; RQTLs; Inter-crossing; Isogenic lines
Introduction
Common bean (Phaseolus vulgaris L.) is the most important grain legume for direct human consumption and used as main food and/or food component in Latin America and eastern and southern Africa. Common bean is seed-propagated and a diploid (2n=2x=22) with a relatively small genome (650 Mb) [1], originated in the Neotropics, with at least two major centers of domestication in Mesoamerica and the Andes [2]. Common bean is believed to have been introduced together with maize via the east coast of Africa by Portuguese and Spanish traders in the sixteenth and seventeenth century [3,4]. In Ethiopia, common bean is the principal food and nutrition security legume crop providing dietary protein and a source of cash income for resource-poor farmers. Among a number of factors that could attribute to the low yield, diseases especially angular leaf spot (ALS) caused by Pseudocercospora griseola and common bacterial blight (CBB) caused by Xanthomonas campestris pv phaseoli cause the most significant harvest losses in common bean in farmer’s field [5]. The impact of disease on crop production in Ethiopia and beyond may be worsening with the current and predicted climate change scenarios of rising temperatures and variability and changes in precipitation. These has been observed in disease incidence and severity of common beans in Ethiopia [5].
Using host resistance has been proven to be the most effective and economical method to control disease in common bean and other crops [6]. Therefore, to obtain a durable and broad-spectrum resistance variety, pyramiding multiple R genes/RQTLs into a recurrent common bean cultivar is an important and practicable breeding strategy to control angular leaf spot and common bacterial blight [7]. The backcrossing approach to deploy one or more genes into an elite line was proposed by Harlan and Pope [8]. Since, backcrossing has become a widely used plant breeding approach in diverse crop species [9,10]. This method is most commonly used to incorporate one or a few highly heritable traits into an adapted or elite variety. In most cases, the elite variety used for backcrossing has a large number of desirable attributes but is deficient in only a few characteristics. The other parent, called the ‘donor parent’, possesses one or more genes controlling an important trait, which is lacking in the elite variety [10]. Traditional backcrossing programs are designed on the assumption that the proportion of the recurrent parent genome is recovered at a rate of 1–(1/2)t+1 for each of t generations of backcrossing. Thus, after four backcrosses, we expect to recover 1–(1/2)5=96.9% of the recurrent parent genome [11]. However, any BC progeny individual will deviate from this expectation due to chance and to linkage between the gene from the donor parent being selected for and nearby genes.
The advent of molecular genetic markers assisted backcrossing has been successfully applied in gene pyramiding programs for targeted transferring and pyramiding resistance loci to create more durable and broad specific resistance in different crops [12]. Successful applications of gene pyramiding research has been reported in different crops by many of authors. In the wheat cultivar “Yang”, [13] successfully combined three powdery mildew resistance gene combinations pm2 +pm4a, pm2 +pm211, and pm4a+pm21 using restriction fragment length polymorphism (RFLP) markers. In soybean for mosaic virus disease resistance (SMV), researchers successfully pyramided three genes Rsv1 , Rsv3 and Rsv4 with the aid of microsatellite markers in order to develop new soybean lines containing multiple resistance genes for soybean mosaic virus (SMV) resistance. Marker-assisted selection (MAS) and gene pyramiding have been reported in common bean research [14-18]. Recently, Deshmukh et al. [19] reported the efficiency and effectiveness of gene pyramiding in improving angular leaf spot resistance in susceptible common bean cultivar.
This specific research aims at cumulating the phg-2 R gene for ALS and two major RQTL’s for CBB resistance into the background of popular common bean cultivar ‘REDWOLAITA’ through the aid of molecular and conventional breeding techniques.
Materials and Methods
Plant materials and breeding strategy
The materials under study included three parents: ‘REDWOLAITA’ (RW) as the recurrent parent and VAX-6 and MEX-54 as sources of disease resistance (Table 1). The RW cultivar from the Mesoamerican gene pool was the most popular and widely grown bean cultivar in the southern parts of Ethiopia. This specific cultivar was highly preferred for its cooking quality, color and high marketability by most of farmers.
| Parents used in MABCP | Gene pool | Seed size & color | Growth Habit/type | Disease Reaction | |
|---|---|---|---|---|---|
| ALS | CBB | ||||
| REDWOLAITA | Mesoamerican | Small red | II | + | + |
| VAX-6 | Mesoamerican | Small pale red | I | + | - |
| MEX-54 | Mesoamerican | Small pink | IV | - | + |
CBB=common bacterial blight, ALS=angular leaf spot, +=compatible(susceptible) disease reaction, -=incompatible (resistance) disease reaction.
