LncRNA Microarray Analysis of Non-small Cell Lung Cancer A549 Cell Lines Treated with Phycocyanin
Received: 11-Dec-2023 / Manuscript No. cmb-23-122475 / Editor assigned: 14-Dec-2023 / PreQC No. cmb-23-122475 / Reviewed: 28-Dec-2023 / QC No. cmb-23-122475 / Revised: 04-Jan-2024 / Manuscript No. cmb-23-122475 / Published Date: 29-Jan-2024
Abstract
Phycocyanin is a type of marine food additive with multiple biological properties, including anticancer activity, but its underlying antineoplastic mechanism in non-small cell lung cancer (NSCLC) remains unclear. To investigate the underlying regulatory mechanism of phycocyanin in NSCLC, an LncRNA microarray analysis was performed using a phycocyanin-treated A549 cell model. The classification and expression of LncRNA s were determined. The profiles of differentially expressed LncRNA s were generated and analyzed using Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) analyses. The results showed that 193 LncRNA s were upregulated and 116 LncRNA s were downregulated in the phycocyanin-treated group compared with the control group, and qRT‒ PCR analysis confirmed the expression of selected LncRNA s. Bioinformatic analysis indicated that the differentially expressed LncRNA s and their target genes were enriched in the extracellular region, epithelium development, NODlike receptor pathway, Notch signaling, and apoptosis process. In addition, coexpression network analysis identified 2,238 LncRNA‒mRNA, LncRNA‒LncRNA, and mRNA‒mRNA pairs. In particular, 72 etc. differentially expressed LncRNA target genes were discovered in the interaction network, which provides insights into the potential mechanism of phycocyanin in A549 cells. Moreover, cell phenotype experiments showed that downregulating the expression of LncRNA ENST00000538717, a LncRNA that is downregulated after phycocyanin treatment, could significantly inhibit the migration and viability of A549 and H460 cells. Consequently, this study lays a theoretical and potential foundation for NSCLC treatment and advances our understanding of the regulatory mechanisms of phycocyanin.
Keywords
A549 cells; Anticancer; LncRNA; Non-small cell lung cancer (NSCLC); Phycocyanin
sIntroduction
Lung cancer remained the leading cause of cancer-related death in 2020, with an estimated 1.8 million deaths [1]. Non-small cell lung cancer (NSCLC), a subtype of lung cancer, accounts for approximately 85% of lung cancer cases and is characterized by high mortality and a low cure rate [2,3]. Currently, most NSCLCs are not effectively diagnosed at the early stage. Although treatments such as radiation therapy, chemotherapy or surgery have improved the survival rates of patients to a certain extent [4], the therapeutic efficacy of treatments for NSCLC is still very poor due to the lack of known biomarkers of NSCLC [4,5]. Therefore, in-depth research to identify specific biomarkers of NSCLC is urgently needed for the development of new drugs that effectively treat NSCLC.
Long noncoding RNAs (LncRNA s) are a class of regulatory RNA molecules that are larger than 200 nucleotides and have no protein-coding ability [6]. Unlike microRNAs and small nucleolar/ nuclear RNAs, LncRNA s have complex higher order structures [7]. It has been shown that the ectopic expression of LncRNA s is relevant to the occurrence and metabolism of lung cancer. Luo et al. reported that LncRNA TRERNA1 could promote the malignant progression of NSCLC by targeting FOXL1 [8]. Zhao et al. found that LncRNA SNHG14 also contributed to the growth of NSCLC via the miR-206/G6PD pathway [9]. In addition, some other LncRNA s, such as FENDRR, could exert antineoplastic effects on NSCLC cells by regulating the miR-761/TIMP2 axis [10]. Therefore, in-depth exploration of the regulatory role of LncRNA s in NSCLC and their mechanism is of great importance for the future treatment of NSCLC.
Phycocyanin, a pigment-protein complex derived from cyanobacteria, is a well-known natural food additive [11]. It has been revealed that phycocyanin, which is a potential natural antineoplastic factor, exerts antitumor effects on NSCLC [12,13]. Previously, we performed a miRNA-seq analysis and discovered that miR-6883- 3p, miR-3150a-3p, miR-642a-5p, and miR-627-5p were involved in the growth inhibitory effect of phycocyanin on NSCLC cells [14,15]. However, few studies have investigated the LncRNA expression profiles in phycocyanin-treated NSCLC cell lines. As LncRNA s are the upstream regulators of miRNAs in cells [16], further exploration of the expression profiles of LncRNA s participating in the phycocyaninmediated antineoplastic process is vital to elucidating the mechanism of phycocyanin-mediated inhibitory effects on NSCLC.
In the present study, to further investigate the precise antineoplastic regulatory mechanism of phycocyanin in NSCLC, a LncRNA-seq was performed to establish the LncRNA profile of NSCLC A549 cells after phycocyanin exposure. The results of this work are expected to explore the anti-tumor mechanism of phycocyanin at the molecular level, which undoubtedly laid a theoretical basis for the potential treatment of NSCLC with phycocyanin and provided insights into the regulatory mechanism of phycocyanin.
Materials and Methods
Study information
This work included experimental techniques such as cell migration and cell viability assays, RNA extraction, RNA-seq assay, qRT‒PCR, and bioinformatics analysis. The present work was performed in the labs of Beijing Engineering and Technology Research Center of Food Additives, Beijing Technology and Business University in 2023.
Cell culture and sample treatment
A549 and H460 cells were cultured in DMEM supplemented with 0.1 mg/mL streptomycin, 100 units/mL penicillin and 10% fetal bovine serum (FBS) in a humidified atmosphere with 5% CO2 at 37 °C (American Type Cell Collection, ATCC, Manassas, VA, USA). Phycocyanin (95% purity) was purchased from Enviroloix (Portland, ME, USA). A549 cells were treated with phycocyanin or PBS as a control, followed by cell lysis and RNA extraction. The control groups were named Con-1, 2 and 3, and the phycocyanin-treated groups were named PC-1, 2 and 3.
RNA extraction and quality evaluation
The High Pure RNA isolation kit was employed for total RNA extraction according to the manufacturer’s specification (Roche, Penzberg, Germany). A Qubit fluorometer (Life Technologies, Eugene, OR, USA) with the Qubit RNA Assay Kit and NanoDrop-1000 instrument were used to evaluate the quality of RNA (Thermo Fisher Scientific Inc., Waltham, USA). The Agilent Bioanalyzer microcapillary electrophoresis system was used to assess RNA quality (Agilent, Santa Clara, CA, USA).
RNA sequencing
Two milligrams of polyA-minus RNA were used for cDNA library establishment. After total RNAs were extracted, mRNA and noncoding RNAs were enriched by removing rRNA, followed by fragmenting into short fragments (200-500 nt) using EpicentreRibo- Zero™ rRNA RemovalKit (Epicentre, WI, USA). RNA-sequencing libraries were generated under the instructions provided by NEB Next Ultra™ Directional RNA Library Prep kit forIllumina (New England BioLabs, Inc.). The Illumina HiSeqTM 2000 Sequencing System by Berry Genomics Inc. (Beijing, China) was employed for sequencing. Differential expression analysis for LncRNA from the database has proceeded. The sequencing reads were mapped to the reference genome using the TopHat program. A coding potential calculator assay was employed for evaluation. The quality control of raw data was performed using SeqPrep (https: //github.com/jstjohn/SeqPrep) and Sickle (https: //github.com/najoshi/sickle). TopHat2 (http: //ccb.jhu.edu/software/ tophat/index.shtml) was used for sequence comparative analysis after quality control. The Cufflinks software (http: //cole-trapnell-lab.github. io/cufflinks/) was employed for transcript assembly.
Analysis of differentially expressed (DE) LncRNA s
The number of mapped tags that was given by the raw expression levels of that particular LncRNA. To correct for differences in tag count between samples, the tag counts were scaled to CPM (count per millions) based on the total number of aligned tags. The analysis of differentially expressed LncRNA between control and phycocyanintreated samples was performed by edgeR. The setting was |logFC| > 1 & FDR < 0.05.
Target genes of LncRNA s and enrichment analysis
KEGG pathway analysis was employed for signal transduction pathway analysis of key genes. The p-value and gene count thresholds of KEGG analysis was p-adjust < 0.05 and 7, respectively. Gene Ontology (GO) analysis was used to identify important biological functions or processes of target genes. The p-value and gene count thresholds of GO analysis was p-adjust < 0.05 and 35, respectively.
qRT‒PCR
qRT‒PCR was performed by the Q6 qPCR system. GAPDH was used as the endogenous control gene. SYBR Green Master Mix was used for signal detection. The 2-ΔΔCT method was used to calculate the expression of each gene. The primer sequences of LncRNA and GAPDH for qRT‒PCR were as follows: forward: 5'-CGTTGCCTACAGTCAAGTCAGT- 3'; reverse: 5'-GCCATGGTTCCCTTACTGATAC-3'. Forward: 5'-TGTTCGTCATGGGTGTGAAC-3'; Reverse: 5'-ATGGCATGGACTGTGGTCAT- 3'.
Cell viability assay
Cells were cultured in 96-well plates, followed by LncRNA inhibitor incubation for 24 h. The MTT method was employed to determine the viability of A549 cells at 24 and 48 h. Briefly, MTT was added to each well for 4 h, followed by HCl-SDS lysis buffer and the cells were incubated for 24 h. The absorbance at 570 nm was recorded using a micro plate reader.
Cell migration assay
Cells were incubated with LncRNA inhibitor and cultured in 6-well plates. A pipette tip was used to scratch the cells and create a "wound". Each wound was observed at three different positions and its width was recorded. The three width values were averaged. The widths of the wounds were observed at 0 and 24 h. The cell migration rate was calculated from the difference in scratch width at 24 h over the original width (0 h).
Statistical Analysis
The data were analyzed by Microsoft Excel and GraphPad Prism. A two-tailed Student’s t test (paired t-test) was used for data analysis, and p < 0.05 indicated a significant difference between the control and test groups.
Results and Discussion
Classification and subgroup analysis of LncRNA s
Phycocyanin has been reported to exert effective antineoplastic effects on NSCLC cells [12,13]. Further investigation of the underlying anticancer mechanism of phycocyanin can help clarify the pathogenesis of NSCLC, laying the theoretical basis for lung cancer treatment. LncRNA s are well-known important molecules that regulate gene expressi
| Sample | Known lncRNA num | Novel lncRNA num | Total |
|---|---|---|---|
| Con-1 | 25,075 | 1,458 | 26,533 |
| Con-2 | 24,543 | 1,464 | 26,007 |
| Con-3 | 25,111 | 1,486 | 26,597 |
| PC-1 | 25,334 | 1,468 | 26,802 |
| PC-2 | 25,836 | 1,481 | 27,317 |
| PC-3 | 25,738 | 1,483 | 27,221 |
Table S1: Statistics of lncRNAs.
Type |
Known lncRNA num | Novel lncRNA num | Total |
|---|---|---|---|
| Antisense | 17768 | 1224 | 18992 |
| Sense intron overlap | 615 | 25 | 640 |
| Intergenic | 36889 | 207 | 37096 |
| Sense exon overlap | 6813 | 362 | 7175 |
| Bidirection | 5147 | 2 | 5149 |
Table S2: Classification and subgroup of lncRNAs.
on at the transcriptional, epigenetic, and posttranscriptional levels [17]. Numerous LncRNA s have been identified by highthroughput technology and are related to growth and development in cancer cells.
Over 26,000 LncRNA s (including known and novel LncRNA s) was identified in the Con and PC groups (Table S1). The chromosomal distribution and transcript types of LncRNA s were determined. In Table S2, of the antisense LncRNA s, the known LncRNA s accounted for 93.5%. Of the sense intron overlap, the known LncRNA s accounted for 96.1%. Of the intergenic LncRNA s, the known LncRNA s accounted for 99.4%. Of the sense exon overlap, the known LncRNA s accounted for 95.0%. Of the bidirectional LncRNA s, the known LncRNA s accounted for 99.9%.
Table S1: Statistics of lncRNAs.
Analysis of LncRNA expression and differentially expressed (DE) LncRNA s
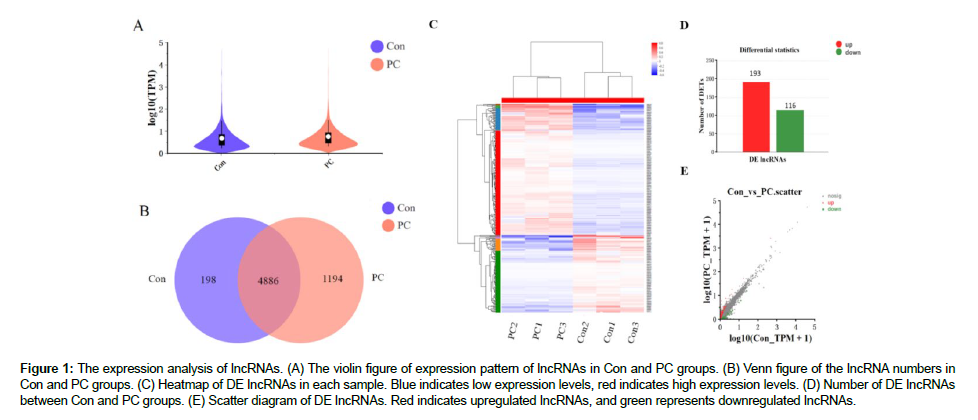
As shown in Figure 1A, the LncRNA expression patterns of the Con and PC samples were similar, indicating the suitable replication of each experiment. Finally, 5,084 and 6,080 LncRNA transcripts were obtained from the Con and PC groups, respectively, after eliminating the single-exon, low-expression, and unreliable fragments. Of these, 4886 LncRNA s were found in both Con and PC samples (Figure 1B). Table S3 presents the top 15 LncRNA s that are expressed in the Con and PC groups based on TPM (transcripts per million reads). Strikingly, some LncRNA s was highly expressed in both the Con and PC groups, including ENST00000602361, ENST00000554988, ENST00000618589, and ENST00000659240. Some LncRNA s exhibited sample-specific expression. For instance, NONHSAT135828.2 and ENST00000508832 were specifically highly expressed in phycocyanin-treated A549 cells, while NONHSAT190639.1 and NONHSAT176665.1 were specifically highly expressed in control cells.
Figure 1: The expression analysis of lncRNAs. (A) The violin figure of expression pattern of lncRNAs in Con and PC groups. (B) Venn figure of the lncRNA numbers in Con and PC groups. (C) Heatmap of DE lncRNAs in each sample. Blue indicates low expression levels, red indicates high expression levels. (D) Number of DE lncRNAs between Con and PC groups. (E) Scatter diagram of DE lncRNAs. Red indicates upregulated lncRNAs, and green represents downregulated lncRNAs.
| Con | PC | ||
|---|---|---|---|
| lncRNAs | TPM | lncRNAs | TPM |
| ENST00000602361 | 58511.15 | ENST00000602361 | 64810.17 |
| ENST00000554988 | 16530.64 | ENST00000554988 | 14324.52 |
| ENST00000618589 | 8645.37 | ENST00000618589 | 7671.39 |
| NONHSAT243766.1 | 6137.81 | NONHSAT243766.1 | 6700.25 |
| ENST00000659240 | 4918.28 | ENST00000659240 | 5387.17 |
| NONHSAT052027.2 | 2032.83 | NONHSAT135828.2 | 2919.21 |
| NONHSAT146442.2 | 1884.87 | ENST00000610851 | 2154.29 |
| ENST00000618227 | 1408.33 | ENST00000618227 | 2010.76 |
| ENST00000616691 | 1106.53 | ENST00000616691 | 1871.71 |
| NONHSAT021728.2 | 1076.28 | NONHSAT052027.2 | 1701.58 |
| ENST00000610851 | 1071.01 | ENST00000508832 | 1288.08 |
| ENST00000541782 | 994.52 | ENST00000534336 | 1157.87 |
| NONHSAT190639.1 | 910.08 | NONHSAT021728.2 | 977.59 |
| ENST00000534336 | 625.55 | ENST00000541782 | 838.71 |
| NONHSAT176665.1 | 594.18 | NONHSAT146442.2 | 707.9 |
Table S3: The top expressed 15 lncRNA transcripts in Con and PC groups.
The heat map of differentially expressed LncRNA s showed that the expression patterns of LncRNA s within Con or PC samples were consistent, while the expression profiles between Con and PC groups exerted extremely significant differences (Figure 1C), indicating the effects of phycocyanin on the expression of LncRNA s in A549 cells. In total, 309 LncRNA s exhibited significant expression differences between the Con and PC groups, including 193 significantly upregulated and 116 significantly downregulated LncRNA s after phycocyanin treatment in A549 cells (Figure 1D). The scatter diagram of DE LncRNA s is shown in Figure 1E. The detailed information of DE LncRNA s was shown in Table S4. This LncRNA s is likely to be key LncRNA s that may act as downstream targets of phycocyanin in NSCLC cells. Table S5 shows the top 10 downregulated and top 10 upregulated LncRNA transcripts.
| lncRNA id | lncRNA gene id | FC(PC/Con) | Log2FC(PC/Con) | Pvalue | Padjust | Significant | Regulate | Con1 | Con2 | Con3 | PC1 | PC2 | PC3 | Con | PC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ENST00000375633 | ENSG00000204387 | 2.216937934 | 1.148568381 | 0.00064 | 0.00839 | yes | up | 4.85 | 2.08 | 4.42 | 9.61 | 10.64 | 8.38 | 3.78333 | 9.54333 |
| ENST00000375642 | ENSG00000204387 | 2.252230455 | 1.171354456 | 0.00246 | 0.02563 | yes | up | 0.82 | 0.84 | 1.31 | 1.79 | 3.19 | 2.62 | 0.99 | 2.53333 |
| ENST00000383808 | ENSG00000206567 | 2.194757374 | 1.134061462 | 0.00024 | 0.00365 | yes | up | 0.19 | 0.21 | 0.14 | 0.46 | 0.49 | 0.43 | 0.18 | 0.46 |
| ENST00000399543 | ENSG00000215068 | 20.55209258 | 4.361213389 | 7.3E-05 | 0.00128 | yes | up | 0 | 0 | 0.04 | 0.3 | 0.46 | 0.28 | 0.01333 | 0.34667 |
| ENST00000399868 | ENSG00000215244 | 74.36631846 | 6.216577447 | 0.00018 | 0.00285 | yes | up | 0 | 0 | 0 | 0.25 | 0.09 | 0.06 | 0 | 0.13333 |
| ENST00000413221 | ENSG00000224032 | 0.497442355 | -1.007398742 | 0.00198 | 0.02144 | yes | down | 0.94 | 1.32 | 1.76 | 0.87 | 0.84 | 0.6 | 1.34 | 0.77 |
| ENST00000416463 | ENSG00000242282 | 58.76533342 | 5.876893433 | 0.00181 | 0.01999 | yes | up | 0 | 0 | 0 | 0.9 | 0.24 | 2.18 | 0 | 1.10667 |
| ENST00000418927 | ENSG00000235269 | 0.0321649 | -4.958369 | 0.0007 | 0.00903 | yes | down | 0.09 | 0.12 | 0.19 | 0 | 0 | 0.01 | 0.13333 | 0.00333 |
| ENST00000422944 | ENSG00000228022 | 74.47820287 | 6.218746356 | 0.00042 | 0.00589 | yes | up | 0 | 0 | 0 | 0.08 | 0.3 | 0.71 | 0 | 0.36333 |
| ENST00000423477 | ENSG00000225439 | 127.2206537 | 6.991189095 | 1.3E-05 | 0.00027 | yes | up | 0 | 0 | 0 | 0.75 | 0.34 | 0.12 | 0 | 0.40333 |
| ENST00000424518 | ENSG00000228630 | 56.7453908 | 5.826431308 | 0.00018 | 0.00285 | yes | up | 0 | 0 | 0 | 0.1 | 0.12 | 0.17 | 0 | 0.13 |
| ENST00000424684 | ENSG00000263590 | 2.676037221 | 1.420098183 | 4.5E-05 | 0.00083 | yes | up | 0.14 | 0.1 | 0.18 | 0.36 | 0.52 | 0.39 | 0.14 | 0.42333 |
| ENST00000426023 | ENSG00000237807 | 0.204739826 | -2.288136334 | 0.00154 | 0.01755 | yes | down | 0.14 | 0.22 | 0.19 | 0.05 | 0.04 | 0.05 | 0.18333 | 0.04667 |
| ENST00000428862 | ENSG00000224596 | 2.373646048 | 1.24710482 | 0.00087 | 0.01084 | yes | up | 0.24 | 0.22 | 0.2 | 0.72 | 0.42 | 0.68 | 0.22 | 0.60667 |
| ENST00000430034 | ENSG00000240801 | 4.690574082 | 2.229764506 | 2.6E-16 | 3.4E-14 | yes | up | 4.07 | 2.77 | 2.9 | 17.97 | 19.01 | 15.26 | 3.24667 | 17.4133 |
| ENST00000430373 | ENSG00000227811 | 135.5234258 | 7.082398439 | 1.2E-07 | 4.1E-06 | yes | up | 0 | 0 | 0 | 0.37 | 0.37 | 0.27 | 0 | 0.33667 |
| ENST00000432735 | ENSG00000215424 | 3.300294589 | 1.722594807 | 0.00066 | 0.00858 | yes | up | 0.24 | 0.22 | 0.72 | 1.49 | 1.73 | 1.26 | 0.39333 | 1.49333 |
| ENST00000434233 | ENSG00000218510 | 0.358038271 | -1.481814289 | 0.00102 | 0.0124 | yes | down | 1.83 | 1.17 | 1.99 | 0.78 | 0.41 | 0.84 | 1.66333 | 0.67667 |
| ENST00000435356 | ENSG00000203392 | 2.104257514 | 1.073311269 | 3.2E-06 | 7.9E-05 | yes | up | 0.8 | 0.7 | 0.67 | 1.68 | 1.56 | 2.01 | 0.72333 | 1.75 |
| ENST00000439559 | ENSG00000224914 | 2.551430127 | 1.351306133 | 2.6E-09 | 1.3E-07 | yes | up | 0.5 | 0.45 | 0.62 | 1.62 | 1.28 | 1.68 | 0.52333 | 1.52667 |
| ENST00000441052 | ENSG00000235314 | 97.64557286 | 6.60948273 | 2.1E-06 | 5.5E-05 | yes | up | 0 | 0 | 0 | 0.25 | 0.2 | 0.17 | 0 | 0.20667 |
| ENST00000443596 | ENSG00000234608 | 0.006900498 | -7.179083786 | 1.8E-06 | 4.7E-05 | yes | down | 1.6 | 0.75 | 0.37 | 0 | 0 | 0 | 0.90667 | 0 |
| ENST00000443993 | ENSG00000227619 | 0.009553142 | -6.709809046 | 0.00024 | 0.00364 | yes | down | 0.02 | 0.31 | 0.31 | 0 | 0 | 0 | 0.21333 | 0 |
| ENST00000444503 | ENSG00000232490 | 0.020634848 | -5.5987734 | 0.00229 | 0.02411 | yes | down | 1.29 | 0.4 | 0.18 | 0 | 0 | 0 | 0.62333 | 0 |
| ENST00000444998 | ENSG00000215424 | 0.043504416 | -4.522694346 | 6.4E-06 | 0.00015 | yes | down | 0.58 | 0.43 | 0.49 | 0.08 | 0 | 0 | 0.5 | 0.02667 |
| ENST00000455395 | ENSG00000230590 | 34.18808119 | 5.095421549 | 2.2E-05 | 0.00045 | yes | up | 0.06 | 0 | 0 | 0.32 | 0.73 | 1.13 | 0.02 | 0.72667 |
| ENST00000456953 | ENSG00000196756 | 0.014481228 | -6.109672284 | 0.00011 | 0.00184 | yes | down | 0.63 | 0.23 | 0.27 | 0 | 0 | 0 | 0.37667 | 0 |
| ENST00000457670 | ENSG00000228412 | 0.017289583 | -5.853953131 | 0.00061 | 0.00804 | yes | down | 0.11 | 0.3 | 0.07 | 0 | 0 | 0 | 0.16 | 0 |
| ENST00000458178 | ENSG00000224086 | 2.081220275 | 1.057429667 | 5.3E-05 | 0.00095 | yes | up | 0.04 | 0.05 | 0.04 | 0.12 | 0.09 | 0.1 | 0.04333 | 0.10333 |
| ENST00000466953 | ENSG00000254154 | 2.455148772 | 1.295810448 | 0.00474 | 0.04321 | yes | up | 0.05 | 0.05 | 0.05 | 0.18 | 0.12 | 0.12 | 0.05 | 0.14 |
| ENST00000475197 | ENSG00000231177 | 39.90459136 | 5.318482845 | 1.3E-37 | 1.1E-34 | yes | up | 0.14 | 0.16 | 0.03 | 5.69 | 4.92 | 4.71 | 0.11 | 5.10667 |
| ENST00000488584 | ENSG00000240875 | 0.003520513 | -8.149998575 | 5.1E-09 | 2.4E-07 | yes | down | 0.52 | 0.16 | 0.65 | 0 | 0 | 0 | 0.44333 | 0 |
| ENST00000498979 | ENSG00000245248 | 0.398254693 | -1.328236734 | 0.00558 | 0.04934 | yes | down | 0.34 | 0.48 | 0.26 | 0.17 | 0.16 | 0.18 | 0.36 | 0.17 |
| ENST00000500698 | ENSG00000245522 | 2.781788251 | 1.476012607 | 0.00048 | 0.0066 | yes | up | 0.19 | 0.16 | 0.12 | 0.46 | 0.56 | 0.49 | 0.15667 | 0.50333 |
| ENST00000501122 | ENSG00000245532 | 2.811058729 | 1.491113595 | 1E-105 | 2E-101 | yes | up | 18.09 | 15.14 | 18.49 | 59.35 | 52.11 | 55 | 17.24 | 55.4867 |
| ENST00000503469 | ENSG00000250041 | 0.027805182 | -5.168502408 | 0.00385 | 0.03684 | yes | down | 0.58 | 0.32 | 0.98 | 0 | 0 | 0 | 0.62667 | 0 |
| ENST00000523313 | ENSG00000249395 | 0.018744053 | -5.737423242 | 0.00097 | 0.01187 | yes | down | 0.49 | 0.23 | 1.17 | 0 | 0 | 0 | 0.63 | 0 |
| ENST00000526566 | ENSG00000255438 | 157.724407 | 7.301262117 | 0.00157 | 0.01777 | yes | up | 0 | 0 | 0 | 0.6 | 0 | 0.46 | 0 | 0.35333 |
| ENST00000534918 | ENSG00000225138 | 3.284735992 | 1.71577742 | 3.8E-08 | 1.5E-06 | yes | up | 0.37 | 0.3 | 0.19 | 1.11 | 1 | 1.12 | 0.28667 | 1.07667 |