Table 1: Characteristics of common bean parental lines which were used in marker assisted gene pyramiding program (MABCP).
The cultivar, although, it was the most preferred by farmers, it is susceptible to common bacterial blight caused by Xanthomonas campestris pv phaseoli X. campestris pv. phaseoli var. fuscans and angular leaf spot caused by Pseudocercospora griseola endemic to Ethiopia. Therefore, this common bean cultivar was selected as recurrent parent to be improved through marker-assisted gene pyramiding program.
The donor parent VAX-6 is with known RQTLs tagged by the SAP6 and SU91 genetic markers on linkage groups 10 and 8, respectively for bacterial blight disease caused by Xanthomonas campestris pv. phaseoli and X. campestris pv. phaseoli var. fuscans. The other donor parent MEX-54 is also with the Phg-2 R gene for ALS caused by Pseudocercospora griseola and tagged by the g796 genetic marker at the linkage group 8 [20-24].
After the BC4 generation, progenies that combined genomic region linked with SAP6 and g796 genetic markers were created and selected from the BC4 inter-crossing of progenies. The parents were tested for marker polymorphism and usefulness for MAS (Tables 2 and 3). To achieve the objective, the parents were hybridized, and an appropriate marker assisted gene pyramiding breeding approach was implemented (Figure 1). Crosses were made and advanced through the application of molecular markers. The resistance gene transfer was confirmed with the aid of molecular marker linked to the R gene/RQTL and through reliable screening techniques.
| Gene/Locus | Linked Molecular markers | linkage group | Primer sequences | Expected band Size/orientation | Reference |
|---|---|---|---|---|---|
| QTL | SAP6 | 10 | F GTCACGTCTCCTTAATAGTA R GTCACGTCTCAATAGGCAAA |
806/cis | [15] |
| QTL | SU91 | 8 | F CCACATCGGTTAACATGAGT R CCACATCGGTGTCAACGTGA |
669/cis | [15] |
| Phg2 | g796 | 8 | F GAGAAACTACGGGCTGTTTTACCC R AATTAAAACACCCACCCACTCCAT |
220 | [24] |
| Phg2 | SN02 | 8 | F ACCAGGGGCATTATGAACAG R ACCAGGGGCAACATACTATG |
890/cis | [17] |
F=Forward, R=Reverse.
Table 2: Polymorphic molecular markers used in gene pyramiding and selection.
| DNA markers | Annealing T°C | RW | VAX-6 | MEX-54 | Description of Marker |
|---|---|---|---|---|---|
| SAP6 | 58 | - | + | - | 1SCAR, Linked to CBB Resistance QTL |
| SU91 | 60 | - | + | - | 2SCAR, Linked to CBB Resistance QTL |
| g796 | 44 | - | - | + | 3STS, linked to phg-2 ALS R gene |
| SN02 | -- | - | - | - | 4STS, linked to phg-2 ALS R gene |
| OPE4 | -- | - | - | + | 5STS, linked to phg-2 ALS R gene |
Table 3: DNA marker validation for selecting polymorphic markers to be used in the marker assisted gene pyramiding.
Molecular markers
Sequence Characterized Amplified Regions (SCAR) markers were used to tag angular leaf spot and common bacterial resistance genes of interest. The original oligonucleotide markers were obtained from Eurofins Genomics (Table 2). A 50/100 bp mixed DNA molecular weight marker (Ladder) specifically designed for determining the size of double strand DNA from 25 to 300bp was used. The presence of SU91700 linked a resistance QTL located on B8, SAP6820 [15] linked to a resistance QTL on B10 whereas the Phg-2 resistance locus were on B10, g796220 [24] were determined using genetic markers.
Marker-assisted selection
DNA extraction and amplifications: Genomic deoxyribonucleic acid (gDNA) was isolated using FTA card matrix technology following the manufacturer’s procedure with minor modification from fresh leaves of 12-day-young plantlets. Common bean progenies were sampled from each succeeding generation, i.e., BC1F1, BC2F1, BC3F1, and BC4F1, and including the progenies created through inter-crossings of BC4s and BC4F2. FTA is a paper-based technology, which was designed for the collection of nucleic acids, either in their purified form or within pressed samples of fresh tissue. Patented chemicals saturated into the paper act to lyse cellular material and fix and preserve DNA within the fibre matrix. As described in the manufacturer’s protocol with minor modification (www.gelifesciences.com/whatman) in which captured nucleic acids were ready for purification when taken with a punch from the FTA card, purification reagents were added, and the paper was washed with TE-1 (10 mM Tris-HCl, 0.1 mM EDTA, pH8) buffer.
The DNA markers SAP6 (829bp), SU91 (700 bp) and g796 (233bp) were used to select plants with linked resistance loci, which were then backcrossed to the recurrent parent. After washing the punched discs, the DNA was tested for its quality using agarose gel (0.98%) for use in PCR.
Polymerase chain reaction (PCR): Sequence characterized amplified region (SCAR) markers used in selection for CBB resistance were dominant and were scored as the presence or absence of a single band on an agarose gel. The INDEL marker used in ALS resistance selection was co-dominant [24]. DNA amplification was performed under ABI 2720 Thermal cycler under the program for SU91, SAP6 and g796 were 34 cycles of 10s at 94°C, 40s at 58°C (for SAB6), 40s at 60°C for SU91 and 30s at 44°C for g796, 2 min at 72°C, and 5 min at 72°C for the final extension (Table 3). PCR results were analyzed using a 1.4% agarose gel stained with ethidium bromide (0.02 μg·mL-1). Bands present on the gel were compared by size to a 100 bp molecular marker.
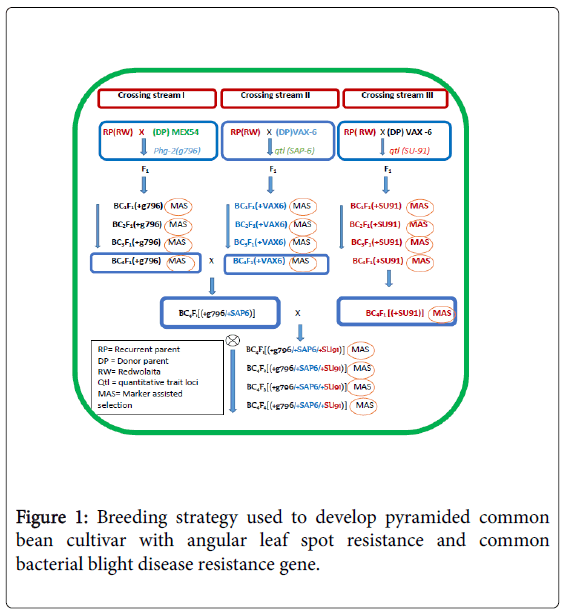
Marker-Assisted back crossing and gene pyramiding: The gene pyramiding and marker-assisted backcrossing breeding selection strategy used is illustrated in Figure 1. The marker-assisted backcrossing program was planned in such a way that three independent crossing streams were undertaken to track R/RQTLs loci tagged by SAB6, SU91 and g796 molecular markers on the linkage groups 10 and 08, respectively (Table 2). The resistance donor parents MEX-54 with R gene (Phg-2) for ALS and VAX-6 (with two RQTLs loci for CBB) were independently crossed with the recurrent parent ‘REDWOLAITA’ common bean cultivar under screening house conditions (Table 3).
The F1 were crossed with the RP to produce the first backcross generation (BC1F1 or just BC1). Markers closely linked to the resistance loci were then used to check targeted genes from each crossing streams of BC1F1 populations. Based on the plan, molecular markers, which included g796 in crossing stream one, SAB6 in crossing stream two, and SU91 in crossing stream three were used. Then, the succeeding backcross generations were made by crossing selected BC1F1plants (that had been screened for the targeted resistance trait (Phg-2 and CBB RQTL loci) from each crossing streams with the RP to produce the BC2F1 populations. Subsequent backcross populations were made by repeatedly crossing the selected backcross (BC) plants with the RP. The backcross progeny with the target trait were selected based on genotype and phenotype during each round of backcrossing. To further identify targeted homozygous plants at the backcross four (BC4) from each backcross streams selected plants were selfed to get targeted homozygous plants. Then, plants with homozygous genotypes for the targeted R genes were selected randomly from each segregating population. BC4F3 seeds were then harvested individually from each selected BC4F2 [RW/RW/ VAX(+SAP6)], [RW/RW/MEX(+g796)] and [RW/RW/VAX(+SU91)] these created lines were monogenic near isogenic lines, constituting MNILSAP6, MNILSU91 and MNILg796 respectively.