| ENST00000538717 | ENSG00000231177 | 0.000963561 | -10.01933648 | 5.9E-17 | 8.4E-15 | yes | down | 4.64 | 3.6 | 4.4 | 0 | 0 | 0 | 4.21333 | 0 |
| ENST00000547387 | ENSG00000258017 | 0.359580402 | -1.475613701 | 4.7E-06 | 0.00011 | yes | down | 3.25 | 3.45 | 3.6 | 1.33 | 1.93 | 0.95 | 3.43333 | 1.40333 |
| ENST00000550290 | ENSG00000257354 | 16.18316295 | 4.016421701 | 8.7E-17 | 1.2E-14 | yes | up | 0.02 | 0.04 | 0.09 | 0.71 | 0.84 | 1.17 | 0.05 | 0.90667 |
| ENST00000555578 | ENSG00000258701 | 120.6971396 | 6.915247676 | 1.1E-06 | 3.1E-05 | yes | up | 0 | 0 | 0 | 0.18 | 0.23 | 0.38 | 0 | 0.26333 |
| ENST00000562992 | ENSG00000260693 | 2.434028955 | 1.28334633 | 5.1E-05 | 0.00093 | yes | up | 1.39 | 1.35 | 0.99 | 3.94 | 3.18 | 3.29 | 1.24333 | 3.47 |
| ENST00000569326 | ENSG00000255182 | 2.01035948 | 1.007453498 | 0.00044 | 0.00616 | yes | up | 0.28 | 0.27 | 0.24 | 0.55 | 0.71 | 0.57 | 0.26333 | 0.61 |
| ENST00000571963 | ENSG00000263072 | 0.012286251 | -6.346811482 | 6.7E-05 | 0.00117 | yes | down | 0.27 | 0.41 | 0.11 | 0 | 0 | 0 | 0.26333 | 0 |
| ENST00000572035 | ENSG00000255135 | 134.2814143 | 7.069115827 | 1.7E-06 | 4.5E-05 | yes | up | 0 | 0 | 0 | 1.5 | 0.51 | 1.61 | 0 | 1.20667 |
| ENST00000576232 | ENSG00000224870 | 59.42880362 | 5.893090433 | 0.00014 | 0.00224 | yes | up | 0 | 0.02 | 0 | 0.88 | 0.82 | 0.47 | 0.00667 | 0.72333 |
| ENST00000602890 | ENSG00000269888 | 0.487903558 | -1.035332091 | 0.00435 | 0.0405 | yes | down | 22.65 | 37.62 | 20.15 | 14.19 | 18.24 | 12.9 | 26.8067 | 15.11 |
| ENST00000606343 | ENSG00000272068 | 2.088855178 | 1.062712473 | 7E-10 | 3.8E-08 | yes | up | 1.09 | 0.85 | 1.21 | 2.54 | 2.32 | 2.69 | 1.05 | 2.51667 |
| ENST00000606892 | ENSG00000231889 | 34.48069722 | 5.107717042 | 0.00037 | 0.0052 | yes | up | 0 | 0 | 0.05 | 0.78 | 0.5 | 0.52 | 0.01667 | 0.6 |
| ENST00000609111 | ENSG00000272734 | 2.419563751 | 1.274746952 | 0.00064 | 0.00843 | yes | up | 0.41 | 0.25 | 0.21 | 0.58 | 1.01 | 0.79 | 0.29 | 0.79333 |
| ENST00000609983 | ENSG00000234608 | 0.290601301 | -1.782886935 | 0.00223 | 0.02364 | yes | down | 2.59 | 3.9 | 3.04 | 0.3 | 1.25 | 1.56 | 3.17667 | 1.03667 |
| ENST00000612722 | ENSG00000275620 | 2.736120192 | 1.452131606 | 2.3E-08 | 9.2E-07 | yes | up | 0.17 | 0.25 | 0.28 | 0.63 | 0.84 | 0.71 | 0.23333 | 0.72667 |
| ENST00000615493 | ENSG00000242086 | 0.007129691 | -7.131944741 | 3.6E-06 | 9E-05 | yes | down | 0.38 | 1.22 | 2.01 | 0 | 0 | 0 | 1.20333 | 0 |
| ENST00000628917 | ENSG00000242086 | 0.010493388 | -6.574375663 | 6.2E-06 | 0.00014 | yes | down | 2 | 1.33 | 0.86 | 0.04 | 0 | 0 | 1.39667 | 0.01333 |
| ENST00000632919 | ENSG00000233871 | 2.338666909 | 1.225686397 | 0.0021 | 0.0225 | yes | up | 0.17 | 0.2 | 0.19 | 0.48 | 0.51 | 0.52 | 0.18667 | 0.50333 |
| ENST00000648209 | ENSG00000255248 | 69.55296901 | 6.120040195 | 6.4E-05 | 0.00113 | yes | up | 0 | 0 | 0 | 0.2 | 0.11 | 0.09 | 0 | 0.13333 |
| ENST00000650025 | ENSG00000236204 | 137.239545 | 7.100552438 | 0.00235 | 0.02466 | yes | up | 0 | 0 | 0 | 0.56 | 0.56 | 0 | 0 | 0.37333 |
| ENST00000651080 | ENSG00000234741 | 0.016455684 | -5.925270221 | 0.00026 | 0.00381 | yes | down | 0.5 | 0.12 | 0.34 | 0 | 0 | 0 | 0.32 | 0 |
| ENST00000651958 | ENSG00000280441 | 2.853676227 | 1.512821658 | 0.00104 | 0.0126 | yes | up | 0.36 | 0.76 | 0.64 | 1.7 | 2.31 | 1.69 | 0.58667 | 1.9 |
| ENST00000652379 | ENSG00000280441 | 2.168376705 | 1.116615413 | 0.00308 | 0.03073 | yes | up | 0.89 | 0.76 | 0.64 | 1.7 | 2.31 | 1.69 | 0.76333 | 1.9 |
| ENST00000652496 | ENSG00000230590 | 2.712508216 | 1.439627507 | 0.00093 | 0.01155 | yes | up | 0.19 | 0.15 | 0.26 | 0.72 | 0.51 | 0.62 | 0.2 | 0.61667 |
| ENST00000652746 | ENSG00000233237 | 0.204178555 | -2.292096749 | 0.00203 | 0.02194 | yes | down | 0.6 | 0.61 | 0.4 | 0.18 | 0.17 | 0.02 | 0.53667 | 0.12333 |
| ENST00000653437 | ENSG00000253190 | 0.014712341 | -6.08682934 | 6.9E-05 | 0.00121 | yes | down | 0.31 | 0.63 | 0.34 | 0 | 0 | 0 | 0.42667 | 0 |
| ENST00000653856 | ENSG00000257261 | 0.015395207 | -6.021374887 | 6.1E-05 | 0.00109 | yes | down | 0.5 | 0.27 | 0.54 | 0 | 0 | 0 | 0.43667 | 0 |
| ENST00000654005 | ENSG00000225470 | 566.8307621 | 9.146774246 | 3E-11 | 2E-09 | yes | up | 0 | 0 | 0 | 1 | 0.22 | 0.43 | 0 | 0.55 |
| ENST00000654166 | ENSG00000183250 | 105.6221692 | 6.722768866 | 0.00499 | 0.04512 | yes | up | 0 | 0 | 0 | 0.25 | 0.34 | 0 | 0 | 0.19667 |
| ENST00000654981 | ENSG00000249406 | 55.73493256 | 5.800509933 | 0.0002 | 0.00313 | yes | up | 0 | 0 | 0 | 0.34 | 0.19 | 0.24 | 0 | 0.25667 |
| ENST00000655003 | ENSG00000229043 | 90.72621219 | 6.503447523 | 0.00118 | 0.01404 | yes | up | 0 | 0 | 0 | 0.23 | 0.02 | 0.62 | 0 | 0.29 |
| ENST00000655030 | ENSG00000231105 | 50.36628587 | 5.654386442 | 0.00067 | 0.00875 | yes | up | 0 | 0 | 0 | 0.23 | 0.07 | 0.12 | 0 | 0.14 |
| ENST00000655308 | ENSG00000286483 | 0.047739545 | -4.388671359 | 0.00455 | 0.04193 | yes | down | 0.53 | 0.25 | 0.29 | 0 | 0 | 0.06 | 0.35667 | 0.02 |
| ENST00000655351 | ENSG00000272695 | 65.0106122 | 6.022603335 | 0.00014 | 0.00218 | yes | up | 0 | 0 | 0 | 0.25 | 0.12 | 0.3 | 0 | 0.22333 |
| ENST00000655606 | ENSG00000223745 | 3.904617781 | 1.965181331 | 1E-06 | 2.8E-05 | yes | up | 0.34 | 0.64 | 0.24 | 1.62 | 1.73 | 2.18 | 0.40667 | 1.84333 |
| ENST00000656647 | ENSG00000261824 | 0.025922263 | -5.269664523 | 0.00426 | 0.03986 | yes | down | 0.31 | 0.1 | 0.07 | 0 | 0 | 0 | 0.16 | 0 |
| ENST00000656756 | ENSG00000259583 | 3.744799819 | 1.9048886 | 0.00067 | 0.00869 | yes | up | 0.13 | 0.21 | 0.61 | 1.74 | 1.09 | 1.23 | 0.31667 | 1.35333 |
| ENST00000657056 | ENSG00000286656 | 2.775175715 | 1.472579121 | 0.00432 | 0.04024 | yes | up | 0.21 | 0.22 | 0.13 | 0.56 | 0.65 | 0.56 | 0.18667 | 0.59 |
| ENST00000657070 | ENSG00000230590 | 0.010255411 | -6.607470867 | 7.9E-05 | 0.00137 | yes | down | 0.08 | 0.52 | 0.26 | 0 | 0 | 0 | 0.28667 | 0 |
| ENST00000657179 | ENSG00000225470 | 154.6829687 | 7.273170548 | 0.00177 | 0.01963 | yes | up | 0 | 0 | 0 | 0.18 | 0 | 0.3 | 0 | 0.16 |
| ENST00000657211 | ENSG00000249859 | 50.0528901 | 5.645381469 | 0.00502 | 0.04536 | yes | up | 0 | 0 | 0 | 0.04 | 0.44 | 0.19 | 0 | 0.22333 |
| ENST00000657677 | ENSG00000261824 | 2.306289108 | 1.205573375 | 0.0045 | 0.04156 | yes | up | 0.43 | 0.24 | 0.3 | 1.12 | 0.41 | 1.04 | 0.32333 | 0.85667 |
| ENST00000657910 | ENSG00000225470 | 0.00355346 | -8.136559685 | 6.5E-10 | 3.5E-08 | yes | down | 1.04 | 0.99 | 0.48 | 0 | 0 | 0 | 0.83667 | 0 |
| ENST00000658183 | ENSG00000255248 | 82.32441279 | 6.363248411 | 1E-05 | 0.00023 | yes | up | 0 | 0 | 0 | 0.46 | 0.42 | 0.29 | 0 | 0.39 |
| ENST00000658188 | ENSG00000228889 | 15.44441864 | 3.949013661 | 0.00047 | 0.00647 | yes | up | 0.06 | 0 | 0 | 0.39 | 0.36 | 0.3 | 0.02 | 0.35 |
| ENST00000658695 | ENSG00000231074 | 0.012796004 | -6.288162859 | 0.00141 | 0.01628 | yes | down | 0 | 0.09 | 0.08 | 0 | 0 | 0 | 0.05667 | 0 |
| ENST00000658813 | ENSG00000223546 | 0.196243661 | -2.349282039 | 0.00128 | 0.01501 | yes | down | 0.61 | 1.07 | 0.55 | 0.25 | 0.02 | 0.24 | 0.74333 | 0.17 |
| ENST00000658859 | ENSG00000224078 | 154.7157098 | 7.273475885 | 3.4E-08 | 1.3E-06 | yes | up | 0 | 0 | 0 | 0.21 | 0.2 | 0.15 | 0 | 0.18667 |
| ENST00000658963 | ENSG00000227388 | 5.104968491 | 2.351902056 | 0.00076 | 0.00972 | yes | up | 0.06 | 0.07 | 0.13 | 0.3 | 0.6 | 0.56 | 0.08667 | 0.48667 |
| ENST00000659892 | ENSG00000249859 | 0.023957439 | -5.383382498 | 0.00124 | 0.01463 | yes | down | 0.2 | 0.31 | 0.45 | 0 | 0 | 0 | 0.32 | 0 |
| ENST00000659899 | ENSG00000241316 | 91.38853589 | 6.513941295 | 4.7E-05 | 0.00085 | yes | up | 0 | 0 | 0 | 0.32 | 0.07 | 0.18 | 0 | 0.19 |
| ENST00000659912 | ENSG00000249859 | 64.67391991 | 6.01511215 | 0.00122 | 0.01447 | yes | up | 0 | 0 | 0 | 0.19 | 0.04 | 0.39 | 0 | 0.20667 |
| ENST00000660010 | ENSG00000257114 | 14.74642391 | 3.88229323 | 0.00116 | 0.0138 | yes | up | 0 | 0.1 | 0 | 0.74 | 0.53 | 0.45 | 0.03333 | 0.57333 |
| ENST00000660030 | ENSG00000263753 | 0.008190606 | -6.931814024 | 2.6E-07 | 8.5E-06 | yes | down | 0.14 | 0.12 | 0.17 | 0 | 0 | 0 | 0.14333 | 0 |
| ENST00000660248 | ENSG00000236144 | 2.746065006 | 1.457365778 | 0.0005 | 0.00677 | yes | up | 0.22 | 0.14 | 0.24 | 0.44 | 0.71 | 0.73 | 0.2 | 0.62667 |
| ENST00000660374 | ENSG00000223960 | 0.012959352 | -6.269862557 | 1.7E-05 | 0.00035 | yes | down | 0.27 | 0.19 | 0.34 | 0 | 0 | 0 | 0.26667 | 0 |
| ENST00000660542 | ENSG00000224078 | 169.7151079 | 7.406971188 | 0.00113 | 0.01355 | yes | up | 0 | 0 | 0 | 0 | 0.18 | 0.18 | 0 | 0.12 |
| ENST00000660547 | ENSG00000255717 | 0.381074532 | -1.3918549 | 0.00032 | 0.00465 | yes | down | 4.02 | 1.66 | 4.28 | 1.11 | 1.52 | 1.67 | 3.32 | 1.43333 |
| ENST00000660608 | ENSG00000232931 | 3.358055155 | 1.747625926 | 1.6E-07 | 5.6E-06 | yes | up | 0.27 | 0.2 | 0.24 | 0.96 | 0.89 | 0.92 | 0.23667 | 0.92333 |
| ENST00000662234 | ENSG00000248932 | 5.299729337 | 2.405918682 | 4.8E-05 | 0.00088 | yes | up | 0.06 | 0.04 | 0.05 | 0.25 | 0.26 | 0.45 | 0.05 | 0.32 |
| ENST00000662245 | ENSG00000203875 | 0.019208769 | -5.702091087 | 1.2E-05 | 0.00025 | yes | down | 1.1 | 1.69 | 0.46 | 0 | 0 | 0.07 | 1.08333 | 0.02333 |
| ENST00000662517 | ENSG00000287300 | 0.049542159 | -4.335199434 | 0.00081 | 0.01027 | yes | down | 0.13 | 0.14 | 0.17 | 0.02 | 0 | 0 | 0.14667 | 0.00667 |
| ENST00000662585 | ENSG00000266903 | 15.17084775 | 3.923229801 | 0.00032 | 0.00463 | yes | up | 0 | 0.01 | 0.06 | 0.49 | 0.56 | 0.21 | 0.02333 | 0.42 |
| ENST00000662995 | ENSG00000233828 | 0.093942816 | -3.412073354 | 0.00538 | 0.04788 | yes | down | 0.18 | 0.16 | 0.23 | 0 | 0 | 0.06 | 0.19 | 0.02 |
| ENST00000663248 | ENSG00000189223 | 67.35267698 | 6.073663383 | 8.4E-05 | 0.00144 | yes | up | 0 | 0 | 0 | 0.06 | 0.05 | 0.02 | 0 | 0.04333 |
| ENST00000663347 | ENSG00000287275 | 2.049996816 | 1.035621669 | 0.00202 | 0.0218 | yes | up | 0.37 | 0.26 | 0.22 | 0.64 | 0.66 | 0.69 | 0.28333 | 0.66333 |
| ENST00000663448 | ENSG00000203875 | 0.067900738 | -3.880428928 | 0.00107 | 0.01291 | yes | down | 0.54 | 0.54 | 0.53 | 0 | 0 | 0.13 | 0.53667 | 0.04333 |
| ENST00000664934 | ENSG00000263753 | 0.007109624 | -7.136010945 | 2.5E-05 | 0.00048 | yes | down | 0.04 | 0.43 | 0.2 | 0 | 0 | 0 | 0.22333 | 0 |
| ENST00000664939 | ENSG00000175061 | 2.783669177 | 1.476987765 | 6E-07 | 1.8E-05 | yes | up | 0.41 | 0.32 | 0.54 | 1.05 | 1.51 | 1.44 | 0.42333 | 1.33333 |
| ENST00000665650 | ENSG00000227373 | 0.027988853 | -5.159003825 | 0.00221 | 0.02345 | yes | down | 0.04 | 0.02 | 0.03 | 0 | 0 | 0 | 0.03 | 0 |
| ENST00000667654 | ENSG00000274265 | 0.402399911 | -1.313298109 | 0.00103 | 0.01248 | yes | down | 1.29 | 2.24 | 1.79 | 0.81 | 0.79 | 0.85 | 1.77333 | 0.81667 |
| ENST00000667709 | ENSG00000247311 | 36.03784189 | 5.171440713 | 0.00456 | 0.04199 | yes | up | 0 | 0 | 0 | 0.22 | 0.11 | 0.39 | 0 | 0.24 |
| ENST00000667751 | ENSG00000288093 | 2.090481995 | 1.063835618 | 0.00187 | 0.02055 | yes | up | 0.37 | 0.31 | 0.3 | 0.77 | 0.89 | 0.71 | 0.32667 | 0.79 |
| ENST00000667900 | ENSG00000243176 | 0.008450125 | -6.886811629 | 1.4E-06 | 3.9E-05 | yes | down | 0.21 | 0.19 | 0.09 | 0 | 0 | 0 | 0.16333 | 0 |
| ENST00000668204 | ENSG00000266401 | 63.8301153 | 5.996165349 | 0.00013 | 0.00205 | yes | up | 0 | 0 | 0 | 0.59 | 0.29 | 0.24 | 0 | 0.37333 |
| ENST00000668253 | ENSG00000248008 | 0.329390681 | -1.602128355 | 0.0008 | 0.01012 | yes | down | 3.28 | 3.81 | 3.6 | 2.35 | 1.2 | 0.57 | 3.56333 | 1.37333 |
| ENST00000669486 | ENSG00000253686 | 41.59225734 | 5.378243081 | 0.00134 | 0.01566 | yes | up | 0 | 0 | 0 | 0.06 | 0.06 | 0.11 | 0 | 0.07667 |
| ENST00000670381 | ENSG00000286715 | 2.058251654 | 1.041419386 | 0.00455 | 0.0419 | yes | up | 1.15 | 1.1 | 0.67 | 2.64 | 2.25 | 2.04 | 0.97333 | 2.31 |
| ENST00000670471 | ENSG00000251138 | 182.0182442 | 7.507939253 | 9.4E-09 | 4.1E-07 | yes | up | 0 | 0 | 0 | 0.23 | 0.24 | 0.32 | 0 | 0.26333 |
| ENST00000671055 | ENSG00000228065 | 75.19950294 | 6.232651221 | 8.1E-05 | 0.0014 | yes | up | 0 | 0 | 0 | 0.14 | 0.42 | 0.18 | 0 | 0.24667 |
| ENST00000671088 | ENSG00000249859 | 0.023354048 | -5.420183557 | 0.00095 | 0.01175 | yes | down | 0.24 | 0.21 | 0.11 | 0 | 0 | 0 | 0.18667 | 0 |
| NONHSAT000917.2 | ENSG00000177000 | 5.349964383 | 2.419529287 | 0.00428 | 0.03993 | yes | up | 0.36 | 0.04 | 0.12 | 0.85 | 1.19 | 1.01 | 0.17333 | 1.01667 |
| NONHSAT002797.2 | ENSG00000234694 | 23.17677341 | 4.534607828 | 0.00395 | 0.03751 | yes | up | 0 | 0 | 0.04 | 0.15 | 0.28 | 0.39 | 0.01333 | 0.27333 |
| NONHSAT003731.2 | ENSG00000162433 | 4.409935348 | 2.140757505 | 3.1E-05 | 0.00059 | yes | up | 0.28 | 0.08 | 0.16 | 0.8 | 0.66 | 1.17 | 0.17333 | 0.87667 |
| NONHSAT004497.2 | ENSG00000223745 | 151.2449954 | 7.240743595 | 0.00169 | 0.01887 | yes | up | 0 | 0 | 0 | 0 | 0.43 | 0.28 | 0 | 0.23667 |
| NONHSAT005858.2 | ENSG00000255148 | 39.11650079 | 5.289705413 | 0.00562 | 0.04968 | yes | up | 0 | 0 | 0 | 1.16 | 0.2 | 0.57 | 0 | 0.64333 |
| NONHSAT010087.2 | ENSG00000223635 | 0.027804151 | -5.168555896 | 0.00198 | 0.02151 | yes | down | 0.11 | 0.1 | 0.06 | 0 | 0 | 0 | 0.09 | 0 |
| NONHSAT011652.2 | ENSG00000165997 | 3.012245344 | 1.590839281 | 0.00414 | 0.03892 | yes | up | 0.1 | 0.48 | 0.3 | 1.04 | 0.79 | 1.3 | 0.29333 | 1.04333 |
| NONHSAT013767.2 | ENSG00000165732 | 0.380525283 | -1.393935782 | 0.00273 | 0.02789 | yes | down | 9.91 | 18.1 | 7.75 | 6.99 | 3.66 | 5.11 | 11.92 | 5.25333 |
| NONHSAT015087.2 | ENSG00000224596 | 9.684439842 | 3.275668604 | 9.3E-05 | 0.00159 | yes | up | 0.04 | 0.09 | 0 | 0.46 | 0.48 | 0.67 | 0.04333 | 0.53667 |
| NONHSAT015626.2 | ENSG00000180628 | 3.929728033 | 1.97442947 | 7.6E-05 | 0.00132 | yes | up | 0.3 | 0.09 | 0.37 | 0.89 | 1.28 | 1.26 | 0.25333 | 1.14333 |
| NONHSAT031960.2 | ENSG00000250208 | 0.005530793 | -7.498298006 | 6.2E-08 | 2.3E-06 | yes | down | 0.09 | 0.03 | 0.06 | 0 | 0 | 0 | 0.06 | 0 |
| NONHSAT032174.2 | ENSG00000271963 | 0.0180873 | -5.788879149 | 3.9E-13 | 3.4E-11 | yes | down | 0.39 | 0.71 | 0.39 | 0.01 | 0.02 | 0 | 0.49667 | 0.01 |
| NONHSAT054508.2 | ENSG00000159217 | 1397.657961 | 10.44879563 | 7.2E-18 | 1.1E-15 | yes | up | 0 | 0 | 0 | 1.67 | 2.48 | 2.6 | 0 | 2.25 |
| NONHSAT055465.2 | ENSG00000278730 | 0.38439749 | -1.379329179 | 1.8E-11 | 1.2E-09 | yes | down | 0.9 | 0.84 | 0.85 | 0.32 | 0.32 | 0.49 | 0.86333 | 0.37667 |
| NONHSAT070449.2 | ENSG00000286728 | 0.329113738 | -1.603341845 | 0.00046 | 0.00631 | yes | down | 0.55 | 0.33 | 0.36 | 0.16 | 0.18 | 0.13 | 0.41333 | 0.15667 |
| NONHSAT075745.2 | ENSG00000223960 | 103.3545509 | 6.691458105 | 0.00528 | 0.04721 | yes | up | 0 | 0 | 0 | 0.09 | 0 | 0.07 | 0 | 0.05333 |
| NONHSAT085106.2 | ENSG00000223695 | 2.023693863 | 1.016991061 | 0.00443 | 0.04105 | yes | up | 0.07 | 0.13 | 0.11 | 0.22 | 0.29 | 0.21 | 0.10333 | 0.24 |
| NONHSAT096667.2 | ENSG00000248479 | 2.234244795 | 1.159787264 | 2.8E-05 | 0.00054 | yes | up | 0.62 | 0.62 | 0.97 | 1.45 | 2.13 | 2.05 | 0.73667 | 1.87667 |
| NONHSAT104164.2 | ENSG00000254363 | 0.006623946 | -7.238093401 | 0.00206 | 0.02215 | yes | down | 0.09 | 0.23 | 0 | 0 | 0 | 0 | 0.10667 | 0 |
| NONHSAT104167.2 | ENSG00000254363 | 433.6863699 | 8.760508292 | 1.9E-11 | 1.3E-09 | yes | up | 0 | 0 | 0 | 0.19 | 0.48 | 0.36 | 0 | 0.34333 |
| NONHSAT106651.2 | ENSG00000250903 | 2.017401454 | 1.012498203 | 0.0049 | 0.04446 | yes | up | 0.49 | 0.27 | 0.3 | 0.89 | 0.84 | 0.75 | 0.35333 | 0.82667 |
| NONHSAT112918.2 | ENSG00000272114 | 3.193587568 | 1.67517801 | 1.4E-11 | 9.7E-10 | yes | up | 3.19 | 2.23 | 3.09 | 8 | 11.31 | 11.69 | 2.83667 | 10.3333 |
| NONHSAT119869.2 | ENSG00000281404 | 0.008213723 | -6.92774801 | 0.004 | 0.03786 | yes | down | 0.08 | 0 | 0.27 | 0 | 0 | 0 | 0.11667 | 0 |
| NONHSAT120520.2 | ENSG00000136275 | 2.660567793 | 1.411734165 | 3.6E-05 | 0.00068 | yes | up | 0.27 | 0.17 | 0.23 | 0.73 | 0.65 | 0.68 | 0.22333 | 0.68667 |
| NONHSAT123250.2 | ENSG00000128595 | 6.693069277 | 2.742667947 | 0.004 | 0.03786 | yes | up | 0.99 | 0 | 1.79 | 6.13 | 10.15 | 4.37 | 0.92667 | 6.88333 |
| NONHSAT124335.2 | ENSG00000273344 | 72.22378437 | 6.174402111 | 2.5E-05 | 0.00049 | yes | up | 0 | 0 | 0 | 0.09 | 0.12 | 0.08 | 0 | 0.09667 |
| NONHSAT124336.2 | ENSG00000273344 | 0.023759464 | -5.395353899 | 4.7E-06 | 0.00011 | yes | down | 0.17 | 0.16 | 0.24 | 0 | 0.01 | 0 | 0.19 | 0.00333 |
| NONHSAT124679.2 | ENSG00000253982 | 0.026652809 | -5.229568602 | 0.00519 | 0.04657 | yes | down | 0.2 | 0.06 | 0.32 | 0 | 0 | 0 | 0.19333 | 0 |
| NONHSAT125770.2 | ENSG00000259366 | 3.012218194 | 1.590826277 | 0.00192 | 0.02101 | yes | up | 0.04 | 0.03 | 0.05 | 0.19 | 0.11 | 0.12 | 0.04 | 0.14 |
| NONHSAT129048.2 | ENSG00000249859 | 0.021647986 | -5.529623406 | 0.00042 | 0.00592 | yes | down | 0.02 | 0.01 | 0.02 | 0 | 0 | 0 | 0.01667 | 0 |
| NONHSAT130177.2 | ENSG00000273056 | 0.318129095 | -1.652315773 | 0.002 | 0.02165 | yes | down | 0.16 | 0.13 | 0.12 | 0.04 | 0.06 | 0.06 | 0.13667 | 0.05333 |
| NONHSAT135554.2 | ENSG00000237886 | 291.9165981 | 8.189412433 | 5.9E-09 | 2.7E-07 | yes | up | 0 | 0 | 0 | 0.09 | 0.05 | 0.18 | 0 | 0.10667 |
| NONHSAT135828.2 | ENSG00000198886 | 4.536264858 | 2.181504877 | 2E-289 | 5E-285 | yes | up | 564.05 | 590.65 | 520.4 | 3067.92 | 2814.19 | 2875.53 | 558.367 | 2919.21 |
| NONHSAT140312.2 | ENSG00000140632 | 2.664694407 | 1.413970091 | 0.00463 | 0.04246 | yes | up | 2.21 | 0.46 | 0.92 | 3.56 | 3.47 | 3.78 | 1.19667 | 3.60333 |
| NONHSAT140811.2 | ENSG00000103489 | 0.144705438 | -2.788808957 | 0.00438 | 0.04066 | yes | down | 0.43 | 0.74 | 0.3 | 0.1 | 0.14 | 0 | 0.49 | 0.08 |
| NONHSAT141673.2 | ENSG00000247735 | 0.285776273 | -1.807041958 | 9.2E-05 | 0.00156 | yes | down | 0.62 | 0.63 | 0.5 | 0.08 | 0.3 | 0.19 | 0.58333 | 0.19 |
| NONHSAT146423.2 | ENSG00000265185 | 0.479451357 | -1.06054364 | 7.2E-08 | 2.6E-06 | yes | down | 216.39 | 300.45 | 192.03 | 126.78 | 138.9 | 127.7 | 236.29 | 131.127 |
| NONHSAT147157.2 | ENSG00000232712 | 0.157583649 | -2.665810244 | 0.00353 | 0.0343 | yes | down | 0.64 | 1.27 | 0.86 | 0.23 | 0.28 | 0 | 0.92333 | 0.17 |
| NONHSAT149933.1 | ENSG00000227741 | 2.580803109 | 1.367820082 | 0.00518 | 0.0465 | yes | up | 0.34 | 0.13 | 0.35 | 0.95 | 0.54 | 0.9 | 0.27333 | 0.79667 |
| NONHSAT151562.1 | ENSG00000225762 | 35.85314706 | 5.164027854 | 0.0032 | 0.03168 | yes | up | 0 | 0 | 0 | 0.12 | 0.25 | 0.13 | 0 | 0.16667 |
| NONHSAT156012.1 | ENSG00000269609 | 0.028668651 | -5.124382147 | 0.00305 | 0.03046 | yes | down | 0.02 | 0.04 | 0.03 | 0 | 0 | 0 | 0.03 | 0 |
| NONHSAT156977.1 | ENSG00000231187 | 17.9481356 | 4.165762084 | 0.00044 | 0.00616 | yes | up | 0 | 0.05 | 0 | 0.23 | 0.49 | 0.35 | 0.01667 | 0.35667 |
| NONHSAT157019.1 | ENSG00000231131 | 2.247688622 | 1.168442189 | 0.00344 | 0.03367 | yes | up | 0.07 | 0.09 | 0.05 | 0.15 | 0.21 | 0.2 | 0.07 | 0.18667 |
| NONHSAT158734.1 | ENSG00000226416 | 2.074086674 | 1.052476184 | 0.0035 | 0.0341 | yes | up | 0.33 | 0.24 | 0.38 | 0.97 | 0.69 | 0.6 | 0.31667 | 0.75333 |
| NONHSAT159591.1 | ENSG00000023445 | 0.213486795 | -2.227781258 | 0.0017 | 0.01891 | yes | down | 1.54 | 3.17 | 1.44 | 0.84 | 0.54 | 0.14 | 2.05 | 0.50667 |
| NONHSAT172961.1 | ENSG00000260086 | 42.10897397 | 5.396055818 | 0.00226 | 0.02383 | yes | up | 0 | 0 | 0 | 0.47 | 0.14 | 0.56 | 0 | 0.39 |
| NONHSAT175847.1 | ENSG00000074370 | 2.476704752 | 1.308421895 | 1.3E-08 | 5.7E-07 | yes | up | 0.86 | 0.73 | 0.82 | 2.71 | 2.21 | 1.96 | 0.80333 | 2.29333 |
| NONHSAT180109.1 | ENSG00000267484 | 0.00826188 | -6.919314194 | 3.3E-06 | 8.3E-05 | yes | down | 1.67 | 0.47 | 0.8 | 0 | 0 | 0 | 0.98 | 0 |
| NONHSAT181266.1 | ENSG00000162976 | 0.363485906 | -1.460028668 | 0.00013 | 0.00211 | yes | down | 3.4 | 2.18 | 2.6 | 0.64 | 1.13 | 1.58 | 2.72667 | 1.11667 |
| NONHSAT183309.1 | ENSG00000235852 | 0.003084757 | -8.340627501 | 7E-08 | 2.6E-06 | yes | down | 0.53 | 0.81 | 5.91 | 0.04 | 0 | 0 | 2.41667 | 0.01333 |
| NONHSAT186058.1 | ENSG00000224063 | 2.032699557 | 1.023396993 | 0.00357 | 0.03463 | yes | up | 0.52 | 0.67 | 0.74 | 1.49 | 1.58 | 1.49 | 0.64333 | 1.52 |
| NONHSAT189966.1 | ENSG00000228705 | 0.010830569 | -6.528747201 | 5.8E-06 | 0.00014 | yes | down | 0.01 | 0.03 | 0.02 | 0 | 0 | 0 | 0.02 | 0 |
| NONHSAT193047.1 | ENSG00000273253 | 0.019694453 | -5.666066855 | 0.00058 | 0.00768 | yes | down | 0.02 | 0.04 | 0.03 | 0 | 0 | 0 | 0.03 | 0 |
| NONHSAT193739.1 | ENSG00000144674 | 5.965786602 | 2.576712373 | 4.4E-06 | 0.00011 | yes | up | 0.58 | 0.16 | 0.28 | 2.6 | 2.74 | 1.77 | 0.34 | 2.37 |
| NONHSAT193792.1 | ENSG00000230084 | 3.021025152 | 1.595038195 | 0.001 | 0.01223 | yes | up | 0.63 | 0.19 | 0.31 | 1.46 | 1.01 | 1.42 | 0.37667 | 1.29667 |
| NONHSAT194109.1 | ENSG00000242516 | 121.7241766 | 6.927471931 | 0.0035 | 0.03412 | yes | up | 0 | 0 | 0 | 0 | 0.09 | 0.04 | 0 | 0.04333 |
| NONHSAT194648.1 | ENSG00000240137 | 0.023386348 | -5.41818963 | 0.00136 | 0.01586 | yes | down | 0.05 | 0.03 | 0.07 | 0 | 0 | 0 | 0.05 | 0 |
| NONHSAT198728.1 | ENSG00000138768 | 9.749899799 | 3.285387392 | 0.00364 | 0.03514 | yes | up | 0.28 | 0 | 0.01 | 1 | 1.5 | 0.81 | 0.09667 | 1.10333 |
| NONHSAT199606.1 | ENSG00000281591 | 2.310923448 | 1.208469469 | 0.00406 | 0.03827 | yes | up | 0.44 | 0.6 | 1.18 | 2.3 | 1.28 | 2.29 | 0.74 | 1.95667 |
| NONHSAT200245.1 | ENSG00000286242 | 0.344068445 | -1.539232509 | 0.00184 | 0.02031 | yes | down | 0.9 | 0.67 | 0.29 | 0.19 | 0.27 | 0.27 | 0.62 | 0.24333 |
| NONHSAT200775.1 | ENSG00000232648 | 171.5461112 | 7.422452611 | 0.00121 | 0.0143 | yes | up | 0 | 0 | 0 | 0.08 | 0 | 0.09 | 0 | 0.05667 |
| NONHSAT208282.1 | ENSG00000287260 | 94.66779203 | 6.564801769 | 0.00197 | 0.02142 | yes | up | 0 | 0.03 | 0 | 0 | 2.17 | 0.49 | 0.01 | 0.88667 |