Then intercrossing was made to further combine the resistance genes into a single background. The pyramid lines with different gene combinations were created combining SAP6 with g796 genes PNILsSAP6/g796, PNILsSAP6/SU91 combining genes SAP6 with SU91, PNILsSAP6/g796 combining genes SAP6 with g796, and further crossing was made to create polygenic line with good agronomic background combining all of the R genes PNILsSAP6/SU91/g796 polygenic lines that combined three R loci linked to SAP6, g796 and SU91 were developed with help of Marker-assisted Selection (MAS).
Selection of near isogenic and polygenic pyramided resistance lines
The isogenic and polygenic near isogenic pyramided lines were evaluated for their reaction to both common bean angular leaf spot and bacterial blight under the screening house using the most virulent pathogens collected from Ethiopia. Fourteen days old plants were inoculated with angular leaf spot suspension 10 × 106 spore concentration and CBB which were virulent to common bean growing areas of Ethiopia. From this study the target was evaluation for R genes and appropriate traits were based on the reaction of selected lines with virulent pathogen and morphological characteristics including growth habit and seed color of the plant were considered.
Results
Marker-assisted backcross breeding
Three independent and separate parallel back crossing schemes were adopted to track resistance loci from two donor parents. From the back-crossing program, three polymorphic DNA-based molecular markers were used during marker-assisted parallel backcrossing (MABC) breeding program to deploy two CBB RQTLs and Phg-2 R gene for ALS disease into the farmer-preferred cultivar but susceptible RW bean cultivar.
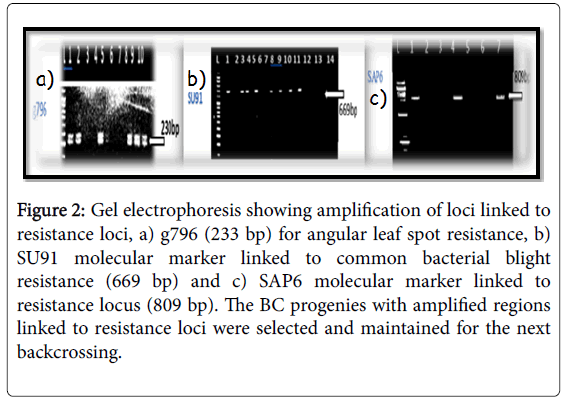
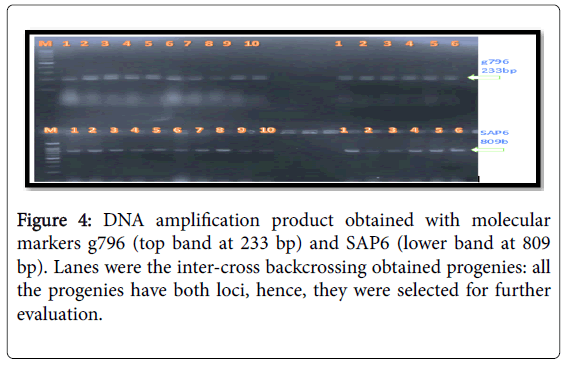
The molecular markers SU91, SAP6 and g796 allowed us to conduct early selection of bean lines with resistance to the fungal and bacterial pathogens (Figures 2-4). Among the pyramided polygenic NILs, the lines that combined three R loci PNILsSAP6/g796/SU91 performed best followed by pyramided lines PNILsSAP6/g796, PNILsSU91/g796, both with two resistance lines in terms of disease reaction, hundred seed weight (HSW) (gm) and seed color.
Figure 2: Gel electrophoresis showing amplification of loci linked to resistance loci, a) g796 (233 bp) for angular leaf spot resistance, b) SU91 molecular marker linked to common bacterial blight resistance (669 bp) and c) SAP6 molecular marker linked to resistance locus (809 bp). The BC progenies with amplified regions linked to resistance loci were selected and maintained for the next backcrossing.
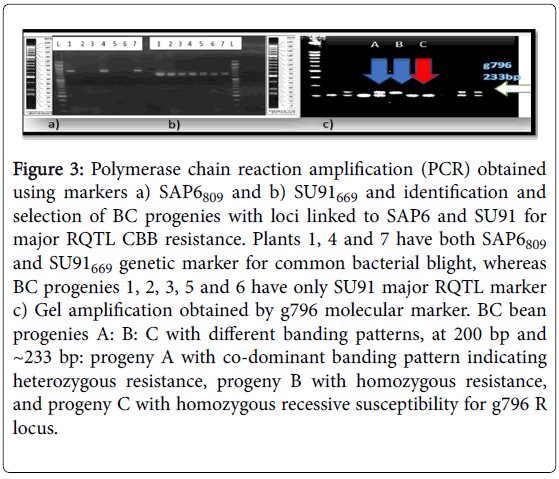
Figure 3: Polymerase chain reaction amplification (PCR) obtained using markers a) SAP6809 and b) SU91669 and identification and selection of BC progenies with loci linked to SAP6 and SU91 for major RQTL CBB resistance. Plants 1, 4 and 7 have both SAP6809 and SU91669 genetic marker for common bacterial blight, whereas BC progenies 1, 2, 3, 5 and 6 have only SU91 major RQTL marker c) Gel amplification obtained by g796 molecular marker. BC bean progenies A: B: C with different banding patterns, at 200 bp and ~233 bp: progeny A with co-dominant banding pattern indicating heterozygous resistance, progeny B with homozygous resistance, and progeny C with homozygous recessive susceptibility for g796 R locus.
The result revealed that successful gene pyramiding of three R genes (Phg-2 R gene for the angular leaf spot and two major RQTLs for CBB resistance) through DNA-based marker-assisted gene pyramiding into the popular and farmer-preferred but susceptible RW common bean cultivar (Table 4).
| Generation | Progenies from Stream one MABC | Target (+SAP6) | Progenies from Stream two MABC | Target (+g796) | Progenies from Stream three MABC | Target +(SU91) |
|---|---|---|---|---|---|---|
| 1 | F1 [RW/VAX] | -- | F1 [RW/MEX] | -- | F1 [RW/VAX] | -- |
| 2 | BC1[RW/VAX] F1 | 03:24 | BC1[RW/MEX] F1 | 07:24 | BC1[RW/MEX] F1 | 2:24 |
| 3 | BC2[RW/VAX] F1 | 05:19 | BC2[RW/MEX] F1 | 07:20 | BC2[RW/MEX] F1 | 11:15 |
| 4 | BC3[RW/VAX] F1 | 05:12 | BC3[RW/MEX] F1 | 03:07 | BC3[RW/MEX] F1 | 5:12 |
| 5 | BC4[RW/VAX] F1 | 05:12 | BC4[RW/MEX] F1 | 08:17 | BC4[RW/VAX] F1 | 5:10 |
| Inter-crossing isogenic lines | Target | |||||
| 6 | BC4[RW/VAX]/BC4[RW/MEX] | (+SAP6/+g796) 5/14 (2 homozygous:3 heterozygous) | ||||
| 7 | BC4[RW/VAX/MEX] F1/BC4[RW/VAX] F1 | (+SAP6/+g796/+SU91) 6:10 (g/SU/SAB)/10:10 (g796+SAP) | ||||
| 8 | BC4[RW/VAX/MEX/VAX] F1 | (+SAP6/+g796/+SU91) | ||||
| 9 | BC4[RW/VAX/MEX/VAX] F2:4 | (+SAP6/+g796/+SU91) | ||||
Table 4: Selected progenies from the successive parallel backcrossing program in each generation based on the target locus linked to molecular marker.
Disease resistance screening for the advanced lines with strains of ALS and CBB showed that developed single gene and poly gene pyramided lines showed high level of resistance to ALS and CBB strains compared to the recurrent parent (Table 5). Among the developed near-isogenic pyramided polygenic line, the PNILsSAP6/SU91 line with two R loci on chromosomes Pv08 and Pv10 for common bacterial blight, performed best under the CBB disease but showed susceptible reaction to angular leaf spot.
| Progenies | Pedigree | ALS (1-9) | CBB (1-9) | Seed HSW (gm) | Seed color |
|---|---|---|---|---|---|
| ETKT01 | BC4 [RW/VAX(+SAP6)] | S | MR | 25.7 | SR |
| ETKT02 | BC4 [RW/VAX (+SU91)] | S | MR | 23.3 | SR |
| ETKT03 | BC4 [RW/MEX-54(+g796)] | R | S | 16.8 | SR |
| ETKT04 | BC4 [RW/VAX(+SAP6/+SU91)] | S | R | 17.4 | SR |
| ETKT05 | BC4 [RW/VAX/MEX(+SAP6/+g796)] | R | MR | 18.4 | SR |
| ETKT06 | BC4 [RW/VAX/MEX(+SU91/+g796)] | R | MR | 11.4 | SR |
| ETKT07 | BC4[RW/VAX/MEX/VAX(+SAP6/+g796/+SU91)] | R | R | 21.3 | SR |
| P1 | RW (RP) | S | S | 18.8 | SR |
| P2 | MEX-54 (DP) | R | S | 28.2 | SPP |
| P3 | VAX-6 (DP) | S | R | 17.2 | SR |
| CV | 3.3 | ||||
| LSD | 1.12 |
MNILs=Monogenic Near Isogenic Lines, PNILs=Polygenic Near Isogenic Lines, R=Resistance, MR=Moderately Resistance, SR=Small Red, SPR=Small Pal Red, SP=Small Pink, HSW=Hundred Seed weight (gm), ALS=Angular Leaf Spot, CBB=Common Bacterial Blight, HSW=Hundred Seed Weight (gm), S=Susceptible.