| NONHSAT212945.1 | ENSG00000186480 | 0.361988653 | -1.465983618 | 0.00318 | 0.03158 | yes | down | 10.28 | 3.99 | 14.14 | 2.4 | 4.04 | 5.24 | 9.47 | 3.89333 |
| NONHSAT215781.1 | ENSG00000228801 | 119.380465 | 6.899422968 | 0.00364 | 0.0352 | yes | up | 0 | 0 | 0 | 0.19 | 0.12 | 0 | 0 | 0.10333 |
| NONHSAT216072.1 | ENSG00000286984 | 2.080442379 | 1.056890331 | 0.00388 | 0.03703 | yes | up | 0.11 | 0.17 | 0.2 | 0.32 | 0.44 | 0.38 | 0.16 | 0.38 |
| NONHSAT216135.1 | ENSG00000132561 | 0.267691701 | -1.901355684 | 0.00147 | 0.01682 | yes | down | 2.16 | 1.65 | 1.03 | 0.24 | 0.83 | 0.43 | 1.61333 | 0.5 |
| NONHSAT216908.1 | ENSG00000254109 | 182.7533019 | 7.513753662 | 1E-07 | 3.6E-06 | yes | up | 0 | 0 | 0 | 0.21 | 0.08 | 0.08 | 0 | 0.12333 |
| NONHSAT217026.1 | ENSG00000023287 | 84.95072614 | 6.408554374 | 7E-05 | 0.00123 | yes | up | 0 | 0 | 0 | 0.25 | 0.62 | 0.15 | 0 | 0.34 |
| NONHSAT219189.1 | ENSG00000225706 | 0.028187945 | -5.148777879 | 0.00357 | 0.03464 | yes | down | 0.06 | 0.02 | 0.02 | 0 | 0 | 0 | 0.03333 | 0 |
| NONHSAT223026.1 | ENSG00000232160 | 2.02154689 | 1.015459668 | 0.00403 | 0.03806 | yes | up | 0.25 | 0.17 | 0.32 | 0.48 | 0.64 | 0.59 | 0.24667 | 0.57 |
| NONHSAT224738.1 | ENSG00000204362 | 15.05086522 | 3.91177452 | 0.0022 | 0.02334 | yes | up | 0 | 0.04 | 0 | 0.32 | 0.2 | 0.11 | 0.01333 | 0.21 |
| NONHSAT225578.1 | ENSG00000237975 | 2.035929582 | 1.025687663 | 1.2E-05 | 0.00026 | yes | up | 0.39 | 0.41 | 0.46 | 1.02 | 0.91 | 1 | 0.42 | 0.97667 |
| NONHSAT225651.1 | ENSG00000272668 | 94.96994058 | 6.569399046 | 3.4E-06 | 8.4E-05 | yes | up | 0 | 0 | 0 | 0.04 | 0.03 | 0.05 | 0 | 0.04 |
| NONHSAT227335.1 | ENSG00000203739 | 0.027050277 | -5.20821284 | 0.00387 | 0.03698 | yes | down | 0.18 | 0.04 | 0.09 | 0 | 0 | 0 | 0.10333 | 0 |
| NONHSAT227914.1 | ENSG00000225269 | 42.89149991 | 5.422619863 | 0.00309 | 0.03079 | yes | up | 0 | 0 | 0 | 0.01 | 0.05 | 0.05 | 0 | 0.03667 |
| NONHSAT228910.1 | ENSG00000254635 | 3.236306059 | 1.694348051 | 0.00437 | 0.04062 | yes | up | 0.07 | 0.02 | 0.15 | 0.38 | 0.17 | 0.32 | 0.08 | 0.29 |
| NONHSAT229151.1 | ENSG00000224596 | 2.164286577 | 1.113891542 | 0.00254 | 0.02626 | yes | up | 0.1 | 0.09 | 0.19 | 0.33 | 0.29 | 0.34 | 0.12667 | 0.32 |
| NONHSAT229860.1 | ENSG00000245532 | 0.009205047 | -6.763359132 | 0.00445 | 0.04122 | yes | down | 0.04 | 0.05 | 0 | 0 | 0 | 0 | 0.03 | 0 |
| NONHSAT230088.1 | ENSG00000226645 | 117.7032953 | 6.879010901 | 0.00417 | 0.03914 | yes | up | 0 | 0 | 0 | 0.25 | 0 | 0.58 | 0 | 0.27667 |
| NONHSAT230538.1 | ENSG00000255557 | 2.40997992 | 1.269021126 | 0.0038 | 0.03649 | yes | up | 0.44 | 0.18 | 0.29 | 0.71 | 0.81 | 0.96 | 0.30333 | 0.82667 |
| NONHSAT230609.1 | ENSG00000248671 | 159.2466858 | 7.315119537 | 0.00163 | 0.01832 | yes | up | 0 | 0 | 0 | 0.53 | 0 | 0.27 | 0 | 0.26667 |
| NONHSAT230637.1 | ENSG00000215841 | 3.072744311 | 1.619527723 | 0.00333 | 0.03276 | yes | up | 0.04 | 0.07 | 0.06 | 0.15 | 0.23 | 0.2 | 0.05667 | 0.19333 |
| NONHSAT230713.1 | ENSG00000255250 | 107.136872 | 6.74331127 | 9.9E-06 | 0.00022 | yes | up | 0 | 0 | 0 | 0.17 | 0.5 | 0.2 | 0 | 0.29 |
| NONHSAT231188.1 | ENSG00000257261 | 2.047208797 | 1.033658252 | 1.3E-06 | 3.5E-05 | yes | up | 0.28 | 0.39 | 0.45 | 0.83 | 0.84 | 0.97 | 0.37333 | 0.88 |
| NONHSAT231246.1 | ENSG00000259887 | 2.706470732 | 1.436412787 | 0.00017 | 0.00272 | yes | up | 0.04 | 0.07 | 0.08 | 0.19 | 0.19 | 0.19 | 0.06333 | 0.19 |
| NONHSAT231312.1 | ENSG00000257354 | 3.43269861 | 1.779343195 | 0.00058 | 0.00771 | yes | up | 1.77 | 2.39 | 0.76 | 7.58 | 7.37 | 4.62 | 1.64 | 6.52333 |
| NONHSAT231324.1 | ENSG00000256268 | 2.068164065 | 1.048350637 | 0.00016 | 0.00249 | yes | up | 0.08 | 0.06 | 0.1 | 0.23 | 0.18 | 0.17 | 0.08 | 0.19333 |
| NONHSAT232890.1 | ENSG00000215417 | 0.472427404 | -1.081835442 | 0.00095 | 0.01177 | yes | down | 0.27 | 0.34 | 0.16 | 0.12 | 0.12 | 0.18 | 0.25667 | 0.14 |
| NONHSAT233171.1 | ENSG00000271216 | 0.011956571 | -6.386052468 | 4.1E-05 | 0.00076 | yes | down | 0.08 | 0.15 | 0.05 | 0 | 0 | 0 | 0.09333 | 0 |
| NONHSAT233307.1 | ENSG00000261105 | 0.228283354 | -2.131102429 | 0.00026 | 0.00387 | yes | down | 0.08 | 0.08 | 0.07 | 0.03 | 0.01 | 0.03 | 0.07667 | 0.02333 |
| NONHSAT233546.1 | ENSG00000258457 | 4.611545751 | 2.205250411 | 0.00139 | 0.01606 | yes | up | 0.28 | 0.25 | 0.02 | 1.14 | 1.02 | 0.84 | 0.18333 | 1 |
| NONHSAT233701.1 | ENSG00000237356 | 152.869183 | 7.256153792 | 1.2E-07 | 4.1E-06 | yes | up | 0 | 0 | 0 | 0.18 | 0.2 | 0.09 | 0 | 0.15667 |
| NONHSAT233705.1 | ENSG00000237356 | 0.009008511 | -6.794495569 | 1.5E-05 | 0.00032 | yes | down | 0.03 | 0.15 | 0.13 | 0 | 0 | 0 | 0.10333 | 0 |
| NONHSAT234256.1 | ENSG00000100528 | 553.4159969 | 9.112220537 | 9.2E-08 | 3.3E-06 | yes | up | 0 | 0 | 0 | 1.22 | 0.04 | 1.59 | 0 | 0.95 |
| NONHSAT235044.1 | ENSG00000285667 | 2.431070444 | 1.281591698 | 0.00424 | 0.03967 | yes | up | 0.25 | 0.3 | 0.18 | 0.77 | 0.58 | 0.73 | 0.24333 | 0.69333 |
| NONHSAT235474.1 | ENSG00000275527 | 0.006464094 | -7.273336099 | 0.00016 | 0.00258 | yes | down | 0.01 | 0.23 | 0.23 | 0 | 0 | 0 | 0.15667 | 0 |
| NONHSAT235679.1 | ENSG00000247809 | 2.286397091 | 1.193075986 | 0.00363 | 0.03509 | yes | up | 0.02 | 0.02 | 0.05 | 0.09 | 0.09 | 0.07 | 0.03 | 0.08333 |
| NONHSAT236426.1 | ENSG00000259925 | 0.011016754 | -6.504157016 | 3.1E-05 | 0.00059 | yes | down | 0.08 | 0.02 | 0.04 | 0 | 0 | 0 | 0.04667 | 0 |
| NONHSAT236849.1 | ENSG00000270124 | 0.009008907 | -6.794432182 | 0.00431 | 0.04012 | yes | down | 0.04 | 0 | 0.02 | 0 | 0 | 0 | 0.02 | 0 |
| NONHSAT237330.1 | ENSG00000265148 | 0.064105542 | -3.963407112 | 0.00051 | 0.00692 | yes | down | 0.12 | 0.16 | 0.17 | 0 | 0 | 0.03 | 0.15 | 0.01 |
| NONHSAT237333.1 | ENSG00000265148 | 110.658463 | 6.78996998 | 0.00468 | 0.0428 | yes | up | 0 | 0 | 0 | 0 | 0.2 | 0.08 | 0 | 0.09333 |
| NONHSAT237464.1 | ENSG00000256124 | 78.28543769 | 6.290672064 | 0.00024 | 0.00365 | yes | up | 0 | 0 | 0 | 0.11 | 0.02 | 0.05 | 0 | 0.06 |
| NONHSAT238003.1 | ENSG00000263766 | 0.009450247 | -6.72543224 | 0.00469 | 0.04285 | yes | down | 0.1 | 0.09 | 0 | 0 | 0 | 0 | 0.06333 | 0 |
| NONHSAT238252.1 | ENSG00000185168 | 3.605269825 | 1.850107237 | 0.00098 | 0.01201 | yes | up | 0.11 | 0.07 | 0.1 | 0.19 | 0.54 | 0.42 | 0.09333 | 0.38333 |
| NONHSAT238261.1 | ENSG00000262877 | 2.313219054 | 1.209901891 | 0.00197 | 0.02144 | yes | up | 0.11 | 0.17 | 0.28 | 0.54 | 0.47 | 0.47 | 0.18667 | 0.49333 |
| NONHSAT239059.1 | ENSG00000226800 | 124.457923 | 6.959514266 | 6.2E-07 | 1.8E-05 | yes | up | 0 | 0 | 0 | 0.07 | 0.03 | 0.07 | 0 | 0.05667 |
| NONHSAT239193.1 | ENSG00000282851 | 0.015118081 | -6.047581204 | 7.1E-05 | 0.00125 | yes | down | 0.1 | 0.05 | 0.06 | 0 | 0 | 0 | 0.07 | 0 |
| NONHSAT239335.1 | ENSG00000233527 | 0.007331842 | -7.091608625 | 0.00285 | 0.02884 | yes | down | 0 | 0.09 | 0.04 | 0 | 0 | 0 | 0.04333 | 0 |
| NONHSAT239403.1 | ENSG00000213904 | 44.63564072 | 5.48012423 | 0.00151 | 0.01725 | yes | up | 0 | 0 | 0 | 0.15 | 0.08 | 0.05 | 0 | 0.09333 |
| NONHSAT239464.1 | ENSG00000286024 | 0.026864041 | -5.218179854 | 0.00243 | 0.0253 | yes | down | 0.07 | 0.03 | 0.03 | 0 | 0 | 0 | 0.04333 | 0 |
| NONHSAT239591.1 | ENSG00000268895 | 2.203771751 | 1.139974809 | 0.00426 | 0.03987 | yes | up | 0.24 | 0.2 | 0.23 | 0.55 | 0.72 | 0.41 | 0.22333 | 0.56 |
| NONHSAT239845.1 | ENSG00000268119 | 0.006006649 | -7.379223946 | 0.00126 | 0.01486 | yes | down | 0.09 | 0.11 | 0 | 0 | 0 | 0 | 0.06667 | 0 |
| NONHSAT239958.1 | ENSG00000271109 | 0.022319915 | -5.485524669 | 0.00047 | 0.00645 | yes | down | 0.29 | 0.29 | 0.28 | 0 | 0 | 0 | 0.28667 | 0 |
| NONHSAT240760.1 | ENSG00000286045 | 0.006444033 | -7.277820493 | 0.00214 | 0.02287 | yes | down | 0.77 | 0.19 | 0 | 0 | 0 | 0 | 0.32 | 0 |
| NONHSAT240792.1 | ENSG00000232504 | 158.3528528 | 7.306999048 | 1.5E-07 | 5E-06 | yes | up | 0 | 0 | 0 | 0.14 | 0.09 | 0.05 | 0 | 0.09333 |
| NONHSAT240922.1 | ENSG00000226508 | 17.47741538 | 4.127419945 | 0.00236 | 0.02473 | yes | up | 0 | 0 | 0.02 | 0.21 | 0.07 | 0.07 | 0.00667 | 0.11667 |
| NONHSAT241672.1 | ENSG00000235092 | 107.0337582 | 6.741922081 | 0.00461 | 0.04236 | yes | up | 0 | 0 | 0 | 0 | 0.1 | 0.16 | 0 | 0.08667 |
| NONHSAT241923.1 | ENSG00000259439 | 14.79533363 | 3.887070324 | 0.00148 | 0.01696 | yes | up | 0 | 0 | 0.06 | 0.37 | 0.22 | 0.3 | 0.02 | 0.29667 |
| NONHSAT242091.1 | ENSG00000226819 | 0.00490735 | -7.670840097 | 2E-09 | 1E-07 | yes | down | 0.08 | 0.06 | 0.08 | 0 | 0 | 0 | 0.07333 | 0 |
| NONHSAT242149.1 | ENSG00000179818 | 0.01067139 | -6.550108082 | 4.3E-06 | 0.0001 | yes | down | 0.13 | 0.18 | 0.23 | 0 | 0 | 0 | 0.18 | 0 |
| NONHSAT242460.1 | ENSG00000270019 | 0.007110442 | -7.135845135 | 1.7E-07 | 5.9E-06 | yes | down | 0.05 | 0.08 | 0.11 | 0 | 0 | 0 | 0.08 | 0 |
| NONHSAT243203.1 | ENSG00000230506 | 3.130015936 | 1.646170002 | 0.00162 | 0.01824 | yes | up | 0.03 | 0.03 | 0.02 | 0.14 | 0.08 | 0.08 | 0.02667 | 0.1 |
| NONHSAT243455.1 | ENSG00000203999 | 0.004698661 | -7.733534492 | 1.9E-07 | 6.5E-06 | yes | down | 0.08 | 0.16 | 0.03 | 0 | 0 | 0 | 0.09 | 0 |
| NONHSAT243602.1 | ENSG00000225377 | 0.008807288 | -6.827086493 | 0.00418 | 0.03917 | yes | down | 0 | 0.07 | 0.1 | 0 | 0 | 0 | 0.05667 | 0 |
| NONHSAT243610.1 | ENSG00000225377 | 0.0084563 | -6.88575773 | 0.00344 | 0.03367 | yes | down | 0.07 | 0 | 0.1 | 0 | 0 | 0 | 0.05667 | 0 |
| NONHSAT243629.1 | ENSG00000286288 | 2.286932684 | 1.1934139 | 0.00021 | 0.00323 | yes | up | 0.04 | 0.04 | 0.04 | 0.13 | 0.1 | 0.09 | 0.04 | 0.10667 |
| NONHSAT244641.1 | ENSG00000230061 | 2.082986514 | 1.058653499 | 0.00227 | 0.0239 | yes | up | 0.1 | 0.14 | 0.14 | 0.3 | 0.18 | 0.43 | 0.12667 | 0.30333 |
| NONHSAT245224.1 | ENSG00000231993 | 2.504652116 | 1.324610234 | 0.00515 | 0.04625 | yes | up | 0.05 | 0.05 | 0.06 | 0.17 | 0.1 | 0.18 | 0.05333 | 0.15 |
| NONHSAT246087.1 | ENSG00000272970 | 133.880924 | 7.064806603 | 2.7E-07 | 8.7E-06 | yes | up | 0 | 0 | 0 | 0.12 | 0.06 | 0.12 | 0 | 0.1 |
| NONHSAT246239.1 | ENSG00000206573 | 0.006597768 | -7.243806151 | 0.00168 | 0.01872 | yes | down | 0.08 | 0.07 | 0 | 0 | 0 | 0 | 0.05 | 0 |
| NONHSAT246539.1 | ENSG00000241472 | 10.8356721 | 3.437716737 | 9.2E-05 | 0.00156 | yes | up | 0.02 | 0 | 0.03 | 0.26 | 0.22 | 0.1 | 0.01667 | 0.19333 |
| NONHSAT246955.1 | ENSG00000240875 | 2.871961171 | 1.522036244 | 0.00566 | 0.04989 | yes | up | 0.06 | 0.04 | 0.05 | 0.24 | 0.12 | 0.14 | 0.05 | 0.16667 |
| NONHSAT247507.1 | ENSG00000248866 | 37.04345612 | 5.211146802 | 0.00045 | 0.00626 | yes | up | 0.01 | 0.01 | 0 | 0.55 | 0.01 | 0.23 | 0.00667 | 0.26333 |
| NONHSAT247904.1 | ENSG00000249675 | 3.956411737 | 1.984192573 | 0.00461 | 0.04231 | yes | up | 0.1 | 0.05 | 0.07 | 0.52 | 0.08 | 0.42 | 0.07333 | 0.34 |
| NONHSAT249286.1 | ENSG00000250320 | 79.6458017 | 6.315526411 | 2.4E-05 | 0.00047 | yes | up | 0 | 0 | 0 | 0.16 | 0.13 | 0.28 | 0 | 0.19 |
| NONHSAT249290.1 | ENSG00000250320 | 0.321343123 | -1.637813496 | 0.00216 | 0.02297 | yes | down | 0.25 | 0.54 | 0.43 | 0.12 | 0.21 | 0.13 | 0.40667 | 0.15333 |
| NONHSAT249568.1 | ENSG00000236714 | 0.006248066 | -7.322374496 | 0.00152 | 0.01732 | yes | down | 0 | 0.06 | 0.06 | 0 | 0 | 0 | 0.04 | 0 |
| NONHSAT249575.1 | ENSG00000236714 | 169.8227814 | 7.407886196 | 0.00307 | 0.03062 | yes | up | 0 | 0 | 0 | 0 | 0.01 | 0.13 | 0 | 0.04667 |
| NONHSAT249579.1 | ENSG00000236714 | 76.96618082 | 6.266152755 | 2.6E-10 | 1.5E-08 | yes | up | 0 | 0 | 0 | 0.14 | 0.15 | 0.07 | 0 | 0.12 |
| NONHSAT249965.1 | ENSG00000271874 | 97.19279718 | 6.602777497 | 0.00017 | 0.00265 | yes | up | 0 | 0 | 0 | 0.11 | 0.02 | 0.19 | 0 | 0.10667 |
| NONHSAT250866.1 | ENSG00000272269 | 0.280629296 | -1.833262471 | 0.0011 | 0.01324 | yes | down | 0.51 | 0.26 | 0.4 | 0.12 | 0.1 | 0.16 | 0.39 | 0.12667 |
| NONHSAT252478.1 | ENSG00000231776 | 55.59101462 | 5.796779809 | 0.00354 | 0.03441 | yes | up | 0 | 0 | 0 | 0 | 0.04 | 0.02 | 0 | 0.02 |
| NONHSAT252787.1 | ENSG00000233452 | 101.5435556 | 6.665954873 | 5E-07 | 1.5E-05 | yes | up | 0 | 0 | 0 | 0.13 | 0.21 | 0.27 | 0 | 0.20333 |
| NONHSAT253164.1 | ENSG00000232949 | 4.620723302 | 2.208118701 | 1.1E-06 | 3.1E-05 | yes | up | 0.04 | 0.11 | 0.07 | 0.41 | 0.3 | 0.46 | 0.07333 | 0.39 |
| NONHSAT253388.1 | ENSG00000223475 | 0.025972887 | -5.266849809 | 0.00455 | 0.04192 | yes | down | 0.08 | 0.03 | 0 | 0 | 0 | 0 | 0.03667 | 0 |
| NONHSAT253802.1 | ENSG00000273344 | 0.00852735 | -6.873686892 | 4E-07 | 1.3E-05 | yes | down | 0.15 | 0.16 | 0.11 | 0 | 0 | 0 | 0.14 | 0 |
| NONHSAT253927.1 | ENSG00000234141 | 0.023818552 | -5.391770505 | 0.00143 | 0.01644 | yes | down | 0.03 | 0.07 | 0.04 | 0 | 0 | 0 | 0.04667 | 0 |
| NONHSAT254537.1 | ENSG00000285106 | 0.303458954 | -1.720426703 | 1.8E-08 | 7.6E-07 | yes | down | 1.93 | 2.88 | 1.88 | 0.76 | 0.62 | 0.96 | 2.23 | 0.78 |
| NONHSAT254980.1 | ENSG00000247134 | 3.32905755 | 1.735113811 | 0.00395 | 0.03757 | yes | up | 0.9 | 0.56 | 0.37 | 1.58 | 2.27 | 2.96 | 0.61 | 2.27 |
| NONHSAT255047.1 | ENSG00000286878 | 43.44093943 | 5.440983397 | 0.00085 | 0.01061 | yes | up | 0 | 0 | 0 | 0.02 | 0.03 | 0.03 | 0 | 0.02667 |
| NONHSAT255521.1 | ENSG00000253931 | 55.21905159 | 5.787094204 | 0.00017 | 0.00266 | yes | up | 0 | 0 | 0 | 0.06 | 0.07 | 0.04 | 0 | 0.05667 |
| NONHSAT255576.1 | ENSG00000255559 | 49.1685731 | 5.619664583 | 0.00053 | 0.00716 | yes | up | 0 | 0 | 0 | 0.1 | 0.09 | 0.18 | 0 | 0.12333 |
| NONHSAT255790.1 | ENSG00000246339 | 62.73450748 | 5.971187319 | 0.0005 | 0.00683 | yes | up | 0 | 0 | 0 | 0.24 | 0.07 | 0.06 | 0 | 0.12333 |
| NONHSAT256271.1 | ENSG00000249375 | 42.08578501 | 5.395261123 | 0.00149 | 0.01704 | yes | up | 0 | 0 | 0 | 0.04 | 0.04 | 0.02 | 0 | 0.03333 |
| NONHSAT256593.1 | ENSG00000281128 | 110.7387624 | 6.791016493 | 0.00467 | 0.04277 | yes | up | 0 | 0 | 0 | 0 | 0.03 | 0.07 | 0 | 0.03333 |
| NONHSAT256625.1 | ENSG00000281649 | 0.035553032 | -4.813883586 | 1.1E-05 | 0.00025 | yes | down | 0.34 | 0.17 | 0.14 | 0 | 0 | 0.02 | 0.21667 | 0.00667 |
| NONHSAT256930.1 | ENSG00000232283 | 116.9582981 | 6.869850414 | 0.00377 | 0.03623 | yes | up | 0 | 0 | 0 | 0.1 | 0.08 | 0 | 0 | 0.06 |
| NONHSAT257109.1 | ENSG00000223478 | 36.03784189 | 5.171440713 | 0.00456 | 0.04199 | yes | up | 0 | 0 | 0 | 0.15 | 0.07 | 0.31 | 0 | 0.17667 |
| NONHSAT257356.1 | ENSG00000171889 | 4.516164546 | 2.175098052 | 2.4E-25 | 7.5E-23 | yes | up | 0.42 | 0.45 | 0.31 | 2.47 | 2.02 | 1.63 | 0.39333 | 2.04 |
| NONHSAT257741.1 | ENSG00000230030 | 0.004100137 | -7.930112322 | 6.3E-11 | 4.1E-09 | yes | down | 0.14 | 0.13 | 0.13 | 0 | 0 | 0 | 0.13333 | 0 |
| NONHSAT257742.1 | ENSG00000230030 | 33.54603354 | 5.068070288 | 1.2E-09 | 6.2E-08 | yes | up | 0 | 0.01 | 0 | 0.24 | 0.21 | 0.18 | 0.00333 | 0.21 |
| NONHSAT258028.1 | ENSG00000236017 | 61.11945067 | 5.933559672 | 7.9E-05 | 0.00137 | yes | up | 0 | 0 | 0 | 0.04 | 0.03 | 0.04 | 0 | 0.03667 |
| NONHSAT258134.1 | ENSG00000267064 | 0.04868287 | -4.360441964 | 0.00105 | 0.01278 | yes | down | 0.07 | 0.07 | 0.04 | 0 | 0.01 | 0 | 0.06 | 0.00333 |
| NONHSAT258180.1 | ENSG00000227486 | 0.018066236 | -5.790560203 | 0.00334 | 0.03284 | yes | down | 0.1 | 0 | 0.08 | 0 | 0 | 0 | 0.06 | 0 |
| NONHSAT258465.1 | ENSG00000266560 | 2.551195189 | 1.351173282 | 2.5E-07 | 8E-06 | yes | up | 0.12 | 0.13 | 0.17 | 0.36 | 0.42 | 0.41 | 0.14 | 0.39667 |
| NONHSAT258467.1 | ENSG00000266560 | 3.412589766 | 1.770866996 | 0.00182 | 0.02007 | yes | up | 0.02 | 0.06 | 0.04 | 0.13 | 0.18 | 0.13 | 0.04 | 0.14667 |
| NONHSAT258657.1 | ENSG00000204904 | 89.57028533 | 6.484948297 | 3.4E-06 | 8.5E-05 | yes | up | 0 | 0 | 0 | 0.29 | 0.27 | 0.26 | 0 | 0.27333 |
| NR_046112.1 | ENSG00000204588 | 68.5301143 | 6.098666188 | 3.2E-05 | 0.00061 | yes | up | 0 | 0 | 0 | 0.17 | 0.15 | 0.19 | 0 | 0.17 |
| NR_152799.1 | ENSG00000245937 | 0.001250998 | -9.642705303 | 5E-13 | 4.3E-11 | yes | down | 1.01 | 0.23 | 0.8 | 0 | 0 | 0 | 0.68 | 0 |
| NR_164126.1 | ENSG00000278915 | 2.26733174 | 1.180995491 | 0.00481 | 0.04378 | yes | up | 0.14 | 0.08 | 0.14 | 0.27 | 0.39 | 0.28 | 0.12 | 0.31333 |
| XR_001738234.1 | ENSG00000229021 | 0.006180584 | -7.338041208 | 0.00147 | 0.01686 | yes | down | 0 | 0.17 | 0.2 | 0 | 0 | 0 | 0.12333 | 0 |
| XR_001739127.1 | ENSG00000230065 | 0.029406753 | -5.087708698 | 0.00098 | 0.012 | yes | down | 0.02 | 0.03 | 0.06 | 0 | 0 | 0 | 0.03667 | 0 |
| XR_001739144.1 | ENSG00000287129 | 2.233729914 | 1.159454757 | 0.00164 | 0.01837 | yes | up | 0.1 | 0.07 | 0.16 | 0.28 | 0.28 | 0.28 | 0.11 | 0.28 |
| XR_001742733.2 | ENSG00000285804 | 135.1827822 | 7.078767601 | 0.00257 | 0.02654 | yes | up | 0 | 0 | 0 | 0.26 | 0 | 0.15 | 0 | 0.13667 |
| XR_001742740.2 | ENSG00000285804 | 0.012837733 | -6.283465776 | 0.00067 | 0.00876 | yes | down | 0.17 | 0.04 | 0.01 | 0 | 0 | 0 | 0.07333 | 0 |
| XR_001744064.1 | ENSG00000285571 | 114.8663108 | 6.843811921 | 0.00459 | 0.04218 | yes | up | 0 | 0 | 0 | 0.11 | 0 | 0.04 | 0 | 0.05 |
| XR_001752115.1 | ENSG00000260219 | 0.024478266 | -5.352354828 | 0.00021 | 0.00323 | yes | down | 0.1 | 0.05 | 0.05 | 0.01 | 0 | 0 | 0.06667 | 0.00333 |
| XR_001752822.1 | ENSG00000238007 | 2.107333437 | 1.075418606 | 0.00399 | 0.0378 | yes | up | 0.11 | 0.09 | 0.09 | 0.23 | 0.19 | 0.26 | 0.09667 | 0.22667 |
| XR_001754470.2 | ENSG00000276649 | 2.118760661 | 1.083220627 | 3E-05 | 0.00058 | yes | up | 0.37 | 0.2 | 0.21 | 0.59 | 0.61 | 0.7 | 0.26 | 0.63333 |
| XR_001755027.1 | ENSG00000231324 | 3.194320445 | 1.675509047 | 0.00014 | 0.00233 | yes | up | 0.41 | 0.15 | 0.35 | 1.35 | 0.86 | 1.1 | 0.30333 | 1.10333 |
| XR_001755114.2 | ENSG00000280145 | 3.417305032 | 1.77285903 | 6.6E-06 | 0.00015 | yes | up | 0.38 | 0.41 | 1.08 | 2.44 | 2.5 | 2.36 | 0.62333 | 2.43333 |
| XR_923345.2 | ENSG00000230065 | 120.7138952 | 6.915447942 | 4.7E-07 | 1.4E-05 | yes | up | 0 | 0 | 0 | 0.09 | 0.08 | 0.05 | 0 | 0.07333 |
| XR_926624.3 | ENSG00000285571 | 118.560275 | 6.889476889 | 0.00408 | 0.03841 | yes | up | 0 | 0 | 0 | 0.13 | 0 | 0.05 | 0 | 0.06 |
| XR_928874.3 | ENSG00000228801 | 0.008891814 | -6.81330658 | 6.3E-07 | 1.9E-05 | yes | down | 0.07 | 0.09 | 0.12 | 0 | 0 | 0 | 0.09333 | 0 |
| XR_929040.2 | ENSG00000253317 | 2.129953248 | 1.090821764 | 0.00262 | 0.02697 | yes | up | 0.16 | 0.14 | 0.21 | 0.38 | 0.48 | 0.39 | 0.17 | 0.41667 |
| XR_931178.2 | ENSG00000286626 | 0.014127013 | -6.145399751 | 5E-05 | 0.0009 | yes | down | 0.2 | 0.27 | 0.11 | 0 | 0 | 0 | 0.19333 | 0 |
| XR_934818.3 | ENSG00000244649 | 0.059669972 | -4.066851102 | 0.00193 | 0.02108 | yes | down | 0.65 | 0.5 | 0.19 | 0 | 0.09 | 0 | 0.44667 | 0.03 |
| XR_936418.3 | ENSG00000268119 | 156.8335821 | 7.2930907 | 0.00158 | 0.01784 | yes | up | 0 | 0 | 0 | 0.11 | 0.11 | 0 | 0 | 0.07333 |
| XR_950885.2 | ENSG00000259925 | 2.076287417 | 1.054006168 | 0.00095 | 0.01176 | yes | up | 0.06 | 0.07 | 0.07 | 0.17 | 0.17 | 0.14 | 0.06667 | 0.16 |
Table S4: The detailed information of DE lncRNAs.
| LncRNA ID | Log2FC(PC/Con) | Regulate |
|---|---|---|
| NONHSAT054508.2 | 10.44 | Up |
| ENST00000654005 | 9.14 | Up |
| NONHSAT234256.1 | 9.11 | Up |
| NONHSAT104167.2 | 8.76 | Up |
| NONHSAT135554.2 | 8.18 | Up |
| NONHSAT216908.1 | 7.51 | Up |
| ENST00000670471 | 7.5 | Up |
| NONHSAT200775.1 | 7.42 | Up |
| NONHSAT249575.1 | 7.4 | Up |
| ENST00000660542 | 7.4 | Up |
| ENST00000538717 | -10.01 | Down |
| NR_152799.1 | -9.64 | Down |
| NONHSAT183309.1 | -8.34 | Down |
| ENST00000488584 | -8.14 | Down |
| ENST00000657910 | -8.13 | Down |
| NONHSAT257741.1 | -7.93 | Down |
| NONHSAT243455.1 | -7.73 | Down |
| NONHSAT242091.1 | -7.67 | Down |
| NONHSAT031960.2 | -7.49 | Down |
| NONHSAT239845.1 | -7.37 | Down |
Table S5: Top 10 upregulated and 10 downregulated lncRNA transcripts.
GO and KEGG enrichment analyses of DE LncRNA s and predicted targets
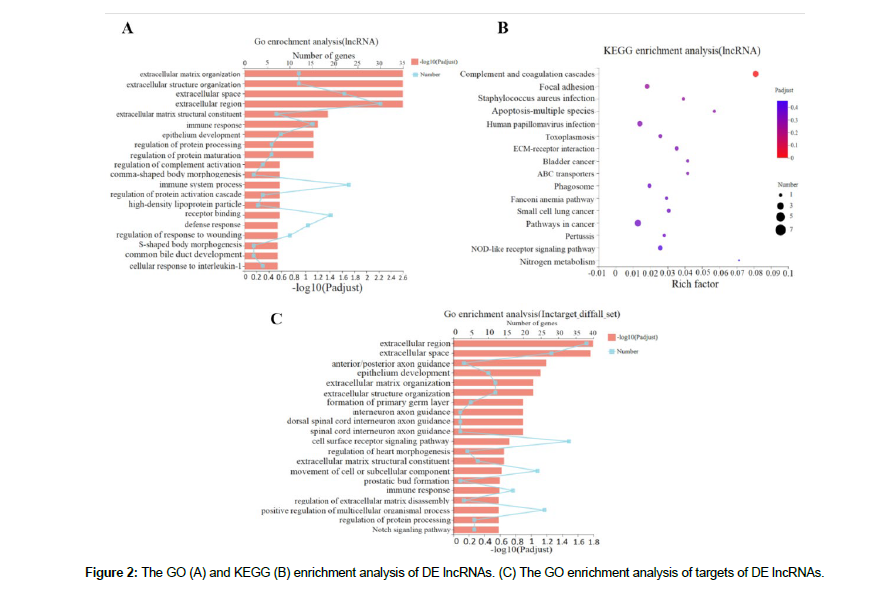
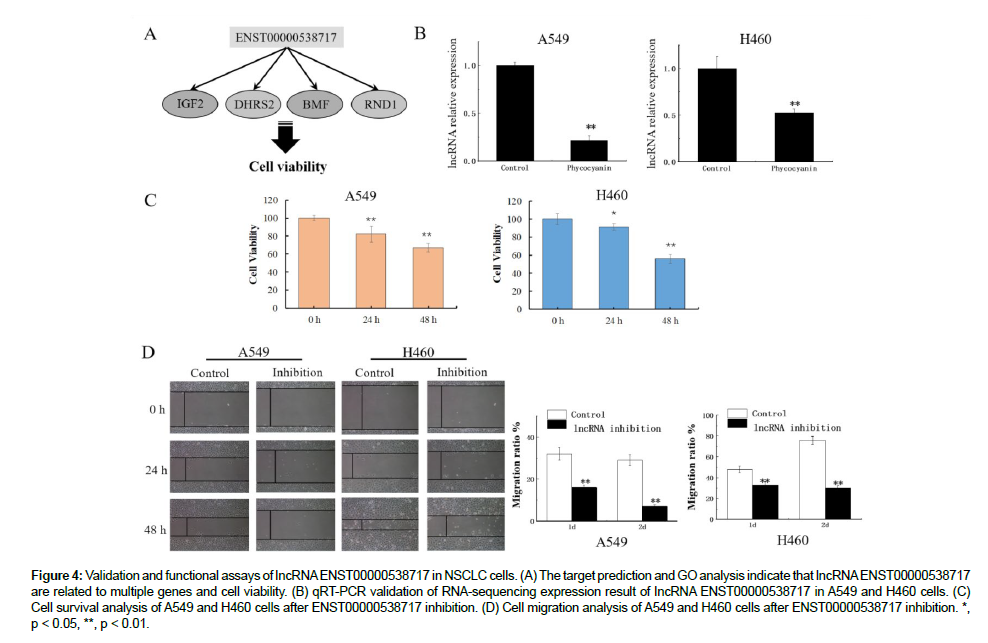
The DE LncRNA s was annotated by KEGG and GO enrichment analyses. Figure 2A shows the top 20 GO entries. Moreover, several of the GO items were related to cell activities, including GO: 0006955 (immune response), GO: 0070613 (regulation of protein processing), GO: 2000257 (regulation of protein activation cascade), GO: 1903034 (regulation of response to wounding), and GO: 0006952 (defense response). Figure 2B shows the KEGG pathway enrichment results.
The DE LncRNA s were shown to be enriched in signaling pathways related to cell growth and metabolism, such as the NOD-like receptor signaling pathway (map04621) and apoptosis (map04215). Strikingly, the DE LncRNA s were enriched in pathways associated with cancer (map05200) and small cell lung cancer (map05222) according to the KEGG analysis, suggesting that the DE LncRNA s affected the growth, apoptosis, and viability of A549 lung cancer cells, which were possibly regulated by phycocyanin.
The potential target genes of DE LncRNA s were predicted by a trans-regulation analysis method to determine the functions of this core LncRNA s. A total of 207 Trans targets (p-adjust < 0.01) for DE LncRNA s were screened and obtained. Detailed information on the potential targets of LncRNA s is shown in Table S6. Next, the target genes were annotated by GO enrichment analysis. In total, 780 GO entries were obtained, and Figure 2C presents the top 20 GO entries. It is worth noting that the DE LncRNA s were enriched in several GO items related to cell movement and proliferation were enriched, including GO: 0007166 (cell surface receptor signaling pathway), GO: 0006928 (movement of cell or subcellular component), GO: 0006955 (immune response), and GO: 0070613 (regulation of protein processing).Moreover, the Notch signaling pathway (GO: 0007219) was predicted to be activated in A549 cells after phycocyanin treatment.