Table 5: Agronomic performance of pyramided MNILs, PNILs and parental lies under screen house.
Hence, this specific which includes monogenic NILs, including NILsSAP6, NILsSU91 and NILsg796 line with more than 76% from RW genetic background recovery and with good agronomic trait will be used for a future gene pyramiding program.
Discussion
Gene pyramiding and cultivar development
Angular leaf spot caused by Pseudocercospora griseola and common bacterial blight caused by Xanthomonas campestris pv phaseoli and X. campestris pv. phaseoli var. fuscans are the major destructive diseases of common bean (Phaseolus vulgarise L.) in Ethiopia. Pyramiding resistance genes/QTLs has been becoming an effective strategy to develop new variety with long-lasting and wide spectrum resistance. Marker-assisted selection (MAS) and gene pyramiding has been reported common bean research [14,15]. Molecular markers linked to major angular leaf spot resistance loci (e.g., Phg-2) genes and common bacterial blight resistance QTLs have been widely reported [15,24,25]. The gene pyramiding approaches of this study complements that of Ragagnin et al. [16] who succeeded using random amplified and polymorphic DNA (RAPD) and sequence-characterized amplified regions (SCAR) markers to pyramid resistance genes Co-4, Co-6, and Co-10 against anthracnose, Phg-1 against angular leaf spot, and Ur-ON for rust into the susceptible ‘carioca’ market class cultivar Ruda.
Ferreira et al. [17] used SCAR, CAPS, and RAPD markers to successfully pyramid Co-2, Co-3/9 anthracnose and I and bc-3 common mosaic virus resistance genes into the ‘fabada’ market class A25 genotype. Ddamulira et al. [26] reported the effectiveness of gene pyramiding in improving angular leaf spot resistance in susceptible common bean cultivar. Recently, Kumar et al. [18] reported marker-assisted pyramiding of bacterial blight and gall midge resistance genes (Gm4 , Gm8 , and Xa21 ) into ‘RPHR-1005’ the restorer line of the popular rice hybrid ‘DRRH-3’ and the variety developed with cumulating thee genes were better yield and increased disease resistance trait. The study also demonstrated that molecular markers can be used to successfully pyramid angular leaf spot and common bacterial blight resistance genes/QTLs into susceptible common bean varieties.
The study introduced Phg-2 and 2 RQTLs into REDWOLAITA resulting in monogenic and polygenic near isogenic lines (MNILs & PNILs) with different gene combinations for the resistance to CBB and ALS. The lines under screen house study showed significantly enhanced levels of resistance. Further inter-crossing and gene pyramiding was conducted in order to combine resistances. Pyramided NILs with R genes/RQTLs linked to SAP6, g796 & SU91 molecular markers including MNILSAP6, MNILSU91 and MNILg796 for the CBB and ALS disease resistance with good agronomic trait were constructed from the RW common bean cultivar genetic background and tested under screening house conditions.
The developed polygene-pyramided isogenic lines (PNILsSAP6/SU91/g796) effectively conferred resistance to most frequently appeared pathotypes (63:59 and 19:33) of angular leaf spot and common bacterial blight pathogens that are endemic to Ethiopia. The developed pyramided lines with different gene combinations showed increased level of disease resistance compared to the parental lines. Monogenic Near Isogenic Lines (MNILs) with R genes linked to SAP6, g796 and SU91 molecular markers were developed. These include MNILSAP6, MNILSU91 & MNILg796 and polygenic PNILsSAP6/g796, PNILsSU91/g796, PNILsSAP6/SU91, PNILsSAP6/g796 and PNILsSAP6/g796/SAP6, with more than 97% RW genetic background were created. The lines will be multiplied and tested under multiple environment and will be tested as a candidate variety for official varietal release. The developed lines were recommended be used for future gene Pyramiding program.
In this particular study, seven resistance lines were developed from the ‘REDWOLAITA’ common bean cultivar to both common bacterial blight and angular leaf spot diseases through marker-assisted gene pyramiding techniques (Table 5). Phenotypic background selection implemented during marker-assisted gene pyramiding associated with the molecular forward selection could be a reliable improvement strategy in the marker-assisted back cross breeding.
Therefore, MABC could be suitable for less well-equipped breeding laboratories, as marker-mediated background selection which is costly strategy. As genetic resistance is an effective strategy for the famers to grow and reduce yield loss due to these economically important diseases and stabilize common bean production.
Conclusion and Implication
Common bean (Phaseolus vulgaris L.) production in Ethiopia is becoming the most and predominantly cultivated pulse crop. It is traditional food and nutritional security crop, and important source of foreign currency and cash as income for smallholder farmers. Whereas, productivity of this important crop under farmer’s field is declining due to the frequent occurrence of the major bacterial and fungal disease. The P. griseola with high pathogenic diversity and its seed born nature, it would be very important to change the common bean improvement strategy in Ethiopia to be able to breed for broad and durable resistance to the pathogen. Conventional breeding is laborious, time consuming and difficult to apply when it comes pyramiding dominant genes with similar reaction [27,28]. Durable resistance based on the major genes has not been effective when resistance genes deployed one at a time.
Therefore, cumulating complimentary resistance genes through marker-assisted gene pyramiding is a strategy that would confer a long-term resistance. In problems with multiple disease infection with different pathogens on common bean affecting its productivity and cause complete crop loss in susceptible varieties. PCR based molecular markers are key to success of MAS and gene pyramiding in common bean improvement. Therefore, gene pyramiding using marker-assisted breeding strategy and back crossing will provide a cost-effective controlling measures to bean diseases. The developed lines with the R genes could be evaluated under multi-location in the future to release best performing lines for the famers. The lines with good genetic background consisting R genes also will be used as parental lines in the future breeding program. The marker-assisted backcrossing (MABC) breeding strategy enabled deploying disease resistance into adapted common bean cultivar ‘REDWOLAITA’, that belongs to the Mesoamerican gene pool.
Acknowledgement
The study was part of the PhD research work for the first author (Yayis Rezene). The authors acknowledge the support, funding of the research and provision of molecular lab facilities from the KIRKHOUSE TRUST. We extend our thanks to the lab technicians Mihiret Tadesse and Bethel Mulugeta. The authors would also like to thank the Southern Agricultural Research Institute (SARI), and the African Bean Consortium (ABC) partner countries.
Conflict of Interest
The Authors declare that there is no conflict of interest.
References
- Broughton WJ, Hernandez G, Blair M, Beebe S, Gepts P, et al. (2003) Beans (Phaseolus spp.)–model food legumes. Plant and Soil 252: 55-128.
- Gepts P (1988) A Middle American and an Andean common bean gene pool. In Genetic resources of Phaseolus Beans, Springer, Dordrecht, pp: 375-390.
- Greenway PJ (1945) Origins of some East African Food Plants: Part III. The East African Agricultural Journal 10: 177-180.
- Gentry HS (1969) Origin of the common bean, Phaseolus vulgaris. Economic Botany 23: 55-69.
- Belete T, Bastas KK (2017) Common bacterial blight (Xanthomonas axonopodis pv. phaseoli) of beans with special focus on Ethiopian condition. J Plant Pathol Microbiol 8: 2.
- Mundt CC (2014) Durable resistance: A key to sustainable management of pathogens and pests. Infection, Genetics and Evolution 27: 446-455.
- De Mendonça HA, Dos Santos JB, Ramalho MAP (2003) Genetic control of common bean reaction to angular leaf spot. Crop Breeding and Applied Biotechnology.
- Harlan HV, Pope MN (1922) The use and value of back-crosses in small-grain breeding. Journal of Heredity 13: 319-322.
- Hasan MM, Rafii MY, Ismail MR, Mahmood M, Rahim HA, et al. (2015) Marker-assisted backcrossing: A useful method for rice improvement. Biotechnology & Biotechnological Equipment 29: 237-254.
- Hanson P, Lu SF, Wang JF, Chen W, Kenyon L, et al. (2016) Conventional and molecular marker-assisted selection and pyramiding of genes for multiple disease resistance in tomato. Scientia Horticulturae 201: 346-354.
- Babu R, Nair SK, Prasanna BM, Gupta HS (2004) Integrating marker-assisted selection in crop breeding–prospects and challenges. Current Science, pp: 607-619.