| lncRNA id | lncRNA gene id | Type | Target gene id | Target gene name | Gene description |
|---|---|---|---|---|---|
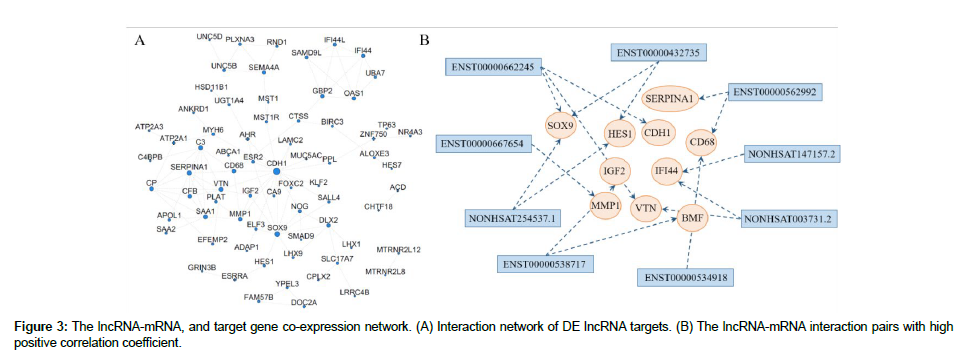
| ENST00000562992 | ENSG00000260693 | known | ENSG00000129226 | CD68 | CD68 molecule [Source:HGNC Symbol;Acc:HGNC:1693] |
| ENST00000562992 | ENSG00000260693 | known | ENSG00000197249 | SERPINA1 | serpin family A member 1 [Source:HGNC Symbol;Acc:HGNC:8941] |
| NONHSAT096667.2 | ENSG00000248479 | known | ENSG00000165821 | SALL2 | spalt like transcription factor 2 [Source:HGNC Symbol;Acc:HGNC:10526] |
| NONHSAT096667.2 | ENSG00000248479 | known | ENSG00000203724 | C1orf53 | chromosome 1 open reading frame 53 [Source:HGNC Symbol;Acc:HGNC:30003] |
| NONHSAT096667.2 | ENSG00000248479 | known | ENSG00000166922 | SCG5 | secretogranin V [Source:HGNC Symbol;Acc:HGNC:10816] |
| NONHSAT216135.1 | ENSG00000132561 | known | ENSG00000123843 | C4BPB | complement component 4 binding protein beta [Source:HGNC Symbol;Acc:HGNC:1328] |
| ENST00000432735 | ENSG00000215424 | known | ENSG00000197774 | EME2 | essential meiotic structure-specific endonuclease subunit 2 [Source:HGNC Symbol;Acc:HGNC:27289] |
| ENST00000606771 | ENSG00000272269 | known | ENSG00000162366 | PDZK1IP1 | PDZK1 interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:16887] |
| ENST00000606771 | ENSG00000272269 | known | ENSG00000167549 | CORO6 | coronin 6 [Source:HGNC Symbol;Acc:HGNC:21356] |
| ENST00000606771 | ENSG00000272269 | known | ENSG00000165029 | ABCA1 | ATP binding cassette subfamily A member 1 [Source:HGNC Symbol;Acc:HGNC:29] |
| ENST00000606771 | ENSG00000272269 | known | ENSG00000023445 | BIRC3 | baculoviral IAP repeat containing 3 [Source:HGNC Symbol;Acc:HGNC:591] |
| ENST00000606771 | ENSG00000272269 | known | ENSG00000134339 | SAA2 | serum amyloid A2 [Source:HGNC Symbol;Acc:HGNC:10514] |
| NONHSAT123250.2 | ENSG00000128595 | known | ENSG00000197774 | EME2 | essential meiotic structure-specific endonuclease subunit 2 [Source:HGNC Symbol;Acc:HGNC:27289] |
| NONHSAT123250.2 | ENSG00000128595 | known | ENSG00000181444 | ZNF467 | zinc finger protein 467 [Source:HGNC Symbol;Acc:HGNC:23154] |
| ENST00000432735 | ENSG00000215424 | known | ENSG00000114315 | HES1 | hes family bHLH transcription factor 1 [Source:HGNC Symbol;Acc:HGNC:5192] |
| ENST00000432735 | ENSG00000215424 | known | ENSG00000125398 | SOX9 | SRY-box transcription factor 9 [Source:HGNC Symbol;Acc:HGNC:11204] |
| ENST00000432735 | ENSG00000215424 | known | ENSG00000181444 | ZNF467 | zinc finger protein 467 [Source:HGNC Symbol;Acc:HGNC:23154] |
| NONHSAT096667.2 | ENSG00000248479 | known | ENSG00000163131 | CTSS | cathepsin S [Source:HGNC Symbol;Acc:HGNC:2545] |
| NONHSAT216135.1 | ENSG00000132561 | known | ENSG00000148677 | ANKRD1 | ankyrin repeat domain 1 [Source:HGNC Symbol;Acc:HGNC:15819] |
| NONHSAT147157.2 | ENSG00000232712 | known | MSTRG.5468 | ||
| NONHSAT147157.2 | ENSG00000232712 | known | ENSG00000130827 | PLXNA3 | plexin A3 [Source:HGNC Symbol;Acc:HGNC:9101] |
| NONHSAT147157.2 | ENSG00000232712 | known | ENSG00000269028 | MTRNR2L12 | MT-RNR2 like 12 [Source:HGNC Symbol;Acc:HGNC:37169] |
| ENST00000656756 | ENSG00000259583 | known | ENSG00000148677 | ANKRD1 | ankyrin repeat domain 1 [Source:HGNC Symbol;Acc:HGNC:15819] |
| ENST00000656756 | ENSG00000259583 | known | ENSG00000123843 | C4BPB | complement component 4 binding protein beta [Source:HGNC Symbol;Acc:HGNC:1328] |
| NONHSAT147157.2 | ENSG00000232712 | known | ENSG00000100027 | YPEL1 | yippee like 1 [Source:HGNC Symbol;Acc:HGNC:12845] |
| NONHSAT147157.2 | ENSG00000232712 | known | ENSG00000137965 | IFI44 | interferon induced protein 44 [Source:HGNC Symbol;Acc:HGNC:16938] |
| ENST00000656756 | ENSG00000259583 | known | ENSG00000104368 | PLAT | plasminogen activator, tissue type [Source:HGNC Symbol;Acc:HGNC:9051] |
| ENST00000656756 | ENSG00000259583 | known | ENSG00000172602 | RND1 | Rho family GTPase 1 [Source:HGNC Symbol;Acc:HGNC:18314] |
| NONHSAT225578.1 | ENSG00000237975 | known | ENSG00000104368 | PLAT | plasminogen activator, tissue type [Source:HGNC Symbol;Acc:HGNC:9051] |
| NONHSAT225578.1 | ENSG00000237975 | known | ENSG00000172602 | RND1 | Rho family GTPase 1 [Source:HGNC Symbol;Acc:HGNC:18314] |
| NONHSAT003731.2 | ENSG00000162433 | known | ENSG00000137965 | IFI44 | interferon induced protein 44 [Source:HGNC Symbol;Acc:HGNC:16938] |
| NONHSAT003731.2 | ENSG00000162433 | known | ENSG00000130827 | PLXNA3 | plexin A3 [Source:HGNC Symbol;Acc:HGNC:9101] |
| NONHSAT003731.2 | ENSG00000162433 | known | ENSG00000269028 | MTRNR2L12 | MT-RNR2 like 12 [Source:HGNC Symbol;Acc:HGNC:37169] |
| NONHSAT225578.1 | ENSG00000237975 | known | ENSG00000148677 | ANKRD1 | ankyrin repeat domain 1 [Source:HGNC Symbol;Acc:HGNC:15819] |
| NONHSAT003731.2 | ENSG00000162433 | known | ENSG00000100027 | YPEL1 | yippee like 1 [Source:HGNC Symbol;Acc:HGNC:12845] |
| NONHSAT225578.1 | ENSG00000237975 | known | ENSG00000123843 | C4BPB | complement component 4 binding protein beta [Source:HGNC Symbol;Acc:HGNC:1328] |
| NONHSAT254537.1 | ENSG00000285106 | known | ENSG00000114315 | HES1 | hes family bHLH transcription factor 1 [Source:HGNC Symbol;Acc:HGNC:5192] |
| NONHSAT254537.1 | ENSG00000285106 | known | ENSG00000125398 | SOX9 | SRY-box transcription factor 9 [Source:HGNC Symbol;Acc:HGNC:11204] |
| NONHSAT254537.1 | ENSG00000285106 | known | ENSG00000181444 | ZNF467 | zinc finger protein 467 [Source:HGNC Symbol;Acc:HGNC:23154] |
| NONHSAT159591.1 | ENSG00000023445 | known | ENSG00000162366 | PDZK1IP1 | PDZK1 interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:16887] |
| NONHSAT216135.1 | ENSG00000132561 | known | ENSG00000104368 | PLAT | plasminogen activator, tissue type [Source:HGNC Symbol;Acc:HGNC:9051] |
| NONHSAT216135.1 | ENSG00000132561 | known | ENSG00000172602 | RND1 | Rho family GTPase 1 [Source:HGNC Symbol;Acc:HGNC:18314] |
| NONHSAT159591.1 | ENSG00000023445 | known | ENSG00000167549 | CORO6 | coronin 6 [Source:HGNC Symbol;Acc:HGNC:21356] |
| NONHSAT159591.1 | ENSG00000023445 | known | ENSG00000165029 | ABCA1 | ATP binding cassette subfamily A member 1 [Source:HGNC Symbol;Acc:HGNC:29] |
| NONHSAT159591.1 | ENSG00000023445 | known | ENSG00000134339 | SAA2 | serum amyloid A2 [Source:HGNC Symbol;Acc:HGNC:10514] |
| NONHSAT186058.1 | ENSG00000224063 | known | ENSG00000160886 | LY6K | lymphocyte antigen 6 family member K [Source:HGNC Symbol;Acc:HGNC:24225] |
| NONHSAT186058.1 | ENSG00000224063 | known | ENSG00000167244 | IGF2 | insulin like growth factor 2 [Source:HGNC Symbol;Acc:HGNC:5466] |
| NONHSAT186058.1 | ENSG00000224063 | known | ENSG00000243649 | CFB | complement factor B [Source:HGNC Symbol;Acc:HGNC:1037] |
| NONHSAT254537.1 | ENSG00000285106 | known | ENSG00000197774 | EME2 | essential meiotic structure-specific endonuclease subunit 2 [Source:HGNC Symbol;Acc:HGNC:27289] |
| ENST00000628917 | ENSG00000242086 | known | ENSG00000145920 | CPLX2 | complexin 2 [Source:HGNC Symbol;Acc:HGNC:2310] |
| ENST00000435356 | ENSG00000203392 | known | ENSG00000125895 | TMEM74B | transmembrane protein 74B [Source:HGNC Symbol;Acc:HGNC:15893] |
| ENST00000435356 | ENSG00000203392 | known | MSTRG.5832 | ||
| ENST00000435356 | ENSG00000203392 | known | ENSG00000117594 | HSD11B1 | hydroxysteroid 11-beta dehydrogenase 1 [Source:HGNC Symbol;Acc:HGNC:5208] |
| ENST00000435356 | ENSG00000203392 | known | ENSG00000173432 | SAA1 | serum amyloid A1 [Source:HGNC Symbol;Acc:HGNC:10513] |
| ENST00000628917 | ENSG00000242086 | known | ENSG00000176692 | FOXC2 | forkhead box C2 [Source:HGNC Symbol;Acc:HGNC:3801] |
| ENST00000628917 | ENSG00000242086 | known | ENSG00000173531 | MST1 | macrophage stimulating 1 [Source:HGNC Symbol;Acc:HGNC:7380] |
| NONHSAT146423.2 | ENSG00000265185 | known | ENSG00000168676 | KCTD19 | potassium channel tetramerization domain containing 19 [Source:HGNC Symbol;Acc:HGNC:24753] |
| NONHSAT146423.2 | ENSG00000265185 | known | ENSG00000008517 | IL32 | interleukin 32 [Source:HGNC Symbol;Acc:HGNC:16830] |
| ENST00000667654 | ENSG00000274265 | known | ENSG00000131409 | LRRC4B | leucine rich repeat containing 4B [Source:HGNC Symbol;Acc:HGNC:25042] |
| ENST00000667654 | ENSG00000274265 | known | ENSG00000107821 | KAZALD1 | Kazal type serine peptidase inhibitor domain 1 [Source:HGNC Symbol;Acc:HGNC:25460] |
| NONHSAT146423.2 | ENSG00000265185 | known | ENSG00000078098 | FAP | fibroblast activation protein alpha [Source:HGNC Symbol;Acc:HGNC:3590] |
| ENST00000651958 | ENSG00000280441 | known | ENSG00000149809 | TM7SF2 | transmembrane 7 superfamily member 2 [Source:HGNC Symbol;Acc:HGNC:11863] |
| NONHSAT146423.2 | ENSG00000265185 | known | ENSG00000101198 | NKAIN4 | sodium/potassium transporting ATPase interacting 4 [Source:HGNC Symbol;Acc:HGNC:16191] |
| NONHSAT015626.2 | ENSG00000180628 | known | ENSG00000166922 | SCG5 | secretogranin V [Source:HGNC Symbol;Acc:HGNC:10816] |
| NONHSAT015626.2 | ENSG00000180628 | known | ENSG00000165821 | SALL2 | spalt like transcription factor 2 [Source:HGNC Symbol;Acc:HGNC:10526] |
| NONHSAT015626.2 | ENSG00000180628 | known | ENSG00000203724 | C1orf53 | chromosome 1 open reading frame 53 [Source:HGNC Symbol;Acc:HGNC:30003] |
| ENST00000658813 | ENSG00000223546 | known | ENSG00000166922 | SCG5 | secretogranin V [Source:HGNC Symbol;Acc:HGNC:10816] |
| NONHSAT015626.2 | ENSG00000180628 | known | ENSG00000163131 | CTSS | cathepsin S [Source:HGNC Symbol;Acc:HGNC:2545] |
| ENST00000658813 | ENSG00000223546 | known | ENSG00000165821 | SALL2 | spalt like transcription factor 2 [Source:HGNC Symbol;Acc:HGNC:10526] |
| ENST00000658813 | ENSG00000223546 | known | ENSG00000203724 | C1orf53 | chromosome 1 open reading frame 53 [Source:HGNC Symbol;Acc:HGNC:30003] |
| ENST00000534918 | ENSG00000225138 | known | ENSG00000125895 | TMEM74B | transmembrane protein 74B [Source:HGNC Symbol;Acc:HGNC:15893] |
| NONHSAT198728.1 | ENSG00000138768 | known | ENSG00000058085 | LAMC2 | laminin subunit gamma 2 [Source:HGNC Symbol;Acc:HGNC:6493] |
| NONHSAT198728.1 | ENSG00000138768 | known | ENSG00000196611 | MMP1 | matrix metallopeptidase 1 [Source:HGNC Symbol;Acc:HGNC:7155] |
| NONHSAT198728.1 | ENSG00000138768 | known | ENSG00000172638 | EFEMP2 | EGF containing fibulin extracellular matrix protein 2 [Source:HGNC Symbol;Acc:HGNC:3219] |
| NONHSAT183309.1 | ENSG00000235852 | known | ENSG00000085831 | TTC39A | tetratricopeptide repeat domain 39A [Source:HGNC Symbol;Acc:HGNC:18657] |
| NONHSAT183309.1 | ENSG00000235852 | known | ENSG00000276600 | RAB7B | RAB7B, member RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:30513] |
| ENST00000534918 | ENSG00000225138 | known | ENSG00000117594 | HSD11B1 | hydroxysteroid 11-beta dehydrogenase 1 [Source:HGNC Symbol;Acc:HGNC:5208] |
| ENST00000534918 | ENSG00000225138 | known | ENSG00000173432 | SAA1 | serum amyloid A1 [Source:HGNC Symbol;Acc:HGNC:10513] |
| ENST00000534918 | ENSG00000225138 | known | MSTRG.5832 | ||
| NONHSAT198728.1 | ENSG00000138768 | known | ENSG00000131409 | LRRC4B | leucine rich repeat containing 4B [Source:HGNC Symbol;Acc:HGNC:25042] |
| NONHSAT198728.1 | ENSG00000138768 | known | ENSG00000107821 | KAZALD1 | Kazal type serine peptidase inhibitor domain 1 [Source:HGNC Symbol;Acc:HGNC:25460] |
| ENST00000667654 | ENSG00000274265 | known | ENSG00000196611 | MMP1 | matrix metallopeptidase 1 [Source:HGNC Symbol;Acc:HGNC:7155] |
| NONHSAT181266.1 | ENSG00000162976 | known | ENSG00000107159 | CA9 | carbonic anhydrase 9 [Source:HGNC Symbol;Acc:HGNC:1383] |
| NONHSAT181266.1 | ENSG00000162976 | known | ENSG00000265972 | TXNIP | thioredoxin interacting protein [Source:HGNC Symbol;Acc:HGNC:16952] |
| NONHSAT181266.1 | ENSG00000162976 | known | ENSG00000179111 | HES7 | hes family bHLH transcription factor 7 [Source:HGNC Symbol;Acc:HGNC:15977] |
| NONHSAT000917.2 | ENSG00000177000 | known | ENSG00000255823 | MTRNR2L8 | MT-RNR2 like 8 [Source:HGNC Symbol;Acc:HGNC:37165] |
| NONHSAT193739.1 | ENSG00000144674 | known | ENSG00000172638 | EFEMP2 | EGF containing fibulin extracellular matrix protein 2 [Source:HGNC Symbol;Acc:HGNC:3219] |
| NONHSAT193739.1 | ENSG00000144674 | known | ENSG00000058085 | LAMC2 | laminin subunit gamma 2 [Source:HGNC Symbol;Acc:HGNC:6493] |
| NONHSAT193739.1 | ENSG00000144674 | known | ENSG00000196611 | MMP1 | matrix metallopeptidase 1 [Source:HGNC Symbol;Acc:HGNC:7155] |
| NONHSAT123250.2 | ENSG00000128595 | known | ENSG00000125398 | SOX9 | SRY-box transcription factor 9 [Source:HGNC Symbol;Acc:HGNC:11204] |
| NONHSAT123250.2 | ENSG00000128595 | known | ENSG00000114315 | HES1 | hes family bHLH transcription factor 1 [Source:HGNC Symbol;Acc:HGNC:5192] |
| NONHSAT000917.2 | ENSG00000177000 | known | ENSG00000169436 | COL22A1 | collagen type XXII alpha 1 chain [Source:HGNC Symbol;Acc:HGNC:22989] |
| NONHSAT000917.2 | ENSG00000177000 | known | ENSG00000283321 | AC019117.3 | novel protein |
| NONHSAT193739.1 | ENSG00000144674 | known | ENSG00000131409 | LRRC4B | leucine rich repeat containing 4B [Source:HGNC Symbol;Acc:HGNC:25042] |
| NONHSAT193739.1 | ENSG00000144674 | known | ENSG00000107821 | KAZALD1 | Kazal type serine peptidase inhibitor domain 1 [Source:HGNC Symbol;Acc:HGNC:25460] |
| NONHSAT257356.1 | ENSG00000171889 | known | ENSG00000102359 | SRPX2 | sushi repeat containing protein X-linked 2 [Source:HGNC Symbol;Acc:HGNC:30668] |
| NONHSAT257356.1 | ENSG00000171889 | known | ENSG00000100342 | APOL1 | apolipoprotein L1 [Source:HGNC Symbol;Acc:HGNC:618] |
| NONHSAT257356.1 | ENSG00000171889 | known | ENSG00000168874 | ATOH8 | atonal bHLH transcription factor 8 [Source:HGNC Symbol;Acc:HGNC:24126] |
| NONHSAT257356.1 | ENSG00000171889 | known | ENSG00000104967 | NOVA2 | NOVA alternative splicing regulator 2 [Source:HGNC Symbol;Acc:HGNC:7887] |
| NONHSAT257356.1 | ENSG00000171889 | known | ENSG00000107159 | CA9 | carbonic anhydrase 9 [Source:HGNC Symbol;Acc:HGNC:1383] |
| ENST00000667654 | ENSG00000274265 | known | ENSG00000172638 | EFEMP2 | EGF containing fibulin extracellular matrix protein 2 [Source:HGNC Symbol;Acc:HGNC:3219] |
| ENST00000667654 | ENSG00000274265 | known | ENSG00000058085 | LAMC2 | laminin subunit gamma 2 [Source:HGNC Symbol;Acc:HGNC:6493] |
| NONHSAT257356.1 | ENSG00000171889 | known | ENSG00000273706 | LHX1 | LIM homeobox 1 [Source:HGNC Symbol;Acc:HGNC:6593] |
| NONHSAT199606.1 | ENSG00000281591 | known | ENSG00000172602 | RND1 | Rho family GTPase 1 [Source:HGNC Symbol;Acc:HGNC:18314] |
| NONHSAT199606.1 | ENSG00000281591 | known | ENSG00000148677 | ANKRD1 | ankyrin repeat domain 1 [Source:HGNC Symbol;Acc:HGNC:15819] |
| NONHSAT199606.1 | ENSG00000281591 | known | ENSG00000123843 | C4BPB | complement component 4 binding protein beta [Source:HGNC Symbol;Acc:HGNC:1328] |
| NONHSAT199606.1 | ENSG00000281591 | known | ENSG00000104368 | PLAT | plasminogen activator, tissue type [Source:HGNC Symbol;Acc:HGNC:9051] |
| XR_001755027.1 | ENSG00000231324 | known | ENSG00000107242 | PIP5K1B | phosphatidylinositol-4-phosphate 5-kinase type 1 beta [Source:HGNC Symbol;Acc:HGNC:8995] |
| XR_001755027.1 | ENSG00000231324 | known | ENSG00000183111 | ARHGEF37 | Rho guanine nucleotide exchange factor 37 [Source:HGNC Symbol;Acc:HGNC:34430] |
| NONHSAT173219.1 | ENSG00000102908 | known | ENSG00000168676 | KCTD19 | potassium channel tetramerization domain containing 19 [Source:HGNC Symbol;Acc:HGNC:24753] |
| NONHSAT173219.1 | ENSG00000102908 | known | ENSG00000008517 | IL32 | interleukin 32 [Source:HGNC Symbol;Acc:HGNC:16830] |
| NONHSAT173219.1 | ENSG00000102908 | known | ENSG00000078098 | FAP | fibroblast activation protein alpha [Source:HGNC Symbol;Acc:HGNC:3590] |
| NONHSAT173219.1 | ENSG00000102908 | known | ENSG00000101198 | NKAIN4 | sodium/potassium transporting ATPase interacting 4 [Source:HGNC Symbol;Acc:HGNC:16191] |
| XR_001755114.2 | ENSG00000280145 | known | ENSG00000243649 | CFB | complement factor B [Source:HGNC Symbol;Acc:HGNC:1037] |
| XR_001755114.2 | ENSG00000280145 | known | ENSG00000167244 | IGF2 | insulin like growth factor 2 [Source:HGNC Symbol;Acc:HGNC:5466] |
| XR_001755114.2 | ENSG00000280145 | known | ENSG00000160886 | LY6K | lymphocyte antigen 6 family member K [Source:HGNC Symbol;Acc:HGNC:24225] |
| NONHSAT003731.2 | ENSG00000162433 | known | MSTRG.5468 | ||
| NONHSAT231312.1 | ENSG00000257354 | known | ENSG00000273706 | LHX1 | LIM homeobox 1 [Source:HGNC Symbol;Acc:HGNC:6593] |
| NONHSAT231312.1 | ENSG00000257354 | known | ENSG00000168874 | ATOH8 | atonal bHLH transcription factor 8 [Source:HGNC Symbol;Acc:HGNC:24126] |
| NONHSAT231312.1 | ENSG00000257354 | known | ENSG00000104967 | NOVA2 | NOVA alternative splicing regulator 2 [Source:HGNC Symbol;Acc:HGNC:7887] |
| NONHSAT231312.1 | ENSG00000257354 | known | ENSG00000107159 | CA9 | carbonic anhydrase 9 [Source:HGNC Symbol;Acc:HGNC:1383] |
| ENST00000434233 | ENSG00000218510 | known | ENSG00000168477 | TNXB | tenascin XB [Source:HGNC Symbol;Acc:HGNC:11976] |
| NONHSAT231312.1 | ENSG00000257354 | known | ENSG00000102359 | SRPX2 | sushi repeat containing protein X-linked 2 [Source:HGNC Symbol;Acc:HGNC:30668] |
| NONHSAT231312.1 | ENSG00000257354 | known | ENSG00000100342 | APOL1 | apolipoprotein L1 [Source:HGNC Symbol;Acc:HGNC:618] |
| ENST00000662245 | ENSG00000203875 | known | ENSG00000187957 | DNER | delta/notch like EGF repeat containing [Source:HGNC Symbol;Acc:HGNC:24456] |
| ENST00000662245 | ENSG00000203875 | known | ENSG00000091262 | ABCC6 | ATP binding cassette subfamily C member 6 [Source:HGNC Symbol;Acc:HGNC:57] |
| ENST00000662245 | ENSG00000203875 | known | ENSG00000101115 | SALL4 | spalt like transcription factor 4 [Source:HGNC Symbol;Acc:HGNC:15924] |
| ENST00000662245 | ENSG00000203875 | known | ENSG00000114315 | HES1 | hes family bHLH transcription factor 1 [Source:HGNC Symbol;Acc:HGNC:5192] |
| ENST00000662245 | ENSG00000203875 | known | ENSG00000167861 | HID1 | HID1 domain containing [Source:HGNC Symbol;Acc:HGNC:15736] |
| ENST00000662245 | ENSG00000203875 | known | ENSG00000181444 | ZNF467 | zinc finger protein 467 [Source:HGNC Symbol;Acc:HGNC:23154] |
| ENST00000662245 | ENSG00000203875 | known | ENSG00000109072 | VTN | vitronectin [Source:HGNC Symbol;Acc:HGNC:12724] |
| ENST00000662245 | ENSG00000203875 | known | ENSG00000125398 | SOX9 | SRY-box transcription factor 9 [Source:HGNC Symbol;Acc:HGNC:11204] |
| ENST00000662245 | ENSG00000203875 | known | ENSG00000039068 | CDH1 | cadherin 1 [Source:HGNC Symbol;Acc:HGNC:1748] |
| NONHSAT013767.2 | ENSG00000165732 | known | ENSG00000163131 | CTSS | cathepsin S [Source:HGNC Symbol;Acc:HGNC:2545] |
| NONHSAT175847.1 | ENSG00000074370 | known | ENSG00000109107 | ALDOC | aldolase, fructose-bisphosphate C [Source:HGNC Symbol;Acc:HGNC:418] |
| NONHSAT013767.2 | ENSG00000165732 | known | ENSG00000166922 | SCG5 | secretogranin V [Source:HGNC Symbol;Acc:HGNC:10816] |
| NONHSAT013767.2 | ENSG00000165732 | known | ENSG00000165821 | SALL2 | spalt like transcription factor 2 [Source:HGNC Symbol;Acc:HGNC:10526] |
| NONHSAT013767.2 | ENSG00000165732 | known | ENSG00000203724 | C1orf53 | chromosome 1 open reading frame 53 [Source:HGNC Symbol;Acc:HGNC:30003] |
| NONHSAT175847.1 | ENSG00000074370 | known | ENSG00000259803 | SLC22A31 | solute carrier family 22 member 31 [Source:HGNC Symbol;Acc:HGNC:27091] |
| ENST00000662245 | ENSG00000203875 | known | ENSG00000197774 | EME2 | essential meiotic structure-specific endonuclease subunit 2 [Source:HGNC Symbol;Acc:HGNC:27289] |
| NONHSAT175847.1 | ENSG00000074370 | known | ENSG00000089127 | OAS1 | 2'-5'-oligoadenylate synthetase 1 [Source:HGNC Symbol;Acc:HGNC:8086] |
| ENST00000375642 | ENSG00000204387 | known | ENSG00000165821 | SALL2 | spalt like transcription factor 2 [Source:HGNC Symbol;Acc:HGNC:10526] |
| NONHSAT193792.1 | ENSG00000230084 | known | ENSG00000183111 | ARHGEF37 | Rho guanine nucleotide exchange factor 37 [Source:HGNC Symbol;Acc:HGNC:34430] |
| NONHSAT193792.1 | ENSG00000230084 | known | ENSG00000107242 | PIP5K1B | phosphatidylinositol-4-phosphate 5-kinase type 1 beta [Source:HGNC Symbol;Acc:HGNC:8995] |
| ENST00000375633 | ENSG00000204387 | known | ENSG00000172638 | EFEMP2 | EGF containing fibulin extracellular matrix protein 2 [Source:HGNC Symbol;Acc:HGNC:3219] |
| ENST00000375633 | ENSG00000204387 | known | ENSG00000058085 | LAMC2 | laminin subunit gamma 2 [Source:HGNC Symbol;Acc:HGNC:6493] |
| ENST00000375633 | ENSG00000204387 | known | ENSG00000196611 | MMP1 | matrix metallopeptidase 1 [Source:HGNC Symbol;Acc:HGNC:7155] |
| ENST00000375633 | ENSG00000204387 | known | ENSG00000131409 | LRRC4B | leucine rich repeat containing 4B [Source:HGNC Symbol;Acc:HGNC:25042] |
| ENST00000375633 | ENSG00000204387 | known | ENSG00000107821 | KAZALD1 | Kazal type serine peptidase inhibitor domain 1 [Source:HGNC Symbol;Acc:HGNC:25460] |
| ENST00000455395 | ENSG00000230590 | known | ENSG00000125730 | C3 | complement C3 [Source:HGNC Symbol;Acc:HGNC:1318] |
| ENST00000455395 | ENSG00000230590 | known | ENSG00000143369 | ECM1 | extracellular matrix protein 1 [Source:HGNC Symbol;Acc:HGNC:3153] |
| ENST00000455395 | ENSG00000230590 | known | ENSG00000243244 | STON1 | stonin 1 [Source:HGNC Symbol;Acc:HGNC:17003] |
| ENST00000609111 | ENSG00000272734 | known | ENSG00000085831 | TTC39A | tetratricopeptide repeat domain 39A [Source:HGNC Symbol;Acc:HGNC:18657] |
| ENST00000609111 | ENSG00000272734 | known | ENSG00000276600 | RAB7B | RAB7B, member RAS oncogene family [Source:HGNC Symbol;Acc:HGNC:30513] |
| ENST00000664939 | ENSG00000175061 | known | ENSG00000166922 | SCG5 | secretogranin V [Source:HGNC Symbol;Acc:HGNC:10816] |
| ENST00000664939 | ENSG00000175061 | known | ENSG00000165821 | SALL2 | spalt like transcription factor 2 [Source:HGNC Symbol;Acc:HGNC:10526] |
| ENST00000664939 | ENSG00000175061 | known | ENSG00000203724 | C1orf53 | chromosome 1 open reading frame 53 [Source:HGNC Symbol;Acc:HGNC:30003] |
| ENST00000430034 | ENSG00000240801 | known | ENSG00000058085 | LAMC2 | laminin subunit gamma 2 [Source:HGNC Symbol;Acc:HGNC:6493] |
| ENST00000430034 | ENSG00000240801 | known | ENSG00000196611 | MMP1 | matrix metallopeptidase 1 [Source:HGNC Symbol;Acc:HGNC:7155] |
| ENST00000430034 | ENSG00000240801 | known | ENSG00000131409 | LRRC4B | leucine rich repeat containing 4B [Source:HGNC Symbol;Acc:HGNC:25042] |
| ENST00000430034 | ENSG00000240801 | known | ENSG00000172638 | EFEMP2 | EGF containing fibulin extracellular matrix protein 2 [Source:HGNC Symbol;Acc:HGNC:3219] |
| ENST00000430034 | ENSG00000240801 | known | ENSG00000107821 | KAZALD1 | Kazal type serine peptidase inhibitor domain 1 [Source:HGNC Symbol;Acc:HGNC:25460] |
| ENST00000668253 | ENSG00000248008 | known | ENSG00000243244 | STON1 | stonin 1 [Source:HGNC Symbol;Acc:HGNC:17003] |
| ENST00000668253 | ENSG00000248008 | known | ENSG00000134339 | SAA2 | serum amyloid A2 [Source:HGNC Symbol;Acc:HGNC:10514] |
| ENST00000668253 | ENSG00000248008 | known | ENSG00000143369 | ECM1 | extracellular matrix protein 1 [Source:HGNC Symbol;Acc:HGNC:3153] |
| ENST00000668253 | ENSG00000248008 | known | ENSG00000125730 | C3 | complement C3 [Source:HGNC Symbol;Acc:HGNC:1318] |
| ENST00000655606 | ENSG00000223745 | known | ENSG00000166924 | NYAP1 | neuronal tyrosine phosphorylated phosphoinositide-3-kinase adaptor 1 [Source:HGNC Symbol;Acc:HGNC:22009] |
| ENST00000547387 | ENSG00000258017 | known | MSTRG.5832 | ||
| ENST00000547387 | ENSG00000258017 | known | ENSG00000117594 | HSD11B1 | hydroxysteroid 11-beta dehydrogenase 1 [Source:HGNC Symbol;Acc:HGNC:5208] |
| ENST00000547387 | ENSG00000258017 | known | ENSG00000173432 | SAA1 | serum amyloid A1 [Source:HGNC Symbol;Acc:HGNC:10513] |
| ENST00000547387 | ENSG00000258017 | known | ENSG00000125895 | TMEM74B | transmembrane protein 74B [Source:HGNC Symbol;Acc:HGNC:15893] |
| ENST00000550290 | ENSG00000257354 | known | ENSG00000173531 | MST1 | macrophage stimulating 1 [Source:HGNC Symbol;Acc:HGNC:7380] |
| ENST00000550290 | ENSG00000257354 | known | ENSG00000176692 | FOXC2 | forkhead box C2 [Source:HGNC Symbol;Acc:HGNC:3801] |
| NONHSAT135828.2 | ENSG00000198886 | known | ENSG00000137878 | GCOM1 | GRINL1A complex locus 1 [Source:HGNC Symbol;Acc:HGNC:26424] |
| NONHSAT135828.2 | ENSG00000198886 | known | ENSG00000105559 | PLEKHA4 | pleckstrin homology domain containing A4 [Source:HGNC Symbol;Acc:HGNC:14339] |
| NONHSAT135828.2 | ENSG00000198886 | known | ENSG00000090238 | YPEL3 | yippee like 3 [Source:HGNC Symbol;Acc:HGNC:18327] |
| ENST00000550290 | ENSG00000257354 | known | ENSG00000162366 | PDZK1IP1 | PDZK1 interacting protein 1 [Source:HGNC Symbol;Acc:HGNC:16887] |
| ENST00000550290 | ENSG00000257354 | known | ENSG00000145920 | CPLX2 | complexin 2 [Source:HGNC Symbol;Acc:HGNC:2310] |
| NONHSAT112918.2 | ENSG00000272114 | known | ENSG00000134339 | SAA2 | serum amyloid A2 [Source:HGNC Symbol;Acc:HGNC:10514] |
| ENST00000652379 | ENSG00000280441 | known | ENSG00000131409 | LRRC4B | leucine rich repeat containing 4B [Source:HGNC Symbol;Acc:HGNC:25042] |
| ENST00000475197 | ENSG00000231177 | known | ENSG00000273706 | LHX1 | LIM homeobox 1 [Source:HGNC Symbol;Acc:HGNC:6593] |
| ENST00000475197 | ENSG00000231177 | known | ENSG00000168874 | ATOH8 | atonal bHLH transcription factor 8 [Source:HGNC Symbol;Acc:HGNC:24126] |
| ENST00000475197 | ENSG00000231177 | known | ENSG00000104967 | NOVA2 | NOVA alternative splicing regulator 2 [Source:HGNC Symbol;Acc:HGNC:7887] |
| ENST00000664939 | ENSG00000175061 | known | ENSG00000163131 | CTSS | cathepsin S [Source:HGNC Symbol;Acc:HGNC:2545] |
| ENST00000652379 | ENSG00000280441 | known | ENSG00000149927 | DOC2A | double C2 domain alpha [Source:HGNC Symbol;Acc:HGNC:2985] |
| ENST00000652379 | ENSG00000280441 | known | ENSG00000166920 | C15orf48 | chromosome 15 open reading frame 48 [Source:HGNC Symbol;Acc:HGNC:29898] |
| ENST00000652379 | ENSG00000280441 | known | ENSG00000140398 | NEIL1 | nei like DNA glycosylase 1 [Source:HGNC Symbol;Acc:HGNC:18448] |
| ENST00000475197 | ENSG00000231177 | known | ENSG00000107159 | CA9 | carbonic anhydrase 9 [Source:HGNC Symbol;Acc:HGNC:1383] |
| NONHSAT112918.2 | ENSG00000272114 | known | ENSG00000125730 | C3 | complement C3 [Source:HGNC Symbol;Acc:HGNC:1318] |
| NONHSAT112918.2 | ENSG00000272114 | known | ENSG00000143369 | ECM1 | extracellular matrix protein 1 [Source:HGNC Symbol;Acc:HGNC:3153] |
| NONHSAT112918.2 | ENSG00000272114 | known | ENSG00000243244 | STON1 | stonin 1 [Source:HGNC Symbol;Acc:HGNC:17003] |
| ENST00000475197 | ENSG00000231177 | known | ENSG00000102359 | SRPX2 | sushi repeat containing protein X-linked 2 [Source:HGNC Symbol;Acc:HGNC:30668] |
| ENST00000475197 | ENSG00000231177 | known | ENSG00000100342 | APOL1 | apolipoprotein L1 [Source:HGNC Symbol;Acc:HGNC:618] |
| ENST00000501122 | ENSG00000245532 | known | ENSG00000168676 | KCTD19 | potassium channel tetramerization domain containing 19 [Source:HGNC Symbol;Acc:HGNC:24753] |
| ENST00000501122 | ENSG00000245532 | known | ENSG00000101198 | NKAIN4 | sodium/potassium transporting ATPase interacting 4 [Source:HGNC Symbol;Acc:HGNC:16191] |
| ENST00000501122 | ENSG00000245532 | known | ENSG00000078098 | FAP | fibroblast activation protein alpha [Source:HGNC Symbol;Acc:HGNC:3590] |
| ENST00000501122 | ENSG00000245532 | known | ENSG00000008517 | IL32 | interleukin 32 [Source:HGNC Symbol;Acc:HGNC:16830] |
| NONHSAT140312.2 | ENSG00000140632 | known | ENSG00000269028 | MTRNR2L12 | MT-RNR2 like 12 [Source:HGNC Symbol;Acc:HGNC:37169] |
| NONHSAT140312.2 | ENSG00000140632 | known | ENSG00000130827 | PLXNA3 | plexin A3 [Source:HGNC Symbol;Acc:HGNC:9101] |
| NONHSAT140312.2 | ENSG00000140632 | known | MSTRG.5468 | ||
| NONHSAT140312.2 | ENSG00000140632 | known | ENSG00000137965 | IFI44 | interferon induced protein 44 [Source:HGNC Symbol;Acc:HGNC:16938] |
| NONHSAT140312.2 | ENSG00000140632 | known | ENSG00000100027 | YPEL1 | yippee like 1 [Source:HGNC Symbol;Acc:HGNC:12845] |
| ENST00000658813 | ENSG00000223546 | known | ENSG00000163131 | CTSS | cathepsin S [Source:HGNC Symbol;Acc:HGNC:2545] |
| ENST00000609983 | ENSG00000234608 | known | ENSG00000089127 | OAS1 | 2'-5'-oligoadenylate synthetase 1 [Source:HGNC Symbol;Acc:HGNC:8086] |
| ENST00000609983 | ENSG00000234608 | known | ENSG00000259803 | SLC22A31 | solute carrier family 22 member 31 [Source:HGNC Symbol;Acc:HGNC:27091] |
| ENST00000609983 | ENSG00000234608 | known | ENSG00000074370 | ATP2A3 | ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 [Source:HGNC Symbol;Acc:HGNC:813] |
| ENST00000609983 | ENSG00000234608 | known | ENSG00000109107 | ALDOC | aldolase, fructose-bisphosphate C [Source:HGNC Symbol;Acc:HGNC:418] |
Table S6: Detailed information for potential targets of DE lncRNAs.