- Joshi RK, Nayak S (2010) Gene pyramiding: A broad spectrum technique for developing durable stress resistance in crops. Biotechnology and Molecular Biology Reviews 5: 51-60.
- Liu J, Liu D, Tao W, Li W, Wang S, et al. (2000) Molecular markerâ€facilitated pyramiding of different genes for powdery mildew resistance in wheat. Plant Breeding 119: 21-24.
- Kelly JD, Gepts P, Miklas PN, Coyne DP (2003) Tagging and mapping of genes and QTL and molecular marker-assisted selection for traits of economic importance in bean and cowpea. Field Crops Research 82: 135-154.
- Miklas PN, Smith JR, Riley R, Grafton KF, Singh SP, et al. (2000) Marker-assisted breeding for pyramided resistance to common bacterial blight in common bean. Annual Report-Bean Improvement Cooperative 43: 39-40.
- Ragagnin VA, De Souza TLPO, Sanglard DA, Arruda KMA, Costa MR, et al. (2009) Development and agronomic performance of common bean lines simultaneously resistant to anthracnose, angular leaf spot and rust. Plant Breeding 128: 156-163.
- Ferreira JJ, Campa A, Pérez-Vega E, RodrÃguez-Suárez C, Giraldez R (2012) Introgression and pyramiding into common bean market class fabada of genes conferring resistance to anthracnose and potyvirus. Theoretical and Applied Genetics 124: 777-788.
- Kumar VA, Balachiranjeevi CH, Naik SB, Rekha G, Rambabu R, et al. (2017) Marker-assisted pyramiding of bacterial blight and gall midge resistance genes into RPHR-1005, the restorer line of the popular rice hybrid DRRH-3. Molecular Breeding 37: 86.
- Deshmukh UC, Verma RK, Saxena RR, Mohan P, Verulkar SB (2017) Marker assisted selection for bacterial leaf blight resistance in segregating populations of Karma Mahsuri. Vegetos 30: 1.
- Caixeta ET, Borém A, Alzate-Marin AL, De Azevedo-Fagundes S, Silva MGDM, et al. (2005) Allelic relationships for genes that confer resistance to angular leaf spot in common bean. Euphytica 145: 237-245.
- Sartorato A, Nietsche S, Barros EG, Moreira MA (2000) RAPD and SCAR markers linked to resistance gene to angular leaf spot in common beans. Fitopatologia Brasileira 25: 637-642.
- Fourie D, Herselman L (2011) Application of molecular markers in breeding for bean common blight resistance in South Africa. African Crop Science Journal 19: 369-376.
- Duncan RW, Gilbertson RL, Singh SP (2012) Direct and marker-assisted selection for resistance to common bacterial blight in common bean. Crop Science 52: 1511-1521.
- Miller T, Gepts P, Kimmo S, Arunga E, Chilagane LA, et al. (2018) Alternative markers linked to the Phg-2 angular leaf spot resistance locus in common bean using the Phaseolus genes marker database. African Journal of Biotechnology 17: 818-828.
- Namayanja A, Buruchara R, Mahuku G, Rubaihayo P, Kimani P, et al. (2006) Inheritance of resistance to angular leaf spot in common bean and validation of the utility of resistance linked markers for marker assisted selection out side the mapping population. Euphytica 151: 361-369.
- Ddamulira G, Mukankusi C, Ochwo-Ssemakula M, Edema R, Sseruwagi P, et al. (2015) Gene pyramiding improved resistance to angular leaf spot in common bean. American Journal of Experimental Agriculture 9: 1-12.
- Kumar GA, Hanchinal RR, Desai S, Biradar S (2017) Marker assisted gene pyramiding of leaf rust resistance genes Lr24 and Lr28 in the background of wheat variety DWR 162 (Triticum aestivum L.). Int J Curr Microbiol App Sci 6: 1883-1893.
- Collard BC, Mackill DJ (2009) Start codon targeted (SCoT) polymorphism: A simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Molecular Biology Reporter 27: 86.
Citation: Rezene Y, Tesfaye K, Mukankusi C, Gepts P (2019) Marker-Assisted Pyramiding Resistance Genes Against Angular Leaf Spot and Common Bacterial Blight Disease into Preferred Common Bean Cultivar “REDWOLAITA”. Adv Crop Sci Tech 7: 416. DOI: 10.4172/2329-8863.1000416
Copyright: © 2019 Rezene Y, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 4819
- [From(publication date): 0-2019 - Apr 19, 2025]
- Breakdown by view type
- HTML page views: 3810
- PDF downloads: 1009