In this work, 193 upregulated and 116 downregulated LncRNA s were identified between phycocyanin-treated and control A549 cells. In addition, the target genes (mRNAs) of DE LncRNA s were identified in PC samples, indicating that the expression of various genes was altered by LncRNA s after phycocyanin treatment. Currently, it has been reported that LncRNA s could be potential biomarkers for multiple cancers, including NSCLC [18,19]; however, the involved regulatory mechanism is not yet clearly understood. In this work, according to the GO enrichment analysis, the biological factors affected most by phycocyanin are extracellular region, extracellular space, and epithelium development, which suggested that NSCLC cell behavior is likely regulated by phycocyanin through LncRNA s and target genes. In addition, KEGG analysis of LncRNA s showed that the NOD-like receptor pathway, Notch pathway, and apoptosis were also enriched after phycocyanin exposure in A549 cells. Alzokaky et al. reported that phycocyanin could protect against ethanol-induced gastric ulcers in rats through multiple signaling pathways, including the HMGB1/ NLRP3/NF-κB and NOD-like receptor pathways [20]. It's worth mentioning that we used mRNA-seq to study the potential mechanism of phycocyanin, which also showed that "apoptosis pathway" and "NOD-like receptor pathway" were significantly enriched after phycocyanin explosure [21]. The similar conclusions of the two studies have reflected the potential regulatory mechanism of phycocyanin on NSCLC cells.
Establishment of the LncRNA ‒mRNA and target gene coexpression networks
As shown in Supplementary Figure S1, a comprehensive LncRNA ‒mRNA coexpression network was constructed. A total of 2,238 mRNA‒mRNA, LncRNA ‒LncRNA, and LncRNA ‒mRNA pairs were identified, showing the potential mutual effects of DE LncRNA s and mRNAs. Detailed information on LncRNA s and mRNAs is presented in Table S7. It is worth noting that ENSG00000130600 (LncRNA) and ENSG00000172638 (mRNA) showed the highest negative correlation coefficient (p-adjust < 0.01), while ENSG00000258479 (LncRNA) and ENSG00000188747 (mRNA) showed the highest positive correlation coefficient (p-adjust < 0.01). In addition, one mRNA with high correlation coefficient (such as ENSG00000188747) might have a regulatory relationship with multiple mRNAs and LncRNA s, including ENSG00000008517 (mRNA), ENSG00000078098 (mRNA), and ENSG00000254815 (LncRNA). The LncRNA ‒mRNA network revealed the underlying complex regulatory mechanisms of phycocyanin in A549 cells, providing a potential theoretical basis for NSCLC treatment.
| Gene id | RNA type | Connected gene id | RNA type | Corr | pvalue | padjust |
|---|---|---|---|---|---|---|
| ENSG00000130600 | lncRNA | ENSG00000244474 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | ENSG00000109107 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | ENSG00000172638 | mRNA | -1 | 0 | 0 |
| ENSG00000130600 | lncRNA | ENSG00000140398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | MSTRG.14266 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | ENSG00000172824 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | ENSG00000255823 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | ENSG00000258908 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | ENSG00000240801 | lncRNA | -1 | 0 | 0 |
| ENSG00000130600 | lncRNA | ENSG00000149927 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | ENSG00000169436 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | ENSG00000288093 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | ENSG00000181444 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | ENSG00000058085 | mRNA | -1 | 0 | 0 |
| ENSG00000130600 | lncRNA | ENSG00000283321 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | ENSG00000237781 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | ENSG00000287929 | lncRNA | -1 | 0 | 0 |
| ENSG00000130600 | lncRNA | ENSG00000196611 | mRNA | -1 | 0 | 0 |
| ENSG00000130600 | lncRNA | ENSG00000166920 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | ENSG00000089127 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | ENSG00000179148 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | ENSG00000107821 | mRNA | -1 | 0 | 0 |
| ENSG00000130600 | lncRNA | ENSG00000131409 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000130600 | lncRNA | ENSG00000259803 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | ENSG00000229921 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | ENSG00000279954 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | ENSG00000114315 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | ENSG00000125398 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | MSTRG.32594 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000130600 | lncRNA | ENSG00000272114 | lncRNA | -1 | 0 | 0 |
| ENSG00000188747 | mRNA | ENSG00000287421 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000104368 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000186862 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000188747 | mRNA | ENSG00000167861 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000183111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000162687 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000266560 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000168676 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000188747 | mRNA | ENSG00000264940 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000245532 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000258479 | lncRNA | 1 | 0 | 0 |
| ENSG00000188747 | mRNA | ENSG00000039068 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000008517 | mRNA | 1 | 0 | 0 |
| ENSG00000188747 | mRNA | ENSG00000275638 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000107242 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000116032 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000172602 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000078098 | mRNA | 1 | 0 | 0 |
| ENSG00000188747 | mRNA | ENSG00000265185 | lncRNA | -1 | 0 | 0 |
| ENSG00000188747 | mRNA | ENSG00000187957 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000254815 | lncRNA | 1 | 0 | 0 |
| ENSG00000188747 | mRNA | ENSG00000101198 | mRNA | 1 | 0 | 0 |
| ENSG00000188747 | mRNA | ENSG00000091262 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000101115 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000123843 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000109072 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000269888 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000148677 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000188747 | mRNA | ENSG00000197558 | mRNA | 1 | 0 | 0 |
| ENSG00000136275 | lncRNA | ENSG00000145920 | mRNA | -1 | 0 | 0 |
| ENSG00000136275 | lncRNA | ENSG00000166750 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000136275 | lncRNA | ENSG00000162366 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000136275 | lncRNA | ENSG00000167549 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000136275 | lncRNA | ENSG00000165029 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000136275 | lncRNA | ENSG00000115844 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000136275 | lncRNA | ENSG00000250708 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000136275 | lncRNA | ENSG00000185168 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000136275 | lncRNA | ENSG00000023445 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000136275 | lncRNA | ENSG00000164078 | mRNA | 1 | 0 | 0 |
| ENSG00000136275 | lncRNA | ENSG00000173531 | mRNA | 1 | 0 | 0 |
| ENSG00000136275 | lncRNA | ENSG00000119508 | mRNA | -1 | 0 | 0 |
| ENSG00000136275 | lncRNA | ENSG00000221571 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000136275 | lncRNA | ENSG00000007516 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000136275 | lncRNA | ENSG00000176692 | mRNA | -1 | 0 | 0 |
| ENSG00000258225 | lncRNA | ENSG00000273706 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258225 | lncRNA | ENSG00000205861 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258225 | lncRNA | ENSG00000140398 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258225 | lncRNA | ENSG00000168874 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258225 | lncRNA | ENSG00000143355 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000258225 | lncRNA | MSTRG.16716 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258225 | lncRNA | ENSG00000149927 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258225 | lncRNA | ENSG00000288093 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258225 | lncRNA | ENSG00000104967 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258225 | lncRNA | ENSG00000278740 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258225 | lncRNA | ENSG00000166920 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258225 | lncRNA | MSTRG.22782 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258225 | lncRNA | MSTRG.26092 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000258225 | lncRNA | ENSG00000107731 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258225 | lncRNA | ENSG00000168477 | mRNA | 1 | 0 | 0 |
| ENSG00000258225 | lncRNA | ENSG00000261080 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258225 | lncRNA | ENSG00000149809 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258225 | lncRNA | ENSG00000183691 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258225 | lncRNA | ENSG00000047457 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258225 | lncRNA | ENSG00000163435 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258225 | lncRNA | ENSG00000267740 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258225 | lncRNA | ENSG00000102359 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258225 | lncRNA | ENSG00000100342 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000109107 | mRNA | 1 | 0 | 0 |
| ENSG00000244474 | mRNA | ENSG00000172638 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | MSTRG.14266 | lncRNA | -1 | 0 | 0 |
| ENSG00000244474 | mRNA | ENSG00000286715 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000240801 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000167861 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000183111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000162687 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000058085 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000287929 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000196611 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000245532 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000089127 | mRNA | 1 | 0 | 0 |
| ENSG00000244474 | mRNA | ENSG00000074370 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000244474 | mRNA | ENSG00000039068 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000179148 | mRNA | 1 | 0 | 0 |
| ENSG00000244474 | mRNA | ENSG00000275638 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000107242 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000116032 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000187957 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000107821 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000259803 | mRNA | 1 | 0 | 0 |
| ENSG00000244474 | mRNA | ENSG00000091262 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000229921 | lncRNA | 1 | 0 | 0 |
| ENSG00000244474 | mRNA | ENSG00000279954 | lncRNA | 1 | 0 | 0 |
| ENSG00000244474 | mRNA | ENSG00000101115 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000109072 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000272114 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000244474 | mRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000197774 | mRNA | ENSG00000172824 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000197774 | mRNA | ENSG00000186862 | mRNA | 0.955882 | 0.002877 | 0.039507 |
| ENSG00000197774 | mRNA | ENSG00000258908 | lncRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000197774 | mRNA | ENSG00000167861 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000197774 | mRNA | ENSG00000162687 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000197774 | mRNA | ENSG00000181444 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000197774 | mRNA | ENSG00000237781 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000197774 | mRNA | ENSG00000168676 | mRNA | 0.955882 | 0.002877 | 0.039507 |
| ENSG00000197774 | mRNA | ENSG00000245532 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000197774 | mRNA | ENSG00000074370 | mRNA | 0.970588 | 0.001285 | 0.032602 |
| ENSG00000197774 | mRNA | ENSG00000039068 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000197774 | mRNA | ENSG00000187957 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000197774 | mRNA | ENSG00000162645 | mRNA | 0.970588 | 0.001285 | 0.032602 |
| ENSG00000197774 | mRNA | ENSG00000091262 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000197774 | mRNA | ENSG00000156687 | mRNA | 0.970588 | 0.001285 | 0.032602 |
| ENSG00000197774 | mRNA | ENSG00000101115 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000197774 | mRNA | ENSG00000114315 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000197774 | mRNA | ENSG00000109072 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000197774 | mRNA | ENSG00000125398 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000197774 | mRNA | MSTRG.32594 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000197774 | mRNA | ENSG00000140009 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000197774 | mRNA | ENSG00000226416 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000109107 | mRNA | ENSG00000172638 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | MSTRG.14266 | lncRNA | -1 | 0 | 0 |
| ENSG00000109107 | mRNA | ENSG00000286715 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | ENSG00000240801 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | ENSG00000167861 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | ENSG00000183111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | ENSG00000162687 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | ENSG00000058085 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | ENSG00000287929 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | ENSG00000196611 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | ENSG00000245532 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | ENSG00000089127 | mRNA | 1 | 0 | 0 |
| ENSG00000109107 | mRNA | ENSG00000074370 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000109107 | mRNA | ENSG00000039068 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | ENSG00000179148 | mRNA | 1 | 0 | 0 |
| ENSG00000109107 | mRNA | ENSG00000275638 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | ENSG00000107242 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | ENSG00000116032 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | ENSG00000187957 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | ENSG00000107821 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | ENSG00000259803 | mRNA | 1 | 0 | 0 |
| ENSG00000109107 | mRNA | ENSG00000091262 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | ENSG00000229921 | lncRNA | 1 | 0 | 0 |
| ENSG00000109107 | mRNA | ENSG00000279954 | lncRNA | 1 | 0 | 0 |
| ENSG00000109107 | mRNA | ENSG00000101115 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | ENSG00000109072 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | ENSG00000272114 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109107 | mRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | ENSG00000140398 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | MSTRG.14266 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | ENSG00000172824 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | ENSG00000255823 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | ENSG00000258908 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | ENSG00000240801 | lncRNA | 1 | 0 | 0 |
| ENSG00000172638 | mRNA | ENSG00000149927 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | ENSG00000169436 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | ENSG00000288093 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | ENSG00000181444 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | ENSG00000058085 | mRNA | 1 | 0 | 0 |
| ENSG00000172638 | mRNA | ENSG00000283321 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | ENSG00000237781 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | ENSG00000287929 | lncRNA | 1 | 0 | 0 |
| ENSG00000172638 | mRNA | ENSG00000196611 | mRNA | 1 | 0 | 0 |
| ENSG00000172638 | mRNA | ENSG00000166920 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | ENSG00000089127 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | ENSG00000179148 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | ENSG00000107821 | mRNA | 1 | 0 | 0 |
| ENSG00000172638 | mRNA | ENSG00000131409 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000172638 | mRNA | ENSG00000259803 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | ENSG00000229921 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | ENSG00000279954 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172638 | mRNA | ENSG00000272114 | lncRNA | 1 | 0 | 0 |
| ENSG00000273706 | mRNA | ENSG00000205861 | lncRNA | -1 | 0 | 0 |
| ENSG00000273706 | mRNA | ENSG00000168874 | mRNA | -1 | 0 | 0 |
| ENSG00000273706 | mRNA | ENSG00000143355 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000273706 | mRNA | ENSG00000286715 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000273706 | mRNA | ENSG00000104967 | mRNA | 1 | 0 | 0 |
| ENSG00000273706 | mRNA | ENSG00000107159 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000273706 | mRNA | ENSG00000130584 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000273706 | mRNA | ENSG00000168477 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000273706 | mRNA | ENSG00000265972 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000273706 | mRNA | ENSG00000266714 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000273706 | mRNA | ENSG00000253317 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000273706 | mRNA | ENSG00000183691 | mRNA | -1 | 0 | 0 |
| ENSG00000273706 | mRNA | ENSG00000145020 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000273706 | mRNA | ENSG00000137878 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000273706 | mRNA | ENSG00000102359 | mRNA | 1 | 0 | 0 |
| ENSG00000273706 | mRNA | ENSG00000105559 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000273706 | mRNA | ENSG00000090238 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000273706 | mRNA | ENSG00000100342 | mRNA | 1 | 0 | 0 |
| ENSG00000273706 | mRNA | ENSG00000179111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000205861 | lncRNA | ENSG00000168874 | mRNA | 1 | 0 | 0 |
| ENSG00000205861 | lncRNA | ENSG00000143355 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000205861 | lncRNA | ENSG00000286715 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000205861 | lncRNA | ENSG00000104967 | mRNA | -1 | 0 | 0 |
| ENSG00000205861 | lncRNA | ENSG00000107159 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000205861 | lncRNA | ENSG00000130584 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000205861 | lncRNA | ENSG00000168477 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000205861 | lncRNA | ENSG00000265972 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000205861 | lncRNA | ENSG00000266714 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000205861 | lncRNA | ENSG00000253317 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000205861 | lncRNA | ENSG00000183691 | mRNA | 1 | 0 | 0 |
| ENSG00000205861 | lncRNA | ENSG00000145020 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000205861 | lncRNA | ENSG00000137878 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000205861 | lncRNA | ENSG00000102359 | mRNA | -1 | 0 | 0 |
| ENSG00000205861 | lncRNA | ENSG00000105559 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000205861 | lncRNA | ENSG00000090238 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000205861 | lncRNA | ENSG00000100342 | mRNA | -1 | 0 | 0 |
| ENSG00000205861 | lncRNA | ENSG00000179111 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000140398 | mRNA | ENSG00000286715 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000140398 | mRNA | ENSG00000240801 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000140398 | mRNA | ENSG00000149927 | mRNA | 1 | 0 | 0 |
| ENSG00000140398 | mRNA | ENSG00000288093 | lncRNA | 1 | 0 | 0 |
| ENSG00000140398 | mRNA | ENSG00000058085 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000140398 | mRNA | ENSG00000287929 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000140398 | mRNA | ENSG00000196611 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000140398 | mRNA | ENSG00000235852 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000140398 | mRNA | ENSG00000166920 | mRNA | 1 | 0 | 0 |
| ENSG00000140398 | mRNA | ENSG00000187688 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000140398 | mRNA | ENSG00000085831 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000140398 | mRNA | ENSG00000168477 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000140398 | mRNA | ENSG00000107821 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000140398 | mRNA | ENSG00000131409 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000140398 | mRNA | ENSG00000276600 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000140398 | mRNA | ENSG00000272114 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000260693 | lncRNA | ENSG00000117594 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000260693 | lncRNA | ENSG00000286715 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000260693 | lncRNA | ENSG00000173432 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000260693 | lncRNA | ENSG00000129226 | mRNA | 1 | 0 | 0 |
| ENSG00000260693 | lncRNA | ENSG00000183111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000260693 | lncRNA | ENSG00000203392 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000260693 | lncRNA | ENSG00000275638 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000260693 | lncRNA | ENSG00000107242 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000260693 | lncRNA | ENSG00000116032 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000260693 | lncRNA | MSTRG.5832 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000260693 | lncRNA | ENSG00000196189 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000260693 | lncRNA | ENSG00000240764 | mRNA | 1 | 0 | 0 |
| ENSG00000260693 | lncRNA | ENSG00000266714 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000260693 | lncRNA | ENSG00000237892 | lncRNA | 1 | 0 | 0 |
| ENSG00000260693 | lncRNA | ENSG00000145020 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000260693 | lncRNA | ENSG00000125895 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000260693 | lncRNA | ENSG00000137878 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000260693 | lncRNA | ENSG00000105559 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000260693 | lncRNA | ENSG00000090238 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000260693 | lncRNA | ENSG00000197249 | mRNA | 1 | 0 | 0 |
| ENSG00000168874 | mRNA | ENSG00000143355 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000168874 | mRNA | ENSG00000286715 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000168874 | mRNA | ENSG00000104967 | mRNA | -1 | 0 | 0 |
| ENSG00000168874 | mRNA | ENSG00000107159 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000168874 | mRNA | ENSG00000130584 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000168874 | mRNA | ENSG00000168477 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000168874 | mRNA | ENSG00000265972 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000168874 | mRNA | ENSG00000266714 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000168874 | mRNA | ENSG00000253317 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000168874 | mRNA | ENSG00000183691 | mRNA | 1 | 0 | 0 |
| ENSG00000168874 | mRNA | ENSG00000145020 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000168874 | mRNA | ENSG00000137878 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000168874 | mRNA | ENSG00000102359 | mRNA | -1 | 0 | 0 |
| ENSG00000168874 | mRNA | ENSG00000105559 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000168874 | mRNA | ENSG00000090238 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000168874 | mRNA | ENSG00000100342 | mRNA | -1 | 0 | 0 |
| ENSG00000168874 | mRNA | ENSG00000179111 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000145920 | mRNA | ENSG00000166750 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000145920 | mRNA | ENSG00000162366 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000145920 | mRNA | ENSG00000167549 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000145920 | mRNA | ENSG00000165029 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000145920 | mRNA | ENSG00000115844 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000145920 | mRNA | ENSG00000250708 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000145920 | mRNA | ENSG00000185168 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000145920 | mRNA | ENSG00000023445 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000145920 | mRNA | ENSG00000164078 | mRNA | -1 | 0 | 0 |
| ENSG00000145920 | mRNA | ENSG00000173531 | mRNA | -1 | 0 | 0 |
| ENSG00000145920 | mRNA | ENSG00000119508 | mRNA | 1 | 0 | 0 |
| ENSG00000145920 | mRNA | ENSG00000221571 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000145920 | mRNA | ENSG00000007516 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000145920 | mRNA | ENSG00000176692 | mRNA | 1 | 0 | 0 |
| ENSG00000105963 | mRNA | ENSG00000117594 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000105963 | mRNA | ENSG00000173432 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000105963 | mRNA | ENSG00000203392 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000105963 | mRNA | ENSG00000232949 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000105963 | mRNA | ENSG00000166924 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000105963 | mRNA | MSTRG.5832 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105963 | mRNA | ENSG00000196189 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000105963 | mRNA | ENSG00000266714 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000105963 | mRNA | ENSG00000120693 | mRNA | 1 | 0 | 0 |
| ENSG00000105963 | mRNA | ENSG00000145020 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000105963 | mRNA | ENSG00000125895 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000105963 | mRNA | ENSG00000137878 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000105963 | mRNA | ENSG00000105559 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000105963 | mRNA | ENSG00000090238 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000286715 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000240801 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000167861 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000183111 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000162687 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000058085 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000287929 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000196611 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000245532 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000089127 | mRNA | -1 | 0 | 0 |
| MSTRG.14266 | lncRNA | ENSG00000074370 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| MSTRG.14266 | lncRNA | ENSG00000039068 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000179148 | mRNA | -1 | 0 | 0 |
| MSTRG.14266 | lncRNA | ENSG00000275638 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000107242 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000116032 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000187957 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000107821 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000259803 | mRNA | -1 | 0 | 0 |
| MSTRG.14266 | lncRNA | ENSG00000091262 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000229921 | lncRNA | -1 | 0 | 0 |
| MSTRG.14266 | lncRNA | ENSG00000279954 | lncRNA | -1 | 0 | 0 |
| MSTRG.14266 | lncRNA | ENSG00000101115 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000109072 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000140009 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000272114 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.14266 | lncRNA | ENSG00000226416 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | MSTRG.5468 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000167549 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000165029 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000250708 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000264940 | lncRNA | -1 | 0 | 0 |
| ENSG00000287421 | lncRNA | ENSG00000258479 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000130827 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000080031 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000231324 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000164182 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000269028 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000185168 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000008517 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000023445 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000198203 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000078098 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000265185 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000255182 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000254815 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000101198 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000221571 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000269888 | lncRNA | -1 | 0 | 0 |
| ENSG00000287421 | lncRNA | ENSG00000100027 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000137965 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287421 | lncRNA | ENSG00000197558 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000143355 | mRNA | ENSG00000104967 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000143355 | mRNA | ENSG00000107159 | mRNA | 0.970588 | 0.001285 | 0.032602 |
| ENSG00000143355 | mRNA | MSTRG.26092 | lncRNA | 0.955882 | 0.002877 | 0.039507 |
| ENSG00000143355 | mRNA | ENSG00000130584 | mRNA | 0.955882 | 0.002877 | 0.039507 |
| ENSG00000143355 | mRNA | ENSG00000168477 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000143355 | mRNA | ENSG00000183691 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000143355 | mRNA | ENSG00000102359 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000143355 | mRNA | ENSG00000100342 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000117594 | mRNA | ENSG00000173432 | mRNA | 1 | 0 | 0 |
| ENSG00000117594 | mRNA | ENSG00000129226 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000117594 | mRNA | ENSG00000238007 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000117594 | mRNA | ENSG00000203392 | lncRNA | 1 | 0 | 0 |
| ENSG00000117594 | mRNA | MSTRG.5468 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000117594 | mRNA | ENSG00000130827 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000117594 | mRNA | ENSG00000080031 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000117594 | mRNA | ENSG00000231324 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000117594 | mRNA | ENSG00000269028 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000117594 | mRNA | ENSG00000287275 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000117594 | mRNA | MSTRG.5832 | mRNA | -1 | 0 | 0 |
| ENSG00000117594 | mRNA | ENSG00000198203 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000117594 | mRNA | ENSG00000196189 | mRNA | 1 | 0 | 0 |
| ENSG00000117594 | mRNA | ENSG00000240764 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000117594 | mRNA | ENSG00000255182 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000117594 | mRNA | ENSG00000159753 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000117594 | mRNA | ENSG00000120693 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000117594 | mRNA | ENSG00000237892 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000117594 | mRNA | ENSG00000125895 | mRNA | 1 | 0 | 0 |
| ENSG00000117594 | mRNA | ENSG00000100027 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000117594 | mRNA | ENSG00000197249 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000117594 | mRNA | ENSG00000137965 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000137959 | mRNA | ENSG00000177409 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000137959 | mRNA | ENSG00000104368 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000137959 | mRNA | ENSG00000266560 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000137959 | mRNA | ENSG00000232949 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000137959 | mRNA | ENSG00000073282 | mRNA | 1 | 0 | 0 |
| ENSG00000137959 | mRNA | ENSG00000100867 | mRNA | 1 | 0 | 0 |
| ENSG00000137959 | mRNA | ENSG00000172602 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000137959 | mRNA | ENSG00000265972 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000137959 | mRNA | ENSG00000266714 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000137959 | mRNA | ENSG00000253317 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000137959 | mRNA | ENSG00000142619 | mRNA | 1 | 0 | 0 |
| ENSG00000137959 | mRNA | ENSG00000145020 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000137959 | mRNA | ENSG00000137878 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000137959 | mRNA | ENSG00000123843 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000137959 | mRNA | ENSG00000105559 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000137959 | mRNA | ENSG00000158825 | mRNA | 1 | 0 | 0 |
| ENSG00000137959 | mRNA | ENSG00000090238 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000137959 | mRNA | ENSG00000148677 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000137959 | mRNA | ENSG00000179111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000137959 | mRNA | ENSG00000122420 | mRNA | 1 | 0 | 0 |
| ENSG00000286715 | lncRNA | ENSG00000129226 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000286715 | lncRNA | ENSG00000149927 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000286715 | lncRNA | ENSG00000288093 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000286715 | lncRNA | ENSG00000104967 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000286715 | lncRNA | ENSG00000166920 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000286715 | lncRNA | ENSG00000089127 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000286715 | lncRNA | ENSG00000179148 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000286715 | lncRNA | ENSG00000240764 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000286715 | lncRNA | ENSG00000183691 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000286715 | lncRNA | ENSG00000259803 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000286715 | lncRNA | ENSG00000237892 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000286715 | lncRNA | ENSG00000229921 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000286715 | lncRNA | ENSG00000279954 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000286715 | lncRNA | ENSG00000102359 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000286715 | lncRNA | ENSG00000100342 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000286715 | lncRNA | ENSG00000197249 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000173432 | mRNA | ENSG00000129226 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000173432 | mRNA | ENSG00000238007 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000173432 | mRNA | ENSG00000203392 | lncRNA | 1 | 0 | 0 |
| ENSG00000173432 | mRNA | MSTRG.5468 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000173432 | mRNA | ENSG00000130827 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000173432 | mRNA | ENSG00000080031 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000173432 | mRNA | ENSG00000231324 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000173432 | mRNA | ENSG00000269028 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000173432 | mRNA | ENSG00000287275 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000173432 | mRNA | MSTRG.5832 | mRNA | -1 | 0 | 0 |
| ENSG00000173432 | mRNA | ENSG00000198203 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000173432 | mRNA | ENSG00000196189 | mRNA | 1 | 0 | 0 |
| ENSG00000173432 | mRNA | ENSG00000240764 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000173432 | mRNA | ENSG00000255182 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000173432 | mRNA | ENSG00000159753 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000173432 | mRNA | ENSG00000120693 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000173432 | mRNA | ENSG00000237892 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000173432 | mRNA | ENSG00000125895 | mRNA | 1 | 0 | 0 |
| ENSG00000173432 | mRNA | ENSG00000100027 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000173432 | mRNA | ENSG00000197249 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000173432 | mRNA | ENSG00000137965 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000129226 | mRNA | ENSG00000183111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000129226 | mRNA | ENSG00000203392 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000129226 | mRNA | ENSG00000275638 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000129226 | mRNA | ENSG00000107242 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000129226 | mRNA | ENSG00000116032 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000129226 | mRNA | MSTRG.5832 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000129226 | mRNA | ENSG00000196189 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000129226 | mRNA | ENSG00000240764 | mRNA | 1 | 0 | 0 |
| ENSG00000129226 | mRNA | ENSG00000266714 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000129226 | mRNA | ENSG00000237892 | lncRNA | 1 | 0 | 0 |
| ENSG00000129226 | mRNA | ENSG00000145020 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000129226 | mRNA | ENSG00000125895 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000129226 | mRNA | ENSG00000137878 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000129226 | mRNA | ENSG00000105559 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000129226 | mRNA | ENSG00000090238 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000129226 | mRNA | ENSG00000197249 | mRNA | 1 | 0 | 0 |
| ENSG00000177409 | mRNA | ENSG00000073282 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000177409 | mRNA | ENSG00000100867 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000177409 | mRNA | ENSG00000107159 | mRNA | 0.970588 | 0.001285 | 0.032602 |
| ENSG00000177409 | mRNA | ENSG00000265972 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000177409 | mRNA | ENSG00000253317 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000177409 | mRNA | ENSG00000142619 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000177409 | mRNA | MSTRG.15840 | mRNA | -0.954864 | 0.00301 | 0.039507 |
| ENSG00000177409 | mRNA | ENSG00000158825 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000177409 | mRNA | ENSG00000179111 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000177409 | mRNA | ENSG00000122420 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000104368 | mRNA | ENSG00000105246 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104368 | mRNA | ENSG00000266560 | lncRNA | 1 | 0 | 0 |
| ENSG00000104368 | mRNA | ENSG00000073282 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104368 | mRNA | ENSG00000118898 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104368 | mRNA | ENSG00000100867 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104368 | mRNA | ENSG00000214353 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000104368 | mRNA | ENSG00000258479 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104368 | mRNA | ENSG00000008517 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104368 | mRNA | ENSG00000172602 | mRNA | 1 | 0 | 0 |
| ENSG00000104368 | mRNA | ENSG00000078098 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104368 | mRNA | ENSG00000265185 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000104368 | mRNA | ENSG00000240219 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104368 | mRNA | ENSG00000104888 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000104368 | mRNA | ENSG00000254815 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104368 | mRNA | ENSG00000101198 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104368 | mRNA | ENSG00000142619 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104368 | mRNA | ENSG00000101197 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104368 | mRNA | ENSG00000123843 | mRNA | -1 | 0 | 0 |
| ENSG00000104368 | mRNA | ENSG00000158825 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104368 | mRNA | ENSG00000148677 | mRNA | -1 | 0 | 0 |
| ENSG00000104368 | mRNA | ENSG00000122420 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104368 | mRNA | ENSG00000197558 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.16716 | mRNA | ENSG00000170044 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.16716 | mRNA | ENSG00000127528 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.16716 | mRNA | ENSG00000146411 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.16716 | mRNA | MSTRG.22782 | mRNA | -1 | 0 | 0 |
| MSTRG.16716 | mRNA | ENSG00000168477 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.16716 | mRNA | ENSG00000243649 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.16716 | mRNA | ENSG00000167244 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.16716 | mRNA | ENSG00000104081 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.16716 | mRNA | ENSG00000117266 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.16716 | mRNA | ENSG00000265972 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.16716 | mRNA | ENSG00000275620 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.16716 | mRNA | ENSG00000253317 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.16716 | mRNA | ENSG00000149809 | mRNA | -1 | 0 | 0 |
| MSTRG.16716 | mRNA | ENSG00000160886 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.16716 | mRNA | ENSG00000047457 | mRNA | -1 | 0 | 0 |
| MSTRG.16716 | mRNA | ENSG00000179111 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000165821 | mRNA | ENSG00000203724 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000165821 | mRNA | ENSG00000166922 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000165821 | mRNA | ENSG00000263590 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000165821 | mRNA | ENSG00000248479 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000165821 | mRNA | ENSG00000162804 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000165821 | mRNA | ENSG00000007516 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000165821 | mRNA | ENSG00000163131 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000203724 | mRNA | ENSG00000172824 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000203724 | mRNA | ENSG00000255823 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000203724 | mRNA | ENSG00000258908 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000203724 | mRNA | ENSG00000169436 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000203724 | mRNA | ENSG00000181444 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000203724 | mRNA | ENSG00000283321 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000203724 | mRNA | ENSG00000237781 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000203724 | mRNA | ENSG00000167549 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000203724 | mRNA | ENSG00000165029 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000203724 | mRNA | ENSG00000250708 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000203724 | mRNA | ENSG00000166922 | mRNA | -1 | 0 | 0 |
| ENSG00000203724 | mRNA | ENSG00000263590 | lncRNA | -1 | 0 | 0 |
| ENSG00000203724 | mRNA | ENSG00000185168 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000203724 | mRNA | ENSG00000023445 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000203724 | mRNA | ENSG00000248479 | lncRNA | -1 | 0 | 0 |
| ENSG00000203724 | mRNA | ENSG00000162804 | mRNA | -1 | 0 | 0 |
| ENSG00000203724 | mRNA | ENSG00000221571 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000203724 | mRNA | ENSG00000007516 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000203724 | mRNA | ENSG00000114315 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000203724 | mRNA | ENSG00000125398 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000203724 | mRNA | MSTRG.32594 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000203724 | mRNA | ENSG00000163131 | mRNA | -1 | 0 | 0 |
| ENSG00000172824 | mRNA | ENSG00000258908 | lncRNA | -1 | 0 | 0 |
| ENSG00000172824 | mRNA | ENSG00000240801 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000167861 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000162687 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000181444 | mRNA | 1 | 0 | 0 |
| ENSG00000172824 | mRNA | ENSG00000058085 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000237781 | lncRNA | 1 | 0 | 0 |
| ENSG00000172824 | mRNA | ENSG00000287929 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000196611 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000166922 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000245532 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000263590 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000039068 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000243649 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000167244 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000117266 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000187957 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000107821 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000275620 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000248479 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000160886 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000091262 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000162804 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000101115 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000114315 | mRNA | -1 | 0 | 0 |
| ENSG00000172824 | mRNA | ENSG00000109072 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000125398 | mRNA | -1 | 0 | 0 |
| ENSG00000172824 | mRNA | MSTRG.32594 | lncRNA | 1 | 0 | 0 |
| ENSG00000172824 | mRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000163131 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000272114 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172824 | mRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | ENSG00000125730 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | ENSG00000240801 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | ENSG00000143369 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | ENSG00000149926 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | ENSG00000169436 | mRNA | -1 | 0 | 0 |
| ENSG00000255823 | mRNA | ENSG00000058085 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | ENSG00000283321 | mRNA | 1 | 0 | 0 |
| ENSG00000255823 | mRNA | MSTRG.12841 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | ENSG00000287929 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | ENSG00000196611 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | ENSG00000235852 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | ENSG00000166922 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | ENSG00000263590 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | MSTRG.34348 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | ENSG00000243244 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | ENSG00000085831 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | ENSG00000107821 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | ENSG00000248479 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | ENSG00000162804 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | ENSG00000129946 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | ENSG00000276600 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | ENSG00000163131 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000255823 | mRNA | ENSG00000272114 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000125730 | mRNA | ENSG00000143369 | mRNA | 1 | 0 | 0 |
| ENSG00000125730 | mRNA | ENSG00000149926 | mRNA | 1 | 0 | 0 |
| ENSG00000125730 | mRNA | ENSG00000169436 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000125730 | mRNA | ENSG00000283321 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000125730 | mRNA | MSTRG.12841 | mRNA | 1 | 0 | 0 |
| ENSG00000125730 | mRNA | MSTRG.5468 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000125730 | mRNA | ENSG00000167549 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000125730 | mRNA | ENSG00000165029 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000125730 | mRNA | ENSG00000250708 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000125730 | mRNA | ENSG00000130827 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000125730 | mRNA | ENSG00000080031 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000125730 | mRNA | ENSG00000231324 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000125730 | mRNA | ENSG00000269028 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000125730 | mRNA | MSTRG.34348 | lncRNA | -1 | 0 | 0 |
| ENSG00000125730 | mRNA | ENSG00000243244 | mRNA | 1 | 0 | 0 |
| ENSG00000125730 | mRNA | ENSG00000185168 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000125730 | mRNA | ENSG00000287275 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000125730 | mRNA | ENSG00000023445 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000125730 | mRNA | ENSG00000198203 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000125730 | mRNA | ENSG00000134339 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000125730 | mRNA | ENSG00000255182 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000125730 | mRNA | ENSG00000159753 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000125730 | mRNA | ENSG00000221571 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000125730 | mRNA | ENSG00000129946 | mRNA | 1 | 0 | 0 |
| ENSG00000125730 | mRNA | ENSG00000100027 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000125730 | mRNA | ENSG00000137965 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000186862 | mRNA | ENSG00000167861 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000186862 | mRNA | ENSG00000162687 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000186862 | mRNA | ENSG00000168676 | mRNA | 1 | 0 | 0 |
| ENSG00000186862 | mRNA | ENSG00000245532 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000186862 | mRNA | ENSG00000258479 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000186862 | mRNA | ENSG00000074370 | mRNA | 0.970588 | 0.001285 | 0.032602 |
| ENSG00000186862 | mRNA | ENSG00000039068 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000186862 | mRNA | ENSG00000008517 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000186862 | mRNA | ENSG00000078098 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000186862 | mRNA | ENSG00000265185 | lncRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000186862 | mRNA | ENSG00000187957 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000186862 | mRNA | ENSG00000162645 | mRNA | 0.970588 | 0.001285 | 0.032602 |
| ENSG00000186862 | mRNA | ENSG00000254815 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000186862 | mRNA | ENSG00000101198 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000186862 | mRNA | ENSG00000091262 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000186862 | mRNA | ENSG00000156687 | mRNA | 0.970588 | 0.001285 | 0.032602 |
| ENSG00000186862 | mRNA | ENSG00000101115 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000186862 | mRNA | ENSG00000109072 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000186862 | mRNA | ENSG00000140009 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000186862 | mRNA | ENSG00000226416 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000186862 | mRNA | ENSG00000197558 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000258908 | lncRNA | ENSG00000240801 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000167861 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000162687 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000181444 | mRNA | -1 | 0 | 0 |
| ENSG00000258908 | lncRNA | ENSG00000058085 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000237781 | lncRNA | -1 | 0 | 0 |
| ENSG00000258908 | lncRNA | ENSG00000287929 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000196611 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000166922 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000245532 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000263590 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000039068 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000243649 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000167244 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000117266 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000187957 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000107821 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000275620 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000248479 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000160886 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000091262 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000162804 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000101115 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000114315 | mRNA | 1 | 0 | 0 |
| ENSG00000258908 | lncRNA | ENSG00000109072 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000125398 | mRNA | 1 | 0 | 0 |
| ENSG00000258908 | lncRNA | MSTRG.32594 | lncRNA | -1 | 0 | 0 |
| ENSG00000258908 | lncRNA | ENSG00000140009 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000163131 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000272114 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258908 | lncRNA | ENSG00000226416 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000166750 | mRNA | ENSG00000170044 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000166750 | mRNA | MSTRG.12194 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000166750 | mRNA | ENSG00000127528 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000166750 | mRNA | ENSG00000232949 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000166750 | mRNA | ENSG00000146411 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000166750 | mRNA | ENSG00000115844 | mRNA | -1 | 0 | 0 |
| ENSG00000166750 | mRNA | ENSG00000166924 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000166750 | mRNA | ENSG00000104081 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000166750 | mRNA | ENSG00000164078 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000166750 | mRNA | ENSG00000173531 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000166750 | mRNA | ENSG00000119508 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000166750 | mRNA | ENSG00000176692 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000240801 | lncRNA | ENSG00000149927 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240801 | lncRNA | ENSG00000169436 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240801 | lncRNA | ENSG00000288093 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240801 | lncRNA | ENSG00000181444 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240801 | lncRNA | ENSG00000058085 | mRNA | 1 | 0 | 0 |
| ENSG00000240801 | lncRNA | ENSG00000283321 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000240801 | lncRNA | ENSG00000237781 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240801 | lncRNA | ENSG00000287929 | lncRNA | 1 | 0 | 0 |
| ENSG00000240801 | lncRNA | ENSG00000196611 | mRNA | 1 | 0 | 0 |
| ENSG00000240801 | lncRNA | ENSG00000166920 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240801 | lncRNA | ENSG00000089127 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240801 | lncRNA | ENSG00000179148 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240801 | lncRNA | ENSG00000107821 | mRNA | 1 | 0 | 0 |
| ENSG00000240801 | lncRNA | ENSG00000131409 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000240801 | lncRNA | ENSG00000259803 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240801 | lncRNA | ENSG00000229921 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240801 | lncRNA | ENSG00000279954 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240801 | lncRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000240801 | lncRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000240801 | lncRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240801 | lncRNA | ENSG00000272114 | lncRNA | 1 | 0 | 0 |
| ENSG00000167861 | mRNA | ENSG00000105246 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167861 | mRNA | ENSG00000162687 | mRNA | 1 | 0 | 0 |
| ENSG00000167861 | mRNA | ENSG00000181444 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167861 | mRNA | ENSG00000237781 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167861 | mRNA | ENSG00000168676 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000167861 | mRNA | ENSG00000118898 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167861 | mRNA | ENSG00000245532 | lncRNA | 1 | 0 | 0 |
| ENSG00000167861 | mRNA | ENSG00000258479 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167861 | mRNA | ENSG00000089127 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167861 | mRNA | ENSG00000074370 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000167861 | mRNA | ENSG00000039068 | mRNA | 1 | 0 | 0 |
| ENSG00000167861 | mRNA | ENSG00000008517 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167861 | mRNA | ENSG00000179148 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167861 | mRNA | ENSG00000078098 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167861 | mRNA | ENSG00000265185 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000167861 | mRNA | ENSG00000240219 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167861 | mRNA | ENSG00000187957 | mRNA | 1 | 0 | 0 |
| ENSG00000167861 | mRNA | ENSG00000259803 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167861 | mRNA | ENSG00000162645 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000167861 | mRNA | ENSG00000254815 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167861 | mRNA | ENSG00000101198 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167861 | mRNA | ENSG00000091262 | mRNA | 1 | 0 | 0 |
| ENSG00000167861 | mRNA | ENSG00000156687 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000167861 | mRNA | ENSG00000229921 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167861 | mRNA | ENSG00000279954 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167861 | mRNA | ENSG00000101115 | mRNA | 1 | 0 | 0 |
| ENSG00000167861 | mRNA | ENSG00000101197 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167861 | mRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000167861 | mRNA | ENSG00000109072 | mRNA | 1 | 0 | 0 |
| ENSG00000167861 | mRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000167861 | mRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167861 | mRNA | ENSG00000140009 | mRNA | 1 | 0 | 0 |
| ENSG00000167861 | mRNA | ENSG00000226416 | lncRNA | 1 | 0 | 0 |
| ENSG00000167861 | mRNA | ENSG00000197558 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000238007 | lncRNA | ENSG00000203392 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000238007 | lncRNA | MSTRG.5468 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000238007 | lncRNA | ENSG00000130827 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000238007 | lncRNA | ENSG00000080031 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000238007 | lncRNA | ENSG00000231324 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000238007 | lncRNA | ENSG00000269028 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000238007 | lncRNA | MSTRG.5832 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000238007 | lncRNA | ENSG00000198203 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000238007 | lncRNA | ENSG00000196189 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000238007 | lncRNA | ENSG00000182179 | mRNA | 0.954864 | 0.00301 | 0.039507 |
| ENSG00000238007 | lncRNA | ENSG00000255182 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000238007 | lncRNA | ENSG00000125895 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000238007 | lncRNA | ENSG00000100027 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000238007 | lncRNA | ENSG00000137965 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000143369 | mRNA | ENSG00000149926 | mRNA | 1 | 0 | 0 |
| ENSG00000143369 | mRNA | ENSG00000169436 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000143369 | mRNA | ENSG00000283321 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000143369 | mRNA | MSTRG.12841 | mRNA | 1 | 0 | 0 |
| ENSG00000143369 | mRNA | MSTRG.5468 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000143369 | mRNA | ENSG00000167549 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000143369 | mRNA | ENSG00000165029 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000143369 | mRNA | ENSG00000250708 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000143369 | mRNA | ENSG00000130827 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000143369 | mRNA | ENSG00000080031 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000143369 | mRNA | ENSG00000231324 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000143369 | mRNA | ENSG00000269028 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000143369 | mRNA | MSTRG.34348 | lncRNA | -1 | 0 | 0 |
| ENSG00000143369 | mRNA | ENSG00000243244 | mRNA | 1 | 0 | 0 |
| ENSG00000143369 | mRNA | ENSG00000185168 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000143369 | mRNA | ENSG00000287275 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000143369 | mRNA | ENSG00000023445 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000143369 | mRNA | ENSG00000198203 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000143369 | mRNA | ENSG00000134339 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000143369 | mRNA | ENSG00000255182 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000143369 | mRNA | ENSG00000159753 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000143369 | mRNA | ENSG00000221571 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000143369 | mRNA | ENSG00000129946 | mRNA | 1 | 0 | 0 |
| ENSG00000143369 | mRNA | ENSG00000100027 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000143369 | mRNA | ENSG00000137965 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149927 | mRNA | ENSG00000288093 | lncRNA | 1 | 0 | 0 |
| ENSG00000149927 | mRNA | ENSG00000058085 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149927 | mRNA | ENSG00000287929 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149927 | mRNA | ENSG00000196611 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149927 | mRNA | ENSG00000235852 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000149927 | mRNA | ENSG00000166920 | mRNA | 1 | 0 | 0 |
| ENSG00000149927 | mRNA | ENSG00000187688 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149927 | mRNA | ENSG00000085831 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149927 | mRNA | ENSG00000168477 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149927 | mRNA | ENSG00000107821 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149927 | mRNA | ENSG00000131409 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000149927 | mRNA | ENSG00000276600 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149927 | mRNA | ENSG00000272114 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149926 | mRNA | ENSG00000169436 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149926 | mRNA | ENSG00000283321 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000149926 | mRNA | MSTRG.12841 | mRNA | 1 | 0 | 0 |
| ENSG00000149926 | mRNA | MSTRG.5468 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000149926 | mRNA | ENSG00000167549 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149926 | mRNA | ENSG00000165029 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149926 | mRNA | ENSG00000250708 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149926 | mRNA | ENSG00000130827 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149926 | mRNA | ENSG00000080031 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149926 | mRNA | ENSG00000231324 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149926 | mRNA | ENSG00000269028 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000149926 | mRNA | MSTRG.34348 | lncRNA | -1 | 0 | 0 |
| ENSG00000149926 | mRNA | ENSG00000243244 | mRNA | 1 | 0 | 0 |
| ENSG00000149926 | mRNA | ENSG00000185168 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149926 | mRNA | ENSG00000287275 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149926 | mRNA | ENSG00000023445 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000149926 | mRNA | ENSG00000198203 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149926 | mRNA | ENSG00000134339 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000149926 | mRNA | ENSG00000255182 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149926 | mRNA | ENSG00000159753 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000149926 | mRNA | ENSG00000221571 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000149926 | mRNA | ENSG00000129946 | mRNA | 1 | 0 | 0 |
| ENSG00000149926 | mRNA | ENSG00000100027 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000149926 | mRNA | ENSG00000137965 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000169436 | mRNA | ENSG00000058085 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000169436 | mRNA | ENSG00000283321 | mRNA | -1 | 0 | 0 |
| ENSG00000169436 | mRNA | MSTRG.12841 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000169436 | mRNA | ENSG00000287929 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000169436 | mRNA | ENSG00000196611 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000169436 | mRNA | ENSG00000235852 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000169436 | mRNA | ENSG00000166922 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000169436 | mRNA | ENSG00000263590 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000169436 | mRNA | MSTRG.34348 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000169436 | mRNA | ENSG00000243244 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000169436 | mRNA | ENSG00000085831 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000169436 | mRNA | ENSG00000107821 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000169436 | mRNA | ENSG00000248479 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000169436 | mRNA | ENSG00000162804 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000169436 | mRNA | ENSG00000129946 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000169436 | mRNA | ENSG00000276600 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000169436 | mRNA | ENSG00000163131 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000169436 | mRNA | ENSG00000272114 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000170044 | mRNA | MSTRG.12194 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000170044 | mRNA | ENSG00000127528 | mRNA | -1 | 0 | 0 |
| ENSG00000170044 | mRNA | ENSG00000278740 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000170044 | mRNA | ENSG00000146411 | mRNA | 1 | 0 | 0 |
| ENSG00000170044 | mRNA | ENSG00000115844 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000170044 | mRNA | MSTRG.22782 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000170044 | mRNA | ENSG00000107731 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000170044 | mRNA | ENSG00000104081 | mRNA | 1 | 0 | 0 |
| ENSG00000170044 | mRNA | ENSG00000261080 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000170044 | mRNA | ENSG00000149809 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000170044 | mRNA | ENSG00000047457 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000170044 | mRNA | ENSG00000163435 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000170044 | mRNA | ENSG00000267740 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000170044 | mRNA | ENSG00000007516 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000162687 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000266560 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000286242 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000118898 | mRNA | 1 | 0 | 0 |
| ENSG00000105246 | mRNA | MSTRG.13291 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000105246 | mRNA | ENSG00000214353 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000105246 | mRNA | ENSG00000245532 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000039068 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000172602 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000243649 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000167244 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000117266 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000265972 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000240219 | lncRNA | 1 | 0 | 0 |
| ENSG00000105246 | mRNA | ENSG00000187957 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000275620 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000253317 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000160886 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000162645 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000105246 | mRNA | ENSG00000104888 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000105246 | mRNA | ENSG00000091262 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000156687 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000105246 | mRNA | ENSG00000101115 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000101197 | mRNA | 1 | 0 | 0 |
| ENSG00000105246 | mRNA | ENSG00000123843 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000109072 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000148677 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000179111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105246 | mRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000288093 | lncRNA | ENSG00000058085 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000288093 | lncRNA | ENSG00000287929 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000288093 | lncRNA | ENSG00000196611 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000288093 | lncRNA | ENSG00000235852 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000288093 | lncRNA | ENSG00000166920 | mRNA | 1 | 0 | 0 |
| ENSG00000288093 | lncRNA | ENSG00000187688 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000288093 | lncRNA | ENSG00000085831 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000288093 | lncRNA | ENSG00000168477 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000288093 | lncRNA | ENSG00000107821 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000288093 | lncRNA | ENSG00000131409 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000288093 | lncRNA | ENSG00000276600 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000288093 | lncRNA | ENSG00000272114 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | MSTRG.5468 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000258479 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000130827 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000080031 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000231324 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000269028 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000089127 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000008517 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000179148 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000275638 | lncRNA | 1 | 0 | 0 |
| ENSG00000183111 | mRNA | ENSG00000107242 | mRNA | -1 | 0 | 0 |
| ENSG00000183111 | mRNA | ENSG00000116032 | mRNA | 1 | 0 | 0 |
| ENSG00000183111 | mRNA | ENSG00000198203 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000078098 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000265185 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000240764 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000255182 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000259803 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000254815 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000101198 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000237892 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000229921 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000279954 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000100027 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000197249 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000137965 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000183111 | mRNA | ENSG00000197558 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104967 | mRNA | ENSG00000107159 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000104967 | mRNA | ENSG00000130584 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000104967 | mRNA | ENSG00000168477 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104967 | mRNA | ENSG00000265972 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104967 | mRNA | ENSG00000266714 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104967 | mRNA | ENSG00000253317 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104967 | mRNA | ENSG00000183691 | mRNA | -1 | 0 | 0 |
| ENSG00000104967 | mRNA | ENSG00000145020 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104967 | mRNA | ENSG00000137878 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104967 | mRNA | ENSG00000102359 | mRNA | 1 | 0 | 0 |
| ENSG00000104967 | mRNA | ENSG00000105559 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104967 | mRNA | ENSG00000090238 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104967 | mRNA | ENSG00000100342 | mRNA | 1 | 0 | 0 |
| ENSG00000104967 | mRNA | ENSG00000179111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000203392 | lncRNA | MSTRG.5468 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000203392 | lncRNA | ENSG00000130827 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000203392 | lncRNA | ENSG00000080031 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000203392 | lncRNA | ENSG00000231324 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000203392 | lncRNA | ENSG00000269028 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000203392 | lncRNA | ENSG00000287275 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000203392 | lncRNA | MSTRG.5832 | mRNA | -1 | 0 | 0 |
| ENSG00000203392 | lncRNA | ENSG00000198203 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000203392 | lncRNA | ENSG00000196189 | mRNA | 1 | 0 | 0 |
| ENSG00000203392 | lncRNA | ENSG00000240764 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000203392 | lncRNA | ENSG00000255182 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000203392 | lncRNA | ENSG00000159753 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000203392 | lncRNA | ENSG00000120693 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000203392 | lncRNA | ENSG00000237892 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000203392 | lncRNA | ENSG00000125895 | mRNA | 1 | 0 | 0 |
| ENSG00000203392 | lncRNA | ENSG00000100027 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000203392 | lncRNA | ENSG00000197249 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000203392 | lncRNA | ENSG00000137965 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162687 | mRNA | ENSG00000181444 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162687 | mRNA | ENSG00000237781 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162687 | mRNA | ENSG00000168676 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000162687 | mRNA | ENSG00000118898 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162687 | mRNA | ENSG00000245532 | lncRNA | 1 | 0 | 0 |
| ENSG00000162687 | mRNA | ENSG00000258479 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162687 | mRNA | ENSG00000089127 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162687 | mRNA | ENSG00000074370 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000162687 | mRNA | ENSG00000039068 | mRNA | 1 | 0 | 0 |
| ENSG00000162687 | mRNA | ENSG00000008517 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162687 | mRNA | ENSG00000179148 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162687 | mRNA | ENSG00000078098 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162687 | mRNA | ENSG00000265185 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000162687 | mRNA | ENSG00000240219 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162687 | mRNA | ENSG00000187957 | mRNA | 1 | 0 | 0 |
| ENSG00000162687 | mRNA | ENSG00000259803 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162687 | mRNA | ENSG00000162645 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000162687 | mRNA | ENSG00000254815 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162687 | mRNA | ENSG00000101198 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162687 | mRNA | ENSG00000091262 | mRNA | 1 | 0 | 0 |
| ENSG00000162687 | mRNA | ENSG00000156687 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000162687 | mRNA | ENSG00000229921 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162687 | mRNA | ENSG00000279954 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162687 | mRNA | ENSG00000101115 | mRNA | 1 | 0 | 0 |
| ENSG00000162687 | mRNA | ENSG00000101197 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162687 | mRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000162687 | mRNA | ENSG00000109072 | mRNA | 1 | 0 | 0 |
| ENSG00000162687 | mRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000162687 | mRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162687 | mRNA | ENSG00000140009 | mRNA | 1 | 0 | 0 |
| ENSG00000162687 | mRNA | ENSG00000226416 | lncRNA | 1 | 0 | 0 |
| ENSG00000162687 | mRNA | ENSG00000197558 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000058085 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000237781 | lncRNA | 1 | 0 | 0 |
| ENSG00000181444 | mRNA | ENSG00000287929 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000196611 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000166922 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000245532 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000263590 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000039068 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000243649 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000167244 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000117266 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000187957 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000107821 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000275620 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000248479 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000160886 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000091262 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000162804 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000101115 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000114315 | mRNA | -1 | 0 | 0 |
| ENSG00000181444 | mRNA | ENSG00000109072 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000125398 | mRNA | -1 | 0 | 0 |
| ENSG00000181444 | mRNA | MSTRG.32594 | lncRNA | 1 | 0 | 0 |
| ENSG00000181444 | mRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000163131 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000272114 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000181444 | mRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162366 | mRNA | ENSG00000167549 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000162366 | mRNA | ENSG00000165029 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000162366 | mRNA | ENSG00000250708 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000162366 | mRNA | ENSG00000185168 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000162366 | mRNA | ENSG00000023445 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000162366 | mRNA | ENSG00000164078 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000162366 | mRNA | ENSG00000173531 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000162366 | mRNA | ENSG00000134339 | mRNA | 0.955882 | 0.002877 | 0.039507 |
| ENSG00000162366 | mRNA | ENSG00000119508 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000162366 | mRNA | ENSG00000221571 | lncRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000162366 | mRNA | ENSG00000176692 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| MSTRG.12194 | mRNA | ENSG00000127528 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| MSTRG.12194 | mRNA | ENSG00000146411 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| MSTRG.12194 | mRNA | ENSG00000115844 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| MSTRG.12194 | mRNA | ENSG00000104081 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| MSTRG.12194 | mRNA | MSTRG.15840 | mRNA | -0.954864 | 0.00301 | 0.039507 |
| ENSG00000058085 | mRNA | ENSG00000283321 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000058085 | mRNA | ENSG00000237781 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000058085 | mRNA | ENSG00000287929 | lncRNA | 1 | 0 | 0 |
| ENSG00000058085 | mRNA | ENSG00000196611 | mRNA | 1 | 0 | 0 |
| ENSG00000058085 | mRNA | ENSG00000166920 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000058085 | mRNA | ENSG00000089127 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000058085 | mRNA | ENSG00000179148 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000058085 | mRNA | ENSG00000107821 | mRNA | 1 | 0 | 0 |
| ENSG00000058085 | mRNA | ENSG00000131409 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000058085 | mRNA | ENSG00000259803 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000058085 | mRNA | ENSG00000229921 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000058085 | mRNA | ENSG00000279954 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000058085 | mRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000058085 | mRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000058085 | mRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000058085 | mRNA | ENSG00000272114 | lncRNA | 1 | 0 | 0 |
| ENSG00000283321 | mRNA | MSTRG.12841 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000283321 | mRNA | ENSG00000287929 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000283321 | mRNA | ENSG00000196611 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000283321 | mRNA | ENSG00000235852 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000283321 | mRNA | ENSG00000166922 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000283321 | mRNA | ENSG00000263590 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000283321 | mRNA | MSTRG.34348 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000283321 | mRNA | ENSG00000243244 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000283321 | mRNA | ENSG00000085831 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000283321 | mRNA | ENSG00000107821 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000283321 | mRNA | ENSG00000248479 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000283321 | mRNA | ENSG00000162804 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000283321 | mRNA | ENSG00000129946 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000283321 | mRNA | ENSG00000276600 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000283321 | mRNA | ENSG00000163131 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000283321 | mRNA | ENSG00000272114 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.12841 | mRNA | MSTRG.5468 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.12841 | mRNA | ENSG00000167549 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.12841 | mRNA | ENSG00000165029 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.12841 | mRNA | ENSG00000250708 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.12841 | mRNA | ENSG00000130827 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.12841 | mRNA | ENSG00000080031 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.12841 | mRNA | ENSG00000231324 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.12841 | mRNA | ENSG00000269028 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.12841 | mRNA | MSTRG.34348 | lncRNA | -1 | 0 | 0 |
| MSTRG.12841 | mRNA | ENSG00000243244 | mRNA | 1 | 0 | 0 |
| MSTRG.12841 | mRNA | ENSG00000185168 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.12841 | mRNA | ENSG00000287275 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.12841 | mRNA | ENSG00000023445 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.12841 | mRNA | ENSG00000198203 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.12841 | mRNA | ENSG00000134339 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| MSTRG.12841 | mRNA | ENSG00000255182 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.12841 | mRNA | ENSG00000159753 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.12841 | mRNA | ENSG00000221571 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.12841 | mRNA | ENSG00000129946 | mRNA | 1 | 0 | 0 |
| MSTRG.12841 | mRNA | ENSG00000100027 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.12841 | mRNA | ENSG00000137965 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000266560 | lncRNA | ENSG00000073282 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000266560 | lncRNA | ENSG00000118898 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000266560 | lncRNA | ENSG00000100867 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000266560 | lncRNA | ENSG00000214353 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000266560 | lncRNA | ENSG00000258479 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000266560 | lncRNA | ENSG00000008517 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000266560 | lncRNA | ENSG00000172602 | mRNA | 1 | 0 | 0 |
| ENSG00000266560 | lncRNA | ENSG00000078098 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000266560 | lncRNA | ENSG00000265185 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000266560 | lncRNA | ENSG00000240219 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000266560 | lncRNA | ENSG00000104888 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000266560 | lncRNA | ENSG00000254815 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000266560 | lncRNA | ENSG00000101198 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000266560 | lncRNA | ENSG00000142619 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000266560 | lncRNA | ENSG00000101197 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000266560 | lncRNA | ENSG00000123843 | mRNA | -1 | 0 | 0 |
| ENSG00000266560 | lncRNA | ENSG00000158825 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000266560 | lncRNA | ENSG00000148677 | mRNA | -1 | 0 | 0 |
| ENSG00000266560 | lncRNA | ENSG00000122420 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000266560 | lncRNA | ENSG00000197558 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000286242 | lncRNA | ENSG00000118898 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000286242 | lncRNA | ENSG00000240219 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000286242 | lncRNA | ENSG00000283761 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000286242 | lncRNA | ENSG00000101197 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000287929 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000196611 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000166922 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000245532 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000263590 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000039068 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000243649 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000167244 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000117266 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000187957 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000107821 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000275620 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000248479 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000160886 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000091262 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000162804 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000101115 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000114315 | mRNA | -1 | 0 | 0 |
| ENSG00000237781 | lncRNA | ENSG00000109072 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000125398 | mRNA | -1 | 0 | 0 |
| ENSG00000237781 | lncRNA | MSTRG.32594 | lncRNA | 1 | 0 | 0 |
| ENSG00000237781 | lncRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000163131 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000272114 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237781 | lncRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287929 | lncRNA | ENSG00000196611 | mRNA | 1 | 0 | 0 |
| ENSG00000287929 | lncRNA | ENSG00000166920 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287929 | lncRNA | ENSG00000089127 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287929 | lncRNA | ENSG00000179148 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287929 | lncRNA | ENSG00000107821 | mRNA | 1 | 0 | 0 |
| ENSG00000287929 | lncRNA | ENSG00000131409 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000287929 | lncRNA | ENSG00000259803 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287929 | lncRNA | ENSG00000229921 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287929 | lncRNA | ENSG00000279954 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287929 | lncRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000287929 | lncRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000287929 | lncRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287929 | lncRNA | ENSG00000272114 | lncRNA | 1 | 0 | 0 |
| ENSG00000127528 | mRNA | ENSG00000278740 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000127528 | mRNA | ENSG00000146411 | mRNA | -1 | 0 | 0 |
| ENSG00000127528 | mRNA | ENSG00000115844 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000127528 | mRNA | MSTRG.22782 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000127528 | mRNA | ENSG00000107731 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000127528 | mRNA | ENSG00000104081 | mRNA | -1 | 0 | 0 |
| ENSG00000127528 | mRNA | ENSG00000261080 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000127528 | mRNA | ENSG00000149809 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000127528 | mRNA | ENSG00000047457 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000127528 | mRNA | ENSG00000163435 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000127528 | mRNA | ENSG00000267740 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000127528 | mRNA | ENSG00000007516 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.5468 | mRNA | ENSG00000264940 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.5468 | mRNA | ENSG00000130827 | mRNA | -1 | 0 | 0 |
| MSTRG.5468 | mRNA | ENSG00000080031 | mRNA | -1 | 0 | 0 |
| MSTRG.5468 | mRNA | ENSG00000231324 | lncRNA | -1 | 0 | 0 |
| MSTRG.5468 | mRNA | ENSG00000269028 | mRNA | 1 | 0 | 0 |
| MSTRG.5468 | mRNA | MSTRG.34348 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.5468 | mRNA | ENSG00000243244 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.5468 | mRNA | ENSG00000275638 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.5468 | mRNA | ENSG00000107242 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.5468 | mRNA | ENSG00000116032 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.5468 | mRNA | MSTRG.5832 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.5468 | mRNA | ENSG00000198203 | mRNA | -1 | 0 | 0 |
| MSTRG.5468 | mRNA | ENSG00000196189 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.5468 | mRNA | ENSG00000255182 | lncRNA | -1 | 0 | 0 |
| MSTRG.5468 | mRNA | ENSG00000125895 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.5468 | mRNA | ENSG00000129946 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.5468 | mRNA | ENSG00000269888 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.5468 | mRNA | ENSG00000100027 | mRNA | -1 | 0 | 0 |
| MSTRG.5468 | mRNA | ENSG00000137965 | mRNA | -1 | 0 | 0 |
| ENSG00000232949 | lncRNA | ENSG00000073282 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000232949 | lncRNA | ENSG00000115844 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000232949 | lncRNA | ENSG00000100867 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000232949 | lncRNA | ENSG00000120693 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000232949 | lncRNA | ENSG00000142619 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000232949 | lncRNA | ENSG00000158825 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000232949 | lncRNA | ENSG00000122420 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000278740 | lncRNA | ENSG00000146411 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000278740 | lncRNA | ENSG00000235852 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000278740 | lncRNA | ENSG00000166924 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000278740 | lncRNA | MSTRG.26092 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000278740 | lncRNA | ENSG00000085831 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000278740 | lncRNA | ENSG00000107731 | mRNA | -1 | 0 | 0 |
| ENSG00000278740 | lncRNA | ENSG00000168477 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000278740 | lncRNA | ENSG00000104081 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000278740 | lncRNA | ENSG00000261080 | lncRNA | 1 | 0 | 0 |
| ENSG00000278740 | lncRNA | ENSG00000163435 | mRNA | 1 | 0 | 0 |
| ENSG00000278740 | lncRNA | ENSG00000267740 | mRNA | -1 | 0 | 0 |
| ENSG00000278740 | lncRNA | ENSG00000276600 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167549 | mRNA | ENSG00000165029 | mRNA | 1 | 0 | 0 |
| ENSG00000167549 | mRNA | ENSG00000250708 | lncRNA | 1 | 0 | 0 |
| ENSG00000167549 | mRNA | ENSG00000264940 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000167549 | mRNA | ENSG00000166922 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167549 | mRNA | ENSG00000263590 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167549 | mRNA | MSTRG.34348 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000167549 | mRNA | ENSG00000243244 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167549 | mRNA | ENSG00000185168 | lncRNA | 1 | 0 | 0 |
| ENSG00000167549 | mRNA | ENSG00000023445 | mRNA | -1 | 0 | 0 |
| ENSG00000167549 | mRNA | ENSG00000164078 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167549 | mRNA | ENSG00000173531 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167549 | mRNA | ENSG00000134339 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000167549 | mRNA | ENSG00000248479 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167549 | mRNA | ENSG00000162804 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167549 | mRNA | ENSG00000119508 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000167549 | mRNA | ENSG00000221571 | lncRNA | -1 | 0 | 0 |
| ENSG00000167549 | mRNA | ENSG00000129946 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167549 | mRNA | ENSG00000269888 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000167549 | mRNA | ENSG00000176692 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000167549 | mRNA | ENSG00000163131 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000165029 | mRNA | ENSG00000250708 | lncRNA | 1 | 0 | 0 |
| ENSG00000165029 | mRNA | ENSG00000264940 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000165029 | mRNA | ENSG00000166922 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000165029 | mRNA | ENSG00000263590 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000165029 | mRNA | MSTRG.34348 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000165029 | mRNA | ENSG00000243244 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000165029 | mRNA | ENSG00000185168 | lncRNA | 1 | 0 | 0 |
| ENSG00000165029 | mRNA | ENSG00000023445 | mRNA | -1 | 0 | 0 |
| ENSG00000165029 | mRNA | ENSG00000164078 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000165029 | mRNA | ENSG00000173531 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000165029 | mRNA | ENSG00000134339 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000165029 | mRNA | ENSG00000248479 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000165029 | mRNA | ENSG00000162804 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000165029 | mRNA | ENSG00000119508 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000165029 | mRNA | ENSG00000221571 | lncRNA | -1 | 0 | 0 |
| ENSG00000165029 | mRNA | ENSG00000129946 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000165029 | mRNA | ENSG00000269888 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000165029 | mRNA | ENSG00000176692 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000165029 | mRNA | ENSG00000163131 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000073282 | mRNA | ENSG00000100867 | mRNA | 1 | 0 | 0 |
| ENSG00000073282 | mRNA | ENSG00000172602 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000073282 | mRNA | ENSG00000265972 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000073282 | mRNA | ENSG00000266714 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000073282 | mRNA | ENSG00000253317 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000073282 | mRNA | ENSG00000142619 | mRNA | 1 | 0 | 0 |
| ENSG00000073282 | mRNA | ENSG00000145020 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000073282 | mRNA | ENSG00000137878 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000073282 | mRNA | ENSG00000123843 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000073282 | mRNA | ENSG00000105559 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000073282 | mRNA | ENSG00000158825 | mRNA | 1 | 0 | 0 |
| ENSG00000073282 | mRNA | ENSG00000090238 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000073282 | mRNA | ENSG00000148677 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000073282 | mRNA | ENSG00000179111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000073282 | mRNA | ENSG00000122420 | mRNA | 1 | 0 | 0 |
| ENSG00000168676 | mRNA | ENSG00000245532 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000168676 | mRNA | ENSG00000258479 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000168676 | mRNA | ENSG00000074370 | mRNA | 0.970588 | 0.001285 | 0.032602 |
| ENSG00000168676 | mRNA | ENSG00000039068 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000168676 | mRNA | ENSG00000008517 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000168676 | mRNA | ENSG00000078098 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000168676 | mRNA | ENSG00000265185 | lncRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000168676 | mRNA | ENSG00000187957 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000168676 | mRNA | ENSG00000162645 | mRNA | 0.970588 | 0.001285 | 0.032602 |
| ENSG00000168676 | mRNA | ENSG00000254815 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000168676 | mRNA | ENSG00000101198 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000168676 | mRNA | ENSG00000091262 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000168676 | mRNA | ENSG00000156687 | mRNA | 0.970588 | 0.001285 | 0.032602 |
| ENSG00000168676 | mRNA | ENSG00000101115 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000168676 | mRNA | ENSG00000109072 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000168676 | mRNA | ENSG00000140009 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000168676 | mRNA | ENSG00000226416 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000168676 | mRNA | ENSG00000197558 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000146411 | mRNA | ENSG00000115844 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000146411 | mRNA | MSTRG.22782 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000146411 | mRNA | ENSG00000107731 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000146411 | mRNA | ENSG00000104081 | mRNA | 1 | 0 | 0 |
| ENSG00000146411 | mRNA | ENSG00000261080 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000146411 | mRNA | ENSG00000149809 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000146411 | mRNA | ENSG00000047457 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000146411 | mRNA | ENSG00000163435 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000146411 | mRNA | ENSG00000267740 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000146411 | mRNA | ENSG00000007516 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000196611 | mRNA | ENSG00000166920 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000196611 | mRNA | ENSG00000089127 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000196611 | mRNA | ENSG00000179148 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000196611 | mRNA | ENSG00000107821 | mRNA | 1 | 0 | 0 |
| ENSG00000196611 | mRNA | ENSG00000131409 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000196611 | mRNA | ENSG00000259803 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000196611 | mRNA | ENSG00000229921 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000196611 | mRNA | ENSG00000279954 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000196611 | mRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000196611 | mRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000196611 | mRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000196611 | mRNA | ENSG00000272114 | lncRNA | 1 | 0 | 0 |
| ENSG00000115844 | mRNA | ENSG00000166924 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000115844 | mRNA | ENSG00000104081 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000115844 | mRNA | ENSG00000164078 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000115844 | mRNA | ENSG00000173531 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000115844 | mRNA | ENSG00000119508 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000115844 | mRNA | ENSG00000176692 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000118898 | mRNA | MSTRG.13291 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000118898 | mRNA | ENSG00000214353 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000118898 | mRNA | ENSG00000245532 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000118898 | mRNA | ENSG00000039068 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000118898 | mRNA | ENSG00000172602 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000118898 | mRNA | ENSG00000243649 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000118898 | mRNA | ENSG00000167244 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000118898 | mRNA | ENSG00000117266 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000118898 | mRNA | ENSG00000265972 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000118898 | mRNA | ENSG00000240219 | lncRNA | 1 | 0 | 0 |
| ENSG00000118898 | mRNA | ENSG00000187957 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000118898 | mRNA | ENSG00000275620 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000118898 | mRNA | ENSG00000253317 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000118898 | mRNA | ENSG00000160886 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000118898 | mRNA | ENSG00000162645 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000118898 | mRNA | ENSG00000104888 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000118898 | mRNA | ENSG00000091262 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000118898 | mRNA | ENSG00000156687 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000118898 | mRNA | ENSG00000101115 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000118898 | mRNA | ENSG00000101197 | mRNA | 1 | 0 | 0 |
| ENSG00000118898 | mRNA | ENSG00000123843 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000118898 | mRNA | ENSG00000109072 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000118898 | mRNA | ENSG00000148677 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000118898 | mRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000118898 | mRNA | ENSG00000179111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000118898 | mRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000100867 | mRNA | ENSG00000172602 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000100867 | mRNA | ENSG00000265972 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000100867 | mRNA | ENSG00000266714 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000100867 | mRNA | ENSG00000253317 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000100867 | mRNA | ENSG00000142619 | mRNA | 1 | 0 | 0 |
| ENSG00000100867 | mRNA | ENSG00000145020 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000100867 | mRNA | ENSG00000137878 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000100867 | mRNA | ENSG00000123843 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000100867 | mRNA | ENSG00000105559 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000100867 | mRNA | ENSG00000158825 | mRNA | 1 | 0 | 0 |
| ENSG00000100867 | mRNA | ENSG00000090238 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000100867 | mRNA | ENSG00000148677 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000100867 | mRNA | ENSG00000179111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000100867 | mRNA | ENSG00000122420 | mRNA | 1 | 0 | 0 |
| ENSG00000235852 | lncRNA | ENSG00000166920 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000235852 | lncRNA | ENSG00000085831 | mRNA | -1 | 0 | 0 |
| ENSG00000235852 | lncRNA | ENSG00000287275 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000235852 | lncRNA | ENSG00000107731 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000235852 | lncRNA | ENSG00000261080 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000235852 | lncRNA | ENSG00000159753 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000235852 | lncRNA | ENSG00000163435 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000235852 | lncRNA | ENSG00000267740 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000235852 | lncRNA | ENSG00000276600 | mRNA | -1 | 0 | 0 |
| ENSG00000250708 | lncRNA | ENSG00000264940 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000250708 | lncRNA | ENSG00000166922 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000250708 | lncRNA | ENSG00000263590 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000250708 | lncRNA | MSTRG.34348 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000250708 | lncRNA | ENSG00000243244 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000250708 | lncRNA | ENSG00000185168 | lncRNA | 1 | 0 | 0 |
| ENSG00000250708 | lncRNA | ENSG00000023445 | mRNA | -1 | 0 | 0 |
| ENSG00000250708 | lncRNA | ENSG00000164078 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000250708 | lncRNA | ENSG00000173531 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000250708 | lncRNA | ENSG00000134339 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000250708 | lncRNA | ENSG00000248479 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000250708 | lncRNA | ENSG00000162804 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000250708 | lncRNA | ENSG00000119508 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000250708 | lncRNA | ENSG00000221571 | lncRNA | -1 | 0 | 0 |
| ENSG00000250708 | lncRNA | ENSG00000129946 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000250708 | lncRNA | ENSG00000269888 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000250708 | lncRNA | ENSG00000176692 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000250708 | lncRNA | ENSG00000163131 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.13291 | mRNA | ENSG00000214353 | lncRNA | -0.955882 | 0.002877 | 0.039507 |
| MSTRG.13291 | mRNA | ENSG00000243649 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| MSTRG.13291 | mRNA | ENSG00000167244 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| MSTRG.13291 | mRNA | ENSG00000117266 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| MSTRG.13291 | mRNA | ENSG00000240219 | lncRNA | -0.985611 | 0.000309 | 0.008045 |
| MSTRG.13291 | mRNA | ENSG00000275620 | lncRNA | -0.985611 | 0.000309 | 0.008045 |
| MSTRG.13291 | mRNA | ENSG00000160886 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| MSTRG.13291 | mRNA | ENSG00000162645 | mRNA | -0.970588 | 0.001285 | 0.032602 |
| MSTRG.13291 | mRNA | ENSG00000104888 | mRNA | -0.955882 | 0.002877 | 0.039507 |
| MSTRG.13291 | mRNA | ENSG00000156687 | mRNA | -0.970588 | 0.001285 | 0.032602 |
| MSTRG.13291 | mRNA | ENSG00000101197 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000214353 | lncRNA | ENSG00000172602 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000214353 | lncRNA | ENSG00000240219 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000214353 | lncRNA | ENSG00000162645 | mRNA | 0.970588 | 0.001285 | 0.032602 |
| ENSG00000214353 | lncRNA | ENSG00000104888 | mRNA | 1 | 0 | 0 |
| ENSG00000214353 | lncRNA | ENSG00000156687 | mRNA | 0.970588 | 0.001285 | 0.032602 |
| ENSG00000214353 | lncRNA | ENSG00000101197 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000214353 | lncRNA | ENSG00000123843 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000214353 | lncRNA | ENSG00000148677 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000264940 | lncRNA | ENSG00000258479 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000264940 | lncRNA | ENSG00000130827 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000264940 | lncRNA | ENSG00000080031 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000264940 | lncRNA | ENSG00000231324 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000264940 | lncRNA | ENSG00000164182 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000264940 | lncRNA | ENSG00000269028 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000264940 | lncRNA | ENSG00000185168 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000264940 | lncRNA | ENSG00000008517 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000264940 | lncRNA | ENSG00000023445 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000264940 | lncRNA | ENSG00000198203 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000264940 | lncRNA | ENSG00000078098 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000264940 | lncRNA | ENSG00000265185 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000264940 | lncRNA | ENSG00000255182 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000264940 | lncRNA | ENSG00000254815 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000264940 | lncRNA | ENSG00000101198 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000264940 | lncRNA | ENSG00000221571 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000264940 | lncRNA | ENSG00000269888 | lncRNA | 1 | 0 | 0 |
| ENSG00000264940 | lncRNA | ENSG00000100027 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000264940 | lncRNA | ENSG00000137965 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000264940 | lncRNA | ENSG00000197558 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000166922 | mRNA | ENSG00000263590 | lncRNA | 1 | 0 | 0 |
| ENSG00000166922 | mRNA | ENSG00000185168 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000166922 | mRNA | ENSG00000023445 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000166922 | mRNA | ENSG00000248479 | lncRNA | 1 | 0 | 0 |
| ENSG00000166922 | mRNA | ENSG00000162804 | mRNA | 1 | 0 | 0 |
| ENSG00000166922 | mRNA | ENSG00000221571 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000166922 | mRNA | ENSG00000007516 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000166922 | mRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000166922 | mRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000166922 | mRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000166922 | mRNA | ENSG00000163131 | mRNA | 1 | 0 | 0 |
| ENSG00000166920 | mRNA | ENSG00000187688 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000166920 | mRNA | ENSG00000085831 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000166920 | mRNA | ENSG00000168477 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000166920 | mRNA | ENSG00000107821 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000166920 | mRNA | ENSG00000131409 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000166920 | mRNA | ENSG00000276600 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000166920 | mRNA | ENSG00000272114 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000166924 | mRNA | ENSG00000287275 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000166924 | mRNA | ENSG00000107731 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000166924 | mRNA | ENSG00000261080 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000166924 | mRNA | ENSG00000159753 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000166924 | mRNA | ENSG00000120693 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000166924 | mRNA | ENSG00000163435 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000166924 | mRNA | ENSG00000267740 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.22782 | mRNA | ENSG00000168477 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.22782 | mRNA | ENSG00000243649 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.22782 | mRNA | ENSG00000167244 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.22782 | mRNA | ENSG00000104081 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.22782 | mRNA | ENSG00000117266 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.22782 | mRNA | ENSG00000265972 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.22782 | mRNA | ENSG00000275620 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.22782 | mRNA | ENSG00000253317 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.22782 | mRNA | ENSG00000149809 | mRNA | 1 | 0 | 0 |
| MSTRG.22782 | mRNA | ENSG00000160886 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.22782 | mRNA | ENSG00000047457 | mRNA | 1 | 0 | 0 |
| MSTRG.22782 | mRNA | ENSG00000179111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000245532 | lncRNA | ENSG00000258479 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000245532 | lncRNA | ENSG00000089127 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000245532 | lncRNA | ENSG00000074370 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000245532 | lncRNA | ENSG00000039068 | mRNA | 1 | 0 | 0 |
| ENSG00000245532 | lncRNA | ENSG00000008517 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000245532 | lncRNA | ENSG00000179148 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000245532 | lncRNA | ENSG00000078098 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000245532 | lncRNA | ENSG00000265185 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000245532 | lncRNA | ENSG00000240219 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000245532 | lncRNA | ENSG00000187957 | mRNA | 1 | 0 | 0 |
| ENSG00000245532 | lncRNA | ENSG00000259803 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000245532 | lncRNA | ENSG00000162645 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000245532 | lncRNA | ENSG00000254815 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000245532 | lncRNA | ENSG00000101198 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000245532 | lncRNA | ENSG00000091262 | mRNA | 1 | 0 | 0 |
| ENSG00000245532 | lncRNA | ENSG00000156687 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000245532 | lncRNA | ENSG00000229921 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000245532 | lncRNA | ENSG00000279954 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000245532 | lncRNA | ENSG00000101115 | mRNA | 1 | 0 | 0 |
| ENSG00000245532 | lncRNA | ENSG00000101197 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000245532 | lncRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000245532 | lncRNA | ENSG00000109072 | mRNA | 1 | 0 | 0 |
| ENSG00000245532 | lncRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000245532 | lncRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000245532 | lncRNA | ENSG00000140009 | mRNA | 1 | 0 | 0 |
| ENSG00000245532 | lncRNA | ENSG00000226416 | lncRNA | 1 | 0 | 0 |
| ENSG00000245532 | lncRNA | ENSG00000197558 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000263590 | lncRNA | ENSG00000185168 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000263590 | lncRNA | ENSG00000023445 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000263590 | lncRNA | ENSG00000248479 | lncRNA | 1 | 0 | 0 |
| ENSG00000263590 | lncRNA | ENSG00000162804 | mRNA | 1 | 0 | 0 |
| ENSG00000263590 | lncRNA | ENSG00000221571 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000263590 | lncRNA | ENSG00000007516 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000263590 | lncRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000263590 | lncRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000263590 | lncRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000263590 | lncRNA | ENSG00000163131 | mRNA | 1 | 0 | 0 |
| ENSG00000258479 | lncRNA | ENSG00000039068 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258479 | lncRNA | ENSG00000008517 | mRNA | 1 | 0 | 0 |
| ENSG00000258479 | lncRNA | ENSG00000275638 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258479 | lncRNA | ENSG00000107242 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258479 | lncRNA | ENSG00000116032 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258479 | lncRNA | ENSG00000172602 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258479 | lncRNA | ENSG00000078098 | mRNA | 1 | 0 | 0 |
| ENSG00000258479 | lncRNA | ENSG00000265185 | lncRNA | -1 | 0 | 0 |
| ENSG00000258479 | lncRNA | ENSG00000187957 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258479 | lncRNA | ENSG00000254815 | lncRNA | 1 | 0 | 0 |
| ENSG00000258479 | lncRNA | ENSG00000101198 | mRNA | 1 | 0 | 0 |
| ENSG00000258479 | lncRNA | ENSG00000091262 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258479 | lncRNA | ENSG00000101115 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258479 | lncRNA | ENSG00000123843 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258479 | lncRNA | ENSG00000109072 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258479 | lncRNA | ENSG00000269888 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258479 | lncRNA | ENSG00000148677 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000258479 | lncRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258479 | lncRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000258479 | lncRNA | ENSG00000197558 | mRNA | 1 | 0 | 0 |
| ENSG00000130827 | mRNA | ENSG00000080031 | mRNA | 1 | 0 | 0 |
| ENSG00000130827 | mRNA | ENSG00000231324 | lncRNA | 1 | 0 | 0 |
| ENSG00000130827 | mRNA | ENSG00000269028 | mRNA | -1 | 0 | 0 |
| ENSG00000130827 | mRNA | MSTRG.34348 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000130827 | mRNA | ENSG00000243244 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000130827 | mRNA | ENSG00000275638 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000130827 | mRNA | ENSG00000107242 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000130827 | mRNA | ENSG00000116032 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000130827 | mRNA | MSTRG.5832 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000130827 | mRNA | ENSG00000198203 | mRNA | 1 | 0 | 0 |
| ENSG00000130827 | mRNA | ENSG00000196189 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000130827 | mRNA | ENSG00000255182 | lncRNA | 1 | 0 | 0 |
| ENSG00000130827 | mRNA | ENSG00000125895 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000130827 | mRNA | ENSG00000129946 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000130827 | mRNA | ENSG00000269888 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000130827 | mRNA | ENSG00000100027 | mRNA | 1 | 0 | 0 |
| ENSG00000130827 | mRNA | ENSG00000137965 | mRNA | 1 | 0 | 0 |
| MSTRG.34052 | mRNA | ENSG00000042980 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.34052 | mRNA | ENSG00000287275 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.34052 | mRNA | ENSG00000159753 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000107159 | mRNA | ENSG00000130584 | mRNA | 0.970588 | 0.001285 | 0.032602 |
| ENSG00000107159 | mRNA | ENSG00000265972 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000107159 | mRNA | ENSG00000253317 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000107159 | mRNA | ENSG00000183691 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000107159 | mRNA | ENSG00000102359 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000107159 | mRNA | ENSG00000100342 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000107159 | mRNA | ENSG00000179111 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| MSTRG.26092 | lncRNA | ENSG00000107731 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| MSTRG.26092 | lncRNA | ENSG00000168477 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| MSTRG.26092 | lncRNA | ENSG00000261080 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| MSTRG.26092 | lncRNA | ENSG00000163435 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| MSTRG.26092 | lncRNA | ENSG00000267740 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000080031 | mRNA | ENSG00000231324 | lncRNA | 1 | 0 | 0 |
| ENSG00000080031 | mRNA | ENSG00000269028 | mRNA | -1 | 0 | 0 |
| ENSG00000080031 | mRNA | MSTRG.34348 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000080031 | mRNA | ENSG00000243244 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000080031 | mRNA | ENSG00000275638 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000080031 | mRNA | ENSG00000107242 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000080031 | mRNA | ENSG00000116032 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000080031 | mRNA | MSTRG.5832 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000080031 | mRNA | ENSG00000198203 | mRNA | 1 | 0 | 0 |
| ENSG00000080031 | mRNA | ENSG00000196189 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000080031 | mRNA | ENSG00000255182 | lncRNA | 1 | 0 | 0 |
| ENSG00000080031 | mRNA | ENSG00000125895 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000080031 | mRNA | ENSG00000129946 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000080031 | mRNA | ENSG00000269888 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000080031 | mRNA | ENSG00000100027 | mRNA | 1 | 0 | 0 |
| ENSG00000080031 | mRNA | ENSG00000137965 | mRNA | 1 | 0 | 0 |
| ENSG00000231324 | lncRNA | ENSG00000269028 | mRNA | -1 | 0 | 0 |
| ENSG00000231324 | lncRNA | MSTRG.34348 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000231324 | lncRNA | ENSG00000243244 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000231324 | lncRNA | ENSG00000275638 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000231324 | lncRNA | ENSG00000107242 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000231324 | lncRNA | ENSG00000116032 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000231324 | lncRNA | MSTRG.5832 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000231324 | lncRNA | ENSG00000198203 | mRNA | 1 | 0 | 0 |
| ENSG00000231324 | lncRNA | ENSG00000196189 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000231324 | lncRNA | ENSG00000255182 | lncRNA | 1 | 0 | 0 |
| ENSG00000231324 | lncRNA | ENSG00000125895 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000231324 | lncRNA | ENSG00000129946 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000231324 | lncRNA | ENSG00000269888 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000231324 | lncRNA | ENSG00000100027 | mRNA | 1 | 0 | 0 |
| ENSG00000231324 | lncRNA | ENSG00000137965 | mRNA | 1 | 0 | 0 |
| ENSG00000130584 | mRNA | ENSG00000266714 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000130584 | mRNA | ENSG00000183691 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000130584 | mRNA | ENSG00000145020 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000130584 | mRNA | ENSG00000137878 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000130584 | mRNA | ENSG00000102359 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000130584 | mRNA | ENSG00000105559 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000130584 | mRNA | ENSG00000090238 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000130584 | mRNA | ENSG00000100342 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000164182 | mRNA | ENSG00000269888 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000269028 | mRNA | MSTRG.34348 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000269028 | mRNA | ENSG00000243244 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000269028 | mRNA | ENSG00000275638 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000269028 | mRNA | ENSG00000107242 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000269028 | mRNA | ENSG00000116032 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000269028 | mRNA | MSTRG.5832 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000269028 | mRNA | ENSG00000198203 | mRNA | -1 | 0 | 0 |
| ENSG00000269028 | mRNA | ENSG00000196189 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000269028 | mRNA | ENSG00000255182 | lncRNA | -1 | 0 | 0 |
| ENSG00000269028 | mRNA | ENSG00000125895 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000269028 | mRNA | ENSG00000129946 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000269028 | mRNA | ENSG00000269888 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000269028 | mRNA | ENSG00000100027 | mRNA | -1 | 0 | 0 |
| ENSG00000269028 | mRNA | ENSG00000137965 | mRNA | -1 | 0 | 0 |
| MSTRG.34348 | lncRNA | ENSG00000243244 | mRNA | -1 | 0 | 0 |
| MSTRG.34348 | lncRNA | ENSG00000185168 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.34348 | lncRNA | ENSG00000287275 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.34348 | lncRNA | ENSG00000023445 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.34348 | lncRNA | ENSG00000198203 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.34348 | lncRNA | ENSG00000134339 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| MSTRG.34348 | lncRNA | ENSG00000255182 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.34348 | lncRNA | ENSG00000159753 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.34348 | lncRNA | ENSG00000221571 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.34348 | lncRNA | ENSG00000129946 | mRNA | -1 | 0 | 0 |
| MSTRG.34348 | lncRNA | ENSG00000100027 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.34348 | lncRNA | ENSG00000137965 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000089127 | mRNA | ENSG00000074370 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000089127 | mRNA | ENSG00000039068 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000089127 | mRNA | ENSG00000179148 | mRNA | 1 | 0 | 0 |
| ENSG00000089127 | mRNA | ENSG00000275638 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000089127 | mRNA | ENSG00000107242 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000089127 | mRNA | ENSG00000116032 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000089127 | mRNA | ENSG00000187957 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000089127 | mRNA | ENSG00000107821 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000089127 | mRNA | ENSG00000259803 | mRNA | 1 | 0 | 0 |
| ENSG00000089127 | mRNA | ENSG00000091262 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000089127 | mRNA | ENSG00000229921 | lncRNA | 1 | 0 | 0 |
| ENSG00000089127 | mRNA | ENSG00000279954 | lncRNA | 1 | 0 | 0 |
| ENSG00000089127 | mRNA | ENSG00000101115 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000089127 | mRNA | ENSG00000109072 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000089127 | mRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000089127 | mRNA | ENSG00000272114 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000089127 | mRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000074370 | mRNA | ENSG00000039068 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000074370 | mRNA | ENSG00000179148 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000074370 | mRNA | ENSG00000187957 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000074370 | mRNA | ENSG00000259803 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000074370 | mRNA | ENSG00000162645 | mRNA | 0.955882 | 0.002877 | 0.039507 |
| ENSG00000074370 | mRNA | ENSG00000091262 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000074370 | mRNA | ENSG00000156687 | mRNA | 0.955882 | 0.002877 | 0.039507 |
| ENSG00000074370 | mRNA | ENSG00000229921 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000074370 | mRNA | ENSG00000279954 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000074370 | mRNA | ENSG00000101115 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000074370 | mRNA | ENSG00000109072 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000074370 | mRNA | ENSG00000140009 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000074370 | mRNA | ENSG00000226416 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000243244 | mRNA | ENSG00000185168 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000243244 | mRNA | ENSG00000287275 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000243244 | mRNA | ENSG00000023445 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000243244 | mRNA | ENSG00000198203 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000243244 | mRNA | ENSG00000134339 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000243244 | mRNA | ENSG00000255182 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000243244 | mRNA | ENSG00000159753 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000243244 | mRNA | ENSG00000221571 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000243244 | mRNA | ENSG00000129946 | mRNA | 1 | 0 | 0 |
| ENSG00000243244 | mRNA | ENSG00000100027 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000243244 | mRNA | ENSG00000137965 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000185168 | lncRNA | ENSG00000023445 | mRNA | -1 | 0 | 0 |
| ENSG00000185168 | lncRNA | ENSG00000164078 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000185168 | lncRNA | ENSG00000173531 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000185168 | lncRNA | ENSG00000134339 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000185168 | lncRNA | ENSG00000248479 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000185168 | lncRNA | ENSG00000162804 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000185168 | lncRNA | ENSG00000119508 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000185168 | lncRNA | ENSG00000221571 | lncRNA | -1 | 0 | 0 |
| ENSG00000185168 | lncRNA | ENSG00000129946 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000185168 | lncRNA | ENSG00000269888 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000185168 | lncRNA | ENSG00000176692 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000185168 | lncRNA | ENSG00000163131 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000039068 | mRNA | ENSG00000008517 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000039068 | mRNA | ENSG00000179148 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000039068 | mRNA | ENSG00000078098 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000039068 | mRNA | ENSG00000265185 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000039068 | mRNA | ENSG00000240219 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000039068 | mRNA | ENSG00000187957 | mRNA | 1 | 0 | 0 |
| ENSG00000039068 | mRNA | ENSG00000259803 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000039068 | mRNA | ENSG00000162645 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000039068 | mRNA | ENSG00000254815 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000039068 | mRNA | ENSG00000101198 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000039068 | mRNA | ENSG00000091262 | mRNA | 1 | 0 | 0 |
| ENSG00000039068 | mRNA | ENSG00000156687 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000039068 | mRNA | ENSG00000229921 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000039068 | mRNA | ENSG00000279954 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000039068 | mRNA | ENSG00000101115 | mRNA | 1 | 0 | 0 |
| ENSG00000039068 | mRNA | ENSG00000101197 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000039068 | mRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000039068 | mRNA | ENSG00000109072 | mRNA | 1 | 0 | 0 |
| ENSG00000039068 | mRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000039068 | mRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000039068 | mRNA | ENSG00000140009 | mRNA | 1 | 0 | 0 |
| ENSG00000039068 | mRNA | ENSG00000226416 | lncRNA | 1 | 0 | 0 |
| ENSG00000039068 | mRNA | ENSG00000197558 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000008517 | mRNA | ENSG00000275638 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000008517 | mRNA | ENSG00000107242 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000008517 | mRNA | ENSG00000116032 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000008517 | mRNA | ENSG00000172602 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000008517 | mRNA | ENSG00000078098 | mRNA | 1 | 0 | 0 |
| ENSG00000008517 | mRNA | ENSG00000265185 | lncRNA | -1 | 0 | 0 |
| ENSG00000008517 | mRNA | ENSG00000187957 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000008517 | mRNA | ENSG00000254815 | lncRNA | 1 | 0 | 0 |
| ENSG00000008517 | mRNA | ENSG00000101198 | mRNA | 1 | 0 | 0 |
| ENSG00000008517 | mRNA | ENSG00000091262 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000008517 | mRNA | ENSG00000101115 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000008517 | mRNA | ENSG00000123843 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000008517 | mRNA | ENSG00000109072 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000008517 | mRNA | ENSG00000269888 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000008517 | mRNA | ENSG00000148677 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000008517 | mRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000008517 | mRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000008517 | mRNA | ENSG00000197558 | mRNA | 1 | 0 | 0 |
| ENSG00000179148 | mRNA | ENSG00000275638 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000179148 | mRNA | ENSG00000107242 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000179148 | mRNA | ENSG00000116032 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000179148 | mRNA | ENSG00000187957 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000179148 | mRNA | ENSG00000107821 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000179148 | mRNA | ENSG00000259803 | mRNA | 1 | 0 | 0 |
| ENSG00000179148 | mRNA | ENSG00000091262 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000179148 | mRNA | ENSG00000229921 | lncRNA | 1 | 0 | 0 |
| ENSG00000179148 | mRNA | ENSG00000279954 | lncRNA | 1 | 0 | 0 |
| ENSG00000179148 | mRNA | ENSG00000101115 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000179148 | mRNA | ENSG00000109072 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000179148 | mRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000179148 | mRNA | ENSG00000272114 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000179148 | mRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000085831 | mRNA | ENSG00000287275 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000085831 | mRNA | ENSG00000107731 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000085831 | mRNA | ENSG00000261080 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000085831 | mRNA | ENSG00000159753 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000085831 | mRNA | ENSG00000163435 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000085831 | mRNA | ENSG00000267740 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000085831 | mRNA | ENSG00000276600 | mRNA | 1 | 0 | 0 |
| ENSG00000275638 | lncRNA | ENSG00000107242 | mRNA | -1 | 0 | 0 |
| ENSG00000275638 | lncRNA | ENSG00000116032 | mRNA | 1 | 0 | 0 |
| ENSG00000275638 | lncRNA | ENSG00000198203 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000275638 | lncRNA | ENSG00000078098 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000275638 | lncRNA | ENSG00000265185 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000275638 | lncRNA | ENSG00000240764 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000275638 | lncRNA | ENSG00000255182 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000275638 | lncRNA | ENSG00000259803 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000275638 | lncRNA | ENSG00000254815 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000275638 | lncRNA | ENSG00000101198 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000275638 | lncRNA | ENSG00000237892 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000275638 | lncRNA | ENSG00000229921 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000275638 | lncRNA | ENSG00000279954 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000275638 | lncRNA | ENSG00000100027 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000275638 | lncRNA | ENSG00000197249 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000275638 | lncRNA | ENSG00000137965 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000275638 | lncRNA | ENSG00000197558 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000107242 | mRNA | ENSG00000116032 | mRNA | -1 | 0 | 0 |
| ENSG00000107242 | mRNA | ENSG00000198203 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000107242 | mRNA | ENSG00000078098 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000107242 | mRNA | ENSG00000265185 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000107242 | mRNA | ENSG00000240764 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000107242 | mRNA | ENSG00000255182 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000107242 | mRNA | ENSG00000259803 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000107242 | mRNA | ENSG00000254815 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000107242 | mRNA | ENSG00000101198 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000107242 | mRNA | ENSG00000237892 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000107242 | mRNA | ENSG00000229921 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000107242 | mRNA | ENSG00000279954 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000107242 | mRNA | ENSG00000100027 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000107242 | mRNA | ENSG00000197249 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000107242 | mRNA | ENSG00000137965 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000107242 | mRNA | ENSG00000197558 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000287275 | lncRNA | MSTRG.5832 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000287275 | lncRNA | ENSG00000196189 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287275 | lncRNA | ENSG00000159753 | mRNA | -1 | 0 | 0 |
| ENSG00000287275 | lncRNA | ENSG00000125895 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287275 | lncRNA | ENSG00000129946 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000287275 | lncRNA | ENSG00000276600 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000116032 | mRNA | ENSG00000198203 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000116032 | mRNA | ENSG00000078098 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000116032 | mRNA | ENSG00000265185 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000116032 | mRNA | ENSG00000240764 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000116032 | mRNA | ENSG00000255182 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000116032 | mRNA | ENSG00000259803 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000116032 | mRNA | ENSG00000254815 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000116032 | mRNA | ENSG00000101198 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000116032 | mRNA | ENSG00000237892 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000116032 | mRNA | ENSG00000229921 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000116032 | mRNA | ENSG00000279954 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000116032 | mRNA | ENSG00000100027 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000116032 | mRNA | ENSG00000197249 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000116032 | mRNA | ENSG00000137965 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000116032 | mRNA | ENSG00000197558 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000107731 | mRNA | ENSG00000168477 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000107731 | mRNA | ENSG00000104081 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000107731 | mRNA | ENSG00000261080 | lncRNA | -1 | 0 | 0 |
| ENSG00000107731 | mRNA | ENSG00000163435 | mRNA | -1 | 0 | 0 |
| ENSG00000107731 | mRNA | ENSG00000267740 | mRNA | 1 | 0 | 0 |
| ENSG00000107731 | mRNA | ENSG00000276600 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000172602 | mRNA | ENSG00000078098 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172602 | mRNA | ENSG00000265185 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000172602 | mRNA | ENSG00000240219 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172602 | mRNA | ENSG00000104888 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000172602 | mRNA | ENSG00000254815 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172602 | mRNA | ENSG00000101198 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172602 | mRNA | ENSG00000142619 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172602 | mRNA | ENSG00000101197 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172602 | mRNA | ENSG00000123843 | mRNA | -1 | 0 | 0 |
| ENSG00000172602 | mRNA | ENSG00000158825 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172602 | mRNA | ENSG00000148677 | mRNA | -1 | 0 | 0 |
| ENSG00000172602 | mRNA | ENSG00000122420 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000172602 | mRNA | ENSG00000197558 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000168477 | mRNA | ENSG00000261080 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000168477 | mRNA | ENSG00000149809 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000168477 | mRNA | ENSG00000183691 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000168477 | mRNA | ENSG00000047457 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000168477 | mRNA | ENSG00000163435 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000168477 | mRNA | ENSG00000267740 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000168477 | mRNA | ENSG00000102359 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000168477 | mRNA | ENSG00000100342 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000023445 | mRNA | ENSG00000164078 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000023445 | mRNA | ENSG00000173531 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000023445 | mRNA | ENSG00000134339 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000023445 | mRNA | ENSG00000248479 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000023445 | mRNA | ENSG00000162804 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000023445 | mRNA | ENSG00000119508 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000023445 | mRNA | ENSG00000221571 | lncRNA | 1 | 0 | 0 |
| ENSG00000023445 | mRNA | ENSG00000129946 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000023445 | mRNA | ENSG00000269888 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000023445 | mRNA | ENSG00000176692 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000023445 | mRNA | ENSG00000163131 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000243649 | mRNA | ENSG00000167244 | mRNA | 1 | 0 | 0 |
| ENSG00000243649 | mRNA | ENSG00000117266 | mRNA | 1 | 0 | 0 |
| ENSG00000243649 | mRNA | ENSG00000240219 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000243649 | mRNA | ENSG00000275620 | lncRNA | 1 | 0 | 0 |
| ENSG00000243649 | mRNA | ENSG00000149809 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000243649 | mRNA | ENSG00000160886 | mRNA | -1 | 0 | 0 |
| ENSG00000243649 | mRNA | ENSG00000283761 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000243649 | mRNA | ENSG00000047457 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000243649 | mRNA | ENSG00000101197 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000243649 | mRNA | ENSG00000007516 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000243649 | mRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000243649 | mRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000243649 | mRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.5832 | mRNA | ENSG00000198203 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.5832 | mRNA | ENSG00000196189 | mRNA | -1 | 0 | 0 |
| MSTRG.5832 | mRNA | ENSG00000240764 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.5832 | mRNA | ENSG00000255182 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.5832 | mRNA | ENSG00000159753 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.5832 | mRNA | ENSG00000120693 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.5832 | mRNA | ENSG00000237892 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.5832 | mRNA | ENSG00000125895 | mRNA | -1 | 0 | 0 |
| MSTRG.5832 | mRNA | ENSG00000100027 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.5832 | mRNA | ENSG00000197249 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| MSTRG.5832 | mRNA | ENSG00000137965 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000167244 | mRNA | ENSG00000117266 | mRNA | 1 | 0 | 0 |
| ENSG00000167244 | mRNA | ENSG00000240219 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167244 | mRNA | ENSG00000275620 | lncRNA | 1 | 0 | 0 |
| ENSG00000167244 | mRNA | ENSG00000149809 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167244 | mRNA | ENSG00000160886 | mRNA | -1 | 0 | 0 |
| ENSG00000167244 | mRNA | ENSG00000283761 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167244 | mRNA | ENSG00000047457 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167244 | mRNA | ENSG00000101197 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167244 | mRNA | ENSG00000007516 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000167244 | mRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000167244 | mRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000167244 | mRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000198203 | mRNA | ENSG00000196189 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000198203 | mRNA | ENSG00000255182 | lncRNA | 1 | 0 | 0 |
| ENSG00000198203 | mRNA | ENSG00000125895 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000198203 | mRNA | ENSG00000129946 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000198203 | mRNA | ENSG00000269888 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000198203 | mRNA | ENSG00000100027 | mRNA | 1 | 0 | 0 |
| ENSG00000198203 | mRNA | ENSG00000137965 | mRNA | 1 | 0 | 0 |
| ENSG00000104081 | mRNA | ENSG00000261080 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104081 | mRNA | ENSG00000149809 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104081 | mRNA | ENSG00000047457 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104081 | mRNA | ENSG00000163435 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000104081 | mRNA | ENSG00000267740 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000104081 | mRNA | ENSG00000007516 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000078098 | mRNA | ENSG00000265185 | lncRNA | -1 | 0 | 0 |
| ENSG00000078098 | mRNA | ENSG00000187957 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000078098 | mRNA | ENSG00000254815 | lncRNA | 1 | 0 | 0 |
| ENSG00000078098 | mRNA | ENSG00000101198 | mRNA | 1 | 0 | 0 |
| ENSG00000078098 | mRNA | ENSG00000091262 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000078098 | mRNA | ENSG00000101115 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000078098 | mRNA | ENSG00000123843 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000078098 | mRNA | ENSG00000109072 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000078098 | mRNA | ENSG00000269888 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000078098 | mRNA | ENSG00000148677 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000078098 | mRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000078098 | mRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000078098 | mRNA | ENSG00000197558 | mRNA | 1 | 0 | 0 |
| ENSG00000117266 | mRNA | ENSG00000240219 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000117266 | mRNA | ENSG00000275620 | lncRNA | 1 | 0 | 0 |
| ENSG00000117266 | mRNA | ENSG00000149809 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000117266 | mRNA | ENSG00000160886 | mRNA | -1 | 0 | 0 |
| ENSG00000117266 | mRNA | ENSG00000283761 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000117266 | mRNA | ENSG00000047457 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000117266 | mRNA | ENSG00000101197 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000117266 | mRNA | ENSG00000007516 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000117266 | mRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000117266 | mRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000117266 | mRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000196189 | mRNA | ENSG00000240764 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000196189 | mRNA | ENSG00000255182 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000196189 | mRNA | ENSG00000159753 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000196189 | mRNA | ENSG00000120693 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000196189 | mRNA | ENSG00000237892 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000196189 | mRNA | ENSG00000125895 | mRNA | 1 | 0 | 0 |
| ENSG00000196189 | mRNA | ENSG00000100027 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000196189 | mRNA | ENSG00000197249 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000196189 | mRNA | ENSG00000137965 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000265972 | mRNA | ENSG00000240219 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000265972 | mRNA | ENSG00000253317 | lncRNA | 1 | 0 | 0 |
| ENSG00000265972 | mRNA | ENSG00000149809 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000265972 | mRNA | ENSG00000183691 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000265972 | mRNA | ENSG00000142619 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000265972 | mRNA | ENSG00000047457 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000265972 | mRNA | ENSG00000101197 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000265972 | mRNA | ENSG00000102359 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000265972 | mRNA | ENSG00000158825 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000265972 | mRNA | ENSG00000100342 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000265972 | mRNA | ENSG00000179111 | mRNA | 1 | 0 | 0 |
| ENSG00000265972 | mRNA | ENSG00000122420 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000164078 | mRNA | ENSG00000173531 | mRNA | 1 | 0 | 0 |
| ENSG00000164078 | mRNA | ENSG00000119508 | mRNA | -1 | 0 | 0 |
| ENSG00000164078 | mRNA | ENSG00000221571 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000164078 | mRNA | ENSG00000007516 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000164078 | mRNA | ENSG00000176692 | mRNA | -1 | 0 | 0 |
| ENSG00000287046 | lncRNA | ENSG00000010295 | mRNA | 0.983739 | 0.000394 | 0.010254 |
| ENSG00000265185 | lncRNA | ENSG00000187957 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000265185 | lncRNA | ENSG00000254815 | lncRNA | -1 | 0 | 0 |
| ENSG00000265185 | lncRNA | ENSG00000101198 | mRNA | -1 | 0 | 0 |
| ENSG00000265185 | lncRNA | ENSG00000091262 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000265185 | lncRNA | ENSG00000101115 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000265185 | lncRNA | ENSG00000123843 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000265185 | lncRNA | ENSG00000109072 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000265185 | lncRNA | ENSG00000269888 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000265185 | lncRNA | ENSG00000148677 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000265185 | lncRNA | ENSG00000140009 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000265185 | lncRNA | ENSG00000226416 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000265185 | lncRNA | ENSG00000197558 | mRNA | -1 | 0 | 0 |
| ENSG00000240219 | lncRNA | ENSG00000187957 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240219 | lncRNA | ENSG00000275620 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240219 | lncRNA | ENSG00000253317 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240219 | lncRNA | ENSG00000160886 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000240219 | lncRNA | ENSG00000162645 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000240219 | lncRNA | ENSG00000104888 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000240219 | lncRNA | ENSG00000091262 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240219 | lncRNA | ENSG00000156687 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000240219 | lncRNA | ENSG00000101115 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240219 | lncRNA | ENSG00000101197 | mRNA | 1 | 0 | 0 |
| ENSG00000240219 | lncRNA | ENSG00000123843 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000240219 | lncRNA | ENSG00000109072 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240219 | lncRNA | ENSG00000148677 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000240219 | lncRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240219 | lncRNA | ENSG00000179111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240219 | lncRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000187957 | mRNA | ENSG00000259803 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000187957 | mRNA | ENSG00000162645 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000187957 | mRNA | ENSG00000254815 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000187957 | mRNA | ENSG00000101198 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000187957 | mRNA | ENSG00000091262 | mRNA | 1 | 0 | 0 |
| ENSG00000187957 | mRNA | ENSG00000156687 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000187957 | mRNA | ENSG00000229921 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000187957 | mRNA | ENSG00000279954 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000187957 | mRNA | ENSG00000101115 | mRNA | 1 | 0 | 0 |
| ENSG00000187957 | mRNA | ENSG00000101197 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000187957 | mRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000187957 | mRNA | ENSG00000109072 | mRNA | 1 | 0 | 0 |
| ENSG00000187957 | mRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000187957 | mRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000187957 | mRNA | ENSG00000140009 | mRNA | 1 | 0 | 0 |
| ENSG00000187957 | mRNA | ENSG00000226416 | lncRNA | 1 | 0 | 0 |
| ENSG00000187957 | mRNA | ENSG00000197558 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000107821 | mRNA | ENSG00000131409 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000107821 | mRNA | ENSG00000259803 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000107821 | mRNA | ENSG00000229921 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000107821 | mRNA | ENSG00000279954 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000107821 | mRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000107821 | mRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000107821 | mRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000107821 | mRNA | ENSG00000272114 | lncRNA | 1 | 0 | 0 |
| ENSG00000240764 | mRNA | ENSG00000266714 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240764 | mRNA | ENSG00000237892 | lncRNA | 1 | 0 | 0 |
| ENSG00000240764 | mRNA | ENSG00000145020 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240764 | mRNA | ENSG00000125895 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240764 | mRNA | ENSG00000137878 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240764 | mRNA | ENSG00000105559 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240764 | mRNA | ENSG00000090238 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000240764 | mRNA | ENSG00000197249 | mRNA | 1 | 0 | 0 |
| ENSG00000266714 | mRNA | ENSG00000183691 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000266714 | mRNA | ENSG00000120693 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000266714 | mRNA | ENSG00000142619 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000266714 | mRNA | ENSG00000237892 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000266714 | mRNA | ENSG00000145020 | mRNA | 1 | 0 | 0 |
| ENSG00000266714 | mRNA | ENSG00000137878 | mRNA | 1 | 0 | 0 |
| ENSG00000266714 | mRNA | ENSG00000102359 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000266714 | mRNA | ENSG00000105559 | mRNA | 1 | 0 | 0 |
| ENSG00000266714 | mRNA | ENSG00000158825 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000266714 | mRNA | ENSG00000090238 | mRNA | 1 | 0 | 0 |
| ENSG00000266714 | mRNA | ENSG00000100342 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000266714 | mRNA | ENSG00000197249 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000266714 | mRNA | ENSG00000122420 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000261080 | lncRNA | ENSG00000163435 | mRNA | 1 | 0 | 0 |
| ENSG00000261080 | lncRNA | ENSG00000267740 | mRNA | -1 | 0 | 0 |
| ENSG00000261080 | lncRNA | ENSG00000276600 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000275620 | lncRNA | ENSG00000149809 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000275620 | lncRNA | ENSG00000160886 | mRNA | -1 | 0 | 0 |
| ENSG00000275620 | lncRNA | ENSG00000283761 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000275620 | lncRNA | ENSG00000047457 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000275620 | lncRNA | ENSG00000101197 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000275620 | lncRNA | ENSG00000007516 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000275620 | lncRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000275620 | lncRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000275620 | lncRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000253317 | lncRNA | ENSG00000149809 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000253317 | lncRNA | ENSG00000183691 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000253317 | lncRNA | ENSG00000142619 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000253317 | lncRNA | ENSG00000047457 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000253317 | lncRNA | ENSG00000101197 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000253317 | lncRNA | ENSG00000102359 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000253317 | lncRNA | ENSG00000158825 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000253317 | lncRNA | ENSG00000100342 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000253317 | lncRNA | ENSG00000179111 | mRNA | 1 | 0 | 0 |
| ENSG00000253317 | lncRNA | ENSG00000122420 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000173531 | mRNA | ENSG00000119508 | mRNA | -1 | 0 | 0 |
| ENSG00000173531 | mRNA | ENSG00000221571 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000173531 | mRNA | ENSG00000007516 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000173531 | mRNA | ENSG00000176692 | mRNA | -1 | 0 | 0 |
| ENSG00000131409 | mRNA | ENSG00000272114 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000134339 | mRNA | ENSG00000221571 | lncRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000134339 | mRNA | ENSG00000129946 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000149809 | mRNA | ENSG00000160886 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000149809 | mRNA | ENSG00000047457 | mRNA | 1 | 0 | 0 |
| ENSG00000149809 | mRNA | ENSG00000179111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000248479 | lncRNA | ENSG00000162804 | mRNA | 1 | 0 | 0 |
| ENSG00000248479 | lncRNA | ENSG00000221571 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000248479 | lncRNA | ENSG00000007516 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000248479 | lncRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000248479 | lncRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000248479 | lncRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000248479 | lncRNA | ENSG00000163131 | mRNA | 1 | 0 | 0 |
| ENSG00000160886 | mRNA | ENSG00000283761 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000160886 | mRNA | ENSG00000047457 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000160886 | mRNA | ENSG00000101197 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000160886 | mRNA | ENSG00000007516 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000160886 | mRNA | ENSG00000114315 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000160886 | mRNA | ENSG00000125398 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000160886 | mRNA | MSTRG.32594 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000183691 | mRNA | ENSG00000145020 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000183691 | mRNA | ENSG00000137878 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000183691 | mRNA | ENSG00000102359 | mRNA | -1 | 0 | 0 |
| ENSG00000183691 | mRNA | ENSG00000105559 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000183691 | mRNA | ENSG00000090238 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000183691 | mRNA | ENSG00000100342 | mRNA | -1 | 0 | 0 |
| ENSG00000183691 | mRNA | ENSG00000179111 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000255182 | lncRNA | ENSG00000125895 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000255182 | lncRNA | ENSG00000129946 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000255182 | lncRNA | ENSG00000269888 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000255182 | lncRNA | ENSG00000100027 | mRNA | 1 | 0 | 0 |
| ENSG00000255182 | lncRNA | ENSG00000137965 | mRNA | 1 | 0 | 0 |
| ENSG00000159753 | mRNA | ENSG00000125895 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000159753 | mRNA | ENSG00000129946 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000159753 | mRNA | ENSG00000276600 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000259803 | mRNA | ENSG00000091262 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000259803 | mRNA | ENSG00000229921 | lncRNA | 1 | 0 | 0 |
| ENSG00000259803 | mRNA | ENSG00000279954 | lncRNA | 1 | 0 | 0 |
| ENSG00000259803 | mRNA | ENSG00000101115 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000259803 | mRNA | ENSG00000109072 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000259803 | mRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000259803 | mRNA | ENSG00000272114 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000259803 | mRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162645 | mRNA | ENSG00000104888 | mRNA | 0.970588 | 0.001285 | 0.032602 |
| ENSG00000162645 | mRNA | ENSG00000091262 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000162645 | mRNA | ENSG00000156687 | mRNA | 1 | 0 | 0 |
| ENSG00000162645 | mRNA | ENSG00000101115 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000162645 | mRNA | ENSG00000101197 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000162645 | mRNA | ENSG00000109072 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000162645 | mRNA | ENSG00000140009 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000162645 | mRNA | ENSG00000226416 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000120693 | mRNA | ENSG00000145020 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000120693 | mRNA | ENSG00000125895 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000120693 | mRNA | ENSG00000137878 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000120693 | mRNA | ENSG00000105559 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000120693 | mRNA | ENSG00000090238 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000104888 | mRNA | ENSG00000156687 | mRNA | 0.970588 | 0.001285 | 0.032602 |
| ENSG00000104888 | mRNA | ENSG00000101197 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000104888 | mRNA | ENSG00000123843 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000104888 | mRNA | ENSG00000148677 | mRNA | -0.985611 | 0.000309 | 0.008045 |
| ENSG00000254815 | lncRNA | ENSG00000101198 | mRNA | 1 | 0 | 0 |
| ENSG00000254815 | lncRNA | ENSG00000091262 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000254815 | lncRNA | ENSG00000101115 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000254815 | lncRNA | ENSG00000123843 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000254815 | lncRNA | ENSG00000109072 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000254815 | lncRNA | ENSG00000269888 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000254815 | lncRNA | ENSG00000148677 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000254815 | lncRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000254815 | lncRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000254815 | lncRNA | ENSG00000197558 | mRNA | 1 | 0 | 0 |
| ENSG00000101198 | mRNA | ENSG00000091262 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000101198 | mRNA | ENSG00000101115 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000101198 | mRNA | ENSG00000123843 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000101198 | mRNA | ENSG00000109072 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000101198 | mRNA | ENSG00000269888 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000101198 | mRNA | ENSG00000148677 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000101198 | mRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000101198 | mRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000101198 | mRNA | ENSG00000197558 | mRNA | 1 | 0 | 0 |
| ENSG00000142619 | mRNA | ENSG00000145020 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000142619 | mRNA | ENSG00000137878 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000142619 | mRNA | ENSG00000123843 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000142619 | mRNA | ENSG00000105559 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000142619 | mRNA | ENSG00000158825 | mRNA | 1 | 0 | 0 |
| ENSG00000142619 | mRNA | ENSG00000090238 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000142619 | mRNA | ENSG00000148677 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000142619 | mRNA | ENSG00000179111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000142619 | mRNA | ENSG00000122420 | mRNA | 1 | 0 | 0 |
| ENSG00000091262 | mRNA | ENSG00000156687 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000091262 | mRNA | ENSG00000229921 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000091262 | mRNA | ENSG00000279954 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000091262 | mRNA | ENSG00000101115 | mRNA | 1 | 0 | 0 |
| ENSG00000091262 | mRNA | ENSG00000101197 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000091262 | mRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000091262 | mRNA | ENSG00000109072 | mRNA | 1 | 0 | 0 |
| ENSG00000091262 | mRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000091262 | mRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000091262 | mRNA | ENSG00000140009 | mRNA | 1 | 0 | 0 |
| ENSG00000091262 | mRNA | ENSG00000226416 | lncRNA | 1 | 0 | 0 |
| ENSG00000091262 | mRNA | ENSG00000197558 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000047457 | mRNA | ENSG00000179111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000163435 | mRNA | ENSG00000267740 | mRNA | -1 | 0 | 0 |
| ENSG00000163435 | mRNA | ENSG00000276600 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237892 | lncRNA | ENSG00000145020 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237892 | lncRNA | ENSG00000125895 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237892 | lncRNA | ENSG00000137878 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237892 | lncRNA | ENSG00000105559 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237892 | lncRNA | ENSG00000090238 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000237892 | lncRNA | ENSG00000197249 | mRNA | 1 | 0 | 0 |
| ENSG00000156687 | mRNA | ENSG00000101115 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000156687 | mRNA | ENSG00000101197 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000156687 | mRNA | ENSG00000109072 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000156687 | mRNA | ENSG00000140009 | mRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000156687 | mRNA | ENSG00000226416 | lncRNA | 0.985611 | 0.000309 | 0.008045 |
| ENSG00000145020 | mRNA | ENSG00000137878 | mRNA | 1 | 0 | 0 |
| ENSG00000145020 | mRNA | ENSG00000102359 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000145020 | mRNA | ENSG00000105559 | mRNA | 1 | 0 | 0 |
| ENSG00000145020 | mRNA | ENSG00000158825 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000145020 | mRNA | ENSG00000090238 | mRNA | 1 | 0 | 0 |
| ENSG00000145020 | mRNA | ENSG00000100342 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000145020 | mRNA | ENSG00000197249 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000145020 | mRNA | ENSG00000122420 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162804 | mRNA | ENSG00000221571 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000162804 | mRNA | ENSG00000007516 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162804 | mRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000162804 | mRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000162804 | mRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000162804 | mRNA | ENSG00000163131 | mRNA | 1 | 0 | 0 |
| ENSG00000119508 | mRNA | ENSG00000221571 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000119508 | mRNA | ENSG00000007516 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000119508 | mRNA | ENSG00000176692 | mRNA | 1 | 0 | 0 |
| ENSG00000229921 | lncRNA | ENSG00000279954 | lncRNA | 1 | 0 | 0 |
| ENSG00000229921 | lncRNA | ENSG00000101115 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000229921 | lncRNA | ENSG00000109072 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000229921 | lncRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000229921 | lncRNA | ENSG00000272114 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000229921 | lncRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000279954 | lncRNA | ENSG00000101115 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000279954 | lncRNA | ENSG00000109072 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000279954 | lncRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000279954 | lncRNA | ENSG00000272114 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000279954 | lncRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000221571 | lncRNA | ENSG00000129946 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000221571 | lncRNA | ENSG00000269888 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000221571 | lncRNA | ENSG00000176692 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000221571 | lncRNA | ENSG00000163131 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000101115 | mRNA | ENSG00000101197 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000101115 | mRNA | ENSG00000114315 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000101115 | mRNA | ENSG00000109072 | mRNA | 1 | 0 | 0 |
| ENSG00000101115 | mRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000101115 | mRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000101115 | mRNA | ENSG00000140009 | mRNA | 1 | 0 | 0 |
| ENSG00000101115 | mRNA | ENSG00000226416 | lncRNA | 1 | 0 | 0 |
| ENSG00000101115 | mRNA | ENSG00000197558 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000125895 | mRNA | ENSG00000100027 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000125895 | mRNA | ENSG00000197249 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000125895 | mRNA | ENSG00000137965 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000101197 | mRNA | ENSG00000123843 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000101197 | mRNA | ENSG00000109072 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000101197 | mRNA | ENSG00000148677 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000101197 | mRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000101197 | mRNA | ENSG00000179111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000101197 | mRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000137878 | mRNA | ENSG00000102359 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000137878 | mRNA | ENSG00000105559 | mRNA | 1 | 0 | 0 |
| ENSG00000137878 | mRNA | ENSG00000158825 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000137878 | mRNA | ENSG00000090238 | mRNA | 1 | 0 | 0 |
| ENSG00000137878 | mRNA | ENSG00000100342 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000137878 | mRNA | ENSG00000197249 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000137878 | mRNA | ENSG00000122420 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000267740 | mRNA | ENSG00000276600 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000007516 | mRNA | ENSG00000176692 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000007516 | mRNA | ENSG00000163131 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000114315 | mRNA | ENSG00000109072 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000114315 | mRNA | ENSG00000125398 | mRNA | 1 | 0 | 0 |
| ENSG00000114315 | mRNA | MSTRG.32594 | lncRNA | -1 | 0 | 0 |
| ENSG00000114315 | mRNA | ENSG00000140009 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000114315 | mRNA | ENSG00000163131 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000114315 | mRNA | ENSG00000272114 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000114315 | mRNA | ENSG00000226416 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000102359 | mRNA | ENSG00000105559 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000102359 | mRNA | ENSG00000090238 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000102359 | mRNA | ENSG00000100342 | mRNA | 1 | 0 | 0 |
| ENSG00000102359 | mRNA | ENSG00000179111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000123843 | mRNA | ENSG00000158825 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000123843 | mRNA | ENSG00000148677 | mRNA | 1 | 0 | 0 |
| ENSG00000123843 | mRNA | ENSG00000122420 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000123843 | mRNA | ENSG00000197558 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000105559 | mRNA | ENSG00000158825 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105559 | mRNA | ENSG00000090238 | mRNA | 1 | 0 | 0 |
| ENSG00000105559 | mRNA | ENSG00000100342 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105559 | mRNA | ENSG00000197249 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000105559 | mRNA | ENSG00000122420 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000158825 | mRNA | ENSG00000090238 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000158825 | mRNA | ENSG00000148677 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000158825 | mRNA | ENSG00000179111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000158825 | mRNA | ENSG00000122420 | mRNA | 1 | 0 | 0 |
| ENSG00000090238 | mRNA | ENSG00000100342 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000090238 | mRNA | ENSG00000197249 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000090238 | mRNA | ENSG00000122420 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109072 | mRNA | ENSG00000125398 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000109072 | mRNA | MSTRG.32594 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000109072 | mRNA | ENSG00000140009 | mRNA | 1 | 0 | 0 |
| ENSG00000109072 | mRNA | ENSG00000226416 | lncRNA | 1 | 0 | 0 |
| ENSG00000109072 | mRNA | ENSG00000197558 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000129946 | mRNA | ENSG00000100027 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000129946 | mRNA | ENSG00000137965 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000125398 | mRNA | MSTRG.32594 | lncRNA | -1 | 0 | 0 |
| ENSG00000125398 | mRNA | ENSG00000140009 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000125398 | mRNA | ENSG00000163131 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000125398 | mRNA | ENSG00000272114 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000125398 | mRNA | ENSG00000226416 | lncRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000269888 | lncRNA | ENSG00000100027 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000269888 | lncRNA | ENSG00000137965 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000269888 | lncRNA | ENSG00000197558 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000100342 | mRNA | ENSG00000179111 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.32594 | lncRNA | ENSG00000140009 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.32594 | lncRNA | ENSG00000163131 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.32594 | lncRNA | ENSG00000272114 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| MSTRG.32594 | lncRNA | ENSG00000226416 | lncRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000148677 | mRNA | ENSG00000122420 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000148677 | mRNA | ENSG00000197558 | mRNA | -0.942857 | 0.004805 | 0.039507 |
| ENSG00000140009 | mRNA | ENSG00000226416 | lncRNA | 1 | 0 | 0 |
| ENSG00000140009 | mRNA | ENSG00000197558 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000100027 | mRNA | ENSG00000137965 | mRNA | 1 | 0 | 0 |
| ENSG00000179111 | mRNA | ENSG00000122420 | mRNA | 0.942857 | 0.004805 | 0.039507 |
| ENSG00000226416 | lncRNA | ENSG00000197558 | mRNA | 0.942857 | 0.004805 | 0.039507 |
Table S7: Detailed information of lncRNA-mRNA co-expression network.
To further investigate the interactions between DE LncRNA s and target genes, an interaction network was constructed. As shown in Figure 3A, 72 target genes were present in the interaction network. Several genes were found to exert high node degree in the network, including cadherin 1 (CDH1, node degree [18], SRY-box transcription Factor 9 (SOX9, node degree G. [10], and serpin family A member 1 (SERPINA1, node degree [8], which suggests that these genes probably play vital roles in phycocyanin-mediated regulatory processes via LncRNA s in A549 cells. Moreover, LncRNA s also regulated multiple target genes in a cross-talk manner (Figure 3B). LncRNA ENST00000662245 was downregulated in phycocyanin-treated A549 cells and regulated the expression of CDH1, SOX9, and VTN (vitronectin). Additionally, ENST00000538717 was downregulated in the PC groups and affected the expression of IGF2 and MBF. Coincidentally, the mRNA-seq in our previous work has discovered multiple pathways and signal molecules involved in phycocyanin-mediated inhibition of NSCLC cells.
Interfering a series of signal pathways and some key node factors such as MMP and p53, phycocyanin influenced the growth of NSCLC cells (21). Interestingly, the MMP protein family both showed significantly differences in present work and our previous study [21], including MMP1(Figure 3A in present work) and MMP3, MMP9, MMP14 (in previous study), indicating LncRNA might suppress cell migration through MMP protein family, which supported the results of previous work. The former study just focused on study the mRNAs levels, which could be regulated by its upstream LncRNAs. So the present work is a further extension and expansion of previous work. In a word, these LncRNA‒mRNA interaction pairs may play a crucial regulatory role in A549 cells after phycocyanin exposure.
The discovery of this LncRNA‒mRNA network deepened the understanding of the pathogenesis of NSCLC and the antineoplastic mechanism of phycocyanin. Zhang et al. revealed a LncRNA‒mRNA coexpression network for myocardial infarction, providing diagnosis and prognosis biomarkers of myocardial infarction by means of specific LncRNA expression profiles [22]. Moreover, Cai et al. constructed a LncRNA‒mRNA ceRNA network, discovering novel prognostic biomarkers in rectal cancer [23]. These reports suggest that LncRNA s could be important regulators of mRNAs and gene expression in various diseases, including cancer. In particular, LncRNA SOX2‐OT was found to reduce the efficacy of therapy and worsen the clinical prognosis of lung cancer patients through AKT/ERK and SOX2/GLI‐1 signaling [24]. It is worth noting that in the current study, the transcription factor SOX9, from the same family as SOX2, was shown to be regulated by two differentially expressed LncRNA s, ENST00000662245, ENST00000432735 and NONHSAT254537.1, revealing the potential regulatory mechanism of phycocyanin in A549 cells. In addition, 2,238 mRNA‒mRNA, LncRNA‒LncRNA, and LncRNA‒mRNA pairs were identified, showing the potential mutual effects of DE LncRNA s and mRNAs in this study. These molecules may be biologically important in the phycocyanin-mediated anticancer effects on NSCLC cells, which will be under investigation in the future.
Validation and functional assays of LncRNA s in NSCLC cells
As shown in Figure 3B, several targets were enriched in the functional category of cell proliferation and movement progress (GO: GO: 0007219). Of these LncRNAs, LncRNA ENST00000538717 was predicted to downregulated (log2FC=-10.02) and regulate multiple genes (Figure 4A), including IGF2 (insulin-like growth Factor 2), DHRS2 (dehydrogenase/reductase 2), BMF (Bcl2 modifying factor), and RND1 (Rho family GTPase 1), indicating that ENST00000538717 may regulate the viability of phycocyanin-treated A549 cells. Next, to verify the RNA-seq results, qRT‒PCR analysis was used to measure the expression of ENST00000538717 in a phycocyanin-treated NSCLC cell model. As shown in Figure 4B, the relative expression of LncRNA s was significantly downregulated after phycocyanin exposure in both H460 and A549 cells, suggesting that the sequencing results are highly reliable. To further estimate whether the ectopic expression of ENST00000538717 affects the growth and migration of NSCLC cells, cell survival and wound healing assays were employed. Figure 4C and 4D show that suppressing the expression of ENST00000538717 could significantly decrease the viability and migration in both A549 and H460 cells. Altogether, the results revealed that ENST00000538717 might be a candidate regulator in phycocyanin-mediated inhibition of NSCLC cell viability.
Figure 4: Validation and functional assays of lncRNA ENST00000538717 in NSCLC cells. (A) The target prediction and GO analysis indicate that lncRNA ENST00000538717 are related to multiple genes and cell viability. (B) qRT-PCR validation of RNA-sequencing expression result of lncRNA ENST00000538717 in A549 and H460 cells. (C) Cell survival analysis of A549 and H460 cells after ENST00000538717 inhibition. (D) Cell migration analysis of A549 and H460 cells after ENST00000538717 inhibition. *, p < 0.05, **, p < 0.01.
Increasing evidence has suggested that LncRNA s could be important prognostic factors for tumors, including lung cancer [18,25,26]. Zhang et al. reported that LncRNA TUC338 was overexpressed in lung cancer and promoted the invasion of lung cancer through the MAPK pathway [27]. In addition, LncRNA s could also act as cancer suppressor genes in NSCLC through LncRNA‒LncRNA interactions [28]. In the current work, a total of 309 LncRNA s, including 116 downregulated and 193 upregulated LncRNA s, exhibited significant expression differences between the Con and PC groups. LncRNA ENST00000538717 was chosen to verify the LncRNA-seq results, and its expression was downregulated after phycocyanin treatment, indicating that it is an inhibitory factor in NSCLC cells. Interestingly, ENST00000538717 performs different functions in different types of tumors. Yi et al. reported that overexpression of LINC00852 promotes prostate cancer cell proliferation and metastasis [29]. However, our results revealed that ENST00000538717 impaired the viability and migration of NSCLC cells, which was consistent with Tuo's study [30]. The data suggested that this LncRNA s may be novel regulatory factors of phycocyanin in NSCLC cells, and the precise functions of LncRNA s remain under further investigation.
In summary, the present work systematically identified the changes in LncRNA s during the process of phycocyanin-mediated inhibition of NSCLC cell activity and further characterized these changes through bioinformatic analyses. A key differential LncRNA was screened and verified, and phycocyanin was found to decrease the activity of NSCLC cells in vitro by inhibiting this LncRNA. Nowadays, competing endogenous RNAs (ceRNAs) hypothesis has become an important research field in the study of cell life activity and tumor regulation [31]. Studies have shown that the LncRNA-mRNA regulatory network is involved in the regulation of various tumor cells [31-33]. As a result, functional study of specific LncRNA in cancer cells has certain limitations. The combination of LncRNA-mRNA network with cell phenotype is the key research direction of ceRNAs. Meanwhile, beside LncRNA, ceRNAs including cirRNA, pseudogene, competing miRNA can also interfere with expression of target mRNAs [34]. Therefore, further explorations are urgently needed to dig out the deep antitumor mechanism of phycocyanin, which is also the topic of our future research.
Conclusion
In this study, differentially expressed LncRNA s and target genes were discovered, and they are involved in cell viability, apoptosis, migration and corresponding signaling pathways. Moreover, the LncRNA ‒mRNA interaction network provided a novel regulatory pattern, which showed potential molecular targets of phycocyanin, molecular therapeutic factors and prognostic markers of NSCLC. Most notably, 72 significant gene nodes were discovered in the target gene interaction network, which are tightly associated with the antineoplastic mechanism of phycocyanin in NSCLC. The results may provide a new strategy for further investigation of antitumor targets and the regulatory process of phycocyanin in NSCLC.
Supplementary materials
Table S1. Statistics of LncRNA s
Table S2. Classification and subgroup of LncRNA s
Table S3. The top expressed 15 LncRNA transcripts in Con and PC groups
Table S4. Detailed information of DE LncRNA s
Table S5. Top 10 downregulated and 10 upregulated LncRNA transcripts
Table S6. Detailed information for potential targets of DE LncRNA s
Table S7. Detailed information of LncRNA-mRNA co-expression network
Figure S1. LncRNA-mRNA co-expression networks.
Acknowledgements
This work was supported by National Natural Science Foundation of China [32272321] and [32072231], the fund of Cultivation Project of Double First-Class Disciplines of Food Science and Engineering, Beijing Technology & Business University [BTBUYXTD202208], and the Project of Cultivation for young top-notch Talents of Beijing Municipal Institution (BPHR202203042).
Author Contributions
Conceptualization, Shuai Hao; Funding acquisition, Shuai Hao; Investigation, Boxiong Wu, Haozhe Cheng, Xinran Li and Wenjing Zhang; Methodology, Boxiong Wu and Shuai Hao; Resources, Shuai Hao; Supervision, Shuai Hao; Validation, Boxiong Wu, Haozhe Cheng and Xinran Li; Writing-original draft, Shuai Hao.
Conflicts of Interest
The authors declare no conflict of interest.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data are available upon request from the corresponding author (haoshuai@btbu.edu.cn).
Informed Consent Statement
Not applicable.
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, et al. (2021) Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 71: 209-249.
- Ettinger DS, Wood DE, Akerley W, Bazhenova LA, Borghaei H, et al. (2016) NCCN Guidelines (R) Insights: Non-Small Cell Lung Cancer, Version 4.2016 Featured Updates to the NCCN Guidelines. J Natl Compr Canc Netw 14: 255-264.
- Molina JR, Yang PG, Cassivi SD, Schild SE and Adjei AA (2008) Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc 83: 584-594.
- Mansoori B, Mohammadi A, Davudian S, Shirjang S and Baradaran B (2017) the Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv Pharm Bull 7: 339-348.
- Guo T, Li J, Zhang L, Hou W, Wang R, et al. (2019) Multidimensional communication of microRNAs and long non-coding RNAs in lung cancer. J Cancer Res Clin 145: 31-48.
- Ransohoff JD, Wei Y and Khavari PA (2018) the functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol 19: 143-157.
- Novikova IV, Hennelly SP and Sanbonmatsu KY (2013) Tackling Structures of Long Noncoding RNAs. Int J Mol Sci 14: 23672-23684.
- Luo DB, Lv HB, Sun XH, Wang Y, Chu JH, et al. (2020) LncRNA TRERNA1 promotes malignant progression of NSCLC through targeting FOXL1. Eur Rev Med Pharmaco 24: 1233-1242.
- Zhao L, Zhang X, Shi Y and Teng T (2020) LncRNA SNHG14 contributes to the progression of NSCLC through miR-206/G6PD pathway. Thorac Cancer 11: 1202-1210.
- 1Zhang G, Wang Q, Zhang X, Ding Z and Liu R (2019) LncRNA FENDRR suppresses the progression of NSCLC via regulating miR-761/TIMP2 axis. Biomed Pharmacother 118.
- Liu Q, Huang Y, Zhang R, Cai T and Cai Y (2016) Medical Application of Spirulina platensis Derived C-Phycocyanin. Evid Based Complementary Altern Med 2016.
- Li B, Gao M-H, Lv C-Y, Yang P and Yin Q-F (2016) Study of the synergistic effects of all-transretinoic acid and C-phycocyanin on the growth and apoptosis of A549 cells. Eur J Cancer Prev 25: 97-101.
- Li B, Gao M-H, Chu X-M, Teng L, Lv C-Y, et al. (2015) The synergistic antitumor effects of all-trans retinoic acid and C-phycocyanin on the lung cancer A549 cells in vitro and in vivo. Eur J Pharmacol 749: 107-114.
- Hao S, Yang Q, Li F, Li Q, Liu Y, et al. (2021) Dysregulated expression of miR-642a-5p and its target receptor-interacting serine/threonine-protein kinase 1 contribute to the phycocyanin-mediated inhibitory function on non-small cell lung cancer. J Funct Foods 85.
- Hao S, Li F, Li S, Li Q, Liu Y, et al. (2022) miR-3150a-3p, miR-6883-3p and miR-627-5p participate in the phycocyanin-mediated growth diminishment of A549 cells, via regulating a common target toll/interleukin 1 receptor domain-containing adaptor protein. J Funct Foods 91.
- Bridges MC, Daulagala AC and Kourtidis A (2021) LNCcation: lncRNA localization and function. J Cell Biol 220.
- Ren C, Deng M, Fan Y, Yang H, Zhang G, et al. (2017) Genome-Wide Analysis Reveals Extensive Changes in LncRNAs during Skeletal Muscle Development in Hu Sheep. Genes 8.
- Zhen Q, Gao L-n, Wang R-f, Chu W-w, Zhang Y-x, et al. (2018) LncRNA DANCR Promotes Lung Cancer by Sequestering miR-216a. Cancer Control 25.
- Khandelwal A, Bacolla A, Vasquez KM and Jain A (2015) long non-coding RNA: A new paradigm for lung cancer. Mol Carcinog 54: 1235-1251.
- Alzokaky AA-m, Abdelkader EM, El-Dessouki AM, Khaleel SA and Raslan NA (2020) C-phycocyanin protects against ethanol-induced gastric ulcers in rats: Role of HMGB1/NLRP3/NF-kappa B pathway. Basic Clinical Pharmacol Toxicol 127: 265-277.
- Hao S, Li S, Wang J, Zhao L, Yan Y, et al. (2018) Transcriptome Analysis of Phycocyanin-Mediated Inhibitory Functions on Non-Small Cell Lung Cancer A549 Cell Growth. Mar Drugs 16.
- Zhang X, Chen Z, Zang J, Yao C, Shi J, et al. (2021) LncRNA-mRNA co-expression analysis discovered the diagnostic and prognostic biomarkers and potential therapeutic agents for myocardial infarction. Aging-Us 13: 8944-8959.
- Cai G, Sun M, Li X and Zhu J (2021) Construction and characterization of rectal cancer-related lncRNA-mRNA ceRNA network reveals prognostic biomarkers in rectal cancer. IET Syst Biol 15: 192-204.
- Marcela Herrera-Solorio A, Peralta-Arrieta I, Armas Lopez L, Hernandez-Cigala N, Mendoza Milla C, et al. (2021) LncRNA SOX2-OT regulates AKT/ERK and SOX2/GLI-1 expression, hinders therapy, and worsens clinical prognosis in malignant lung diseases. Mol Oncol 15: 1110-1129.
- Loewen G, Jayawickramarajah J, Zhuo Y and Shan B (2014) Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol 7.
- Huang JW, Luo XY, Li ZH and Lang BP (2020) LncRNA NNT-AS1 regulates the progression of lung cancer through the NNT-AS1/miR-3666/E2F2 axis. Riv Eur Sci Med Farmacol 24: 238-248.
- Zhang YX, Yuan J, Gao ZM and Zhang ZG (2018) LncRNA TUC338 promotes invasion of lung cancer by activating MAPK pathway. Riv Eur Sci Med Farmacol 22: 443-449.
- Wan Y, Yao D, Fang F, Wang Y, Wu G, et al. (2021) LncRNA WT1-AS downregulates lncRNA UCA1 to suppress non-small cell lung cancer and predicts poor survival. Bmc Cancer 21.
- Yi S, Li G and Sun B (2021) Overexpression of LINC00852 promotes prostate cancer cell proliferation and metastasis. Asia-Pacific J Clin Oncol 17: 435-441.
- Tuo Z, Liang L and Zhou R (2021) LINC00852 is associated with poor prognosis in non-small cell lung cancer patients and its inhibition suppresses cancer cell proliferation and chemoresistance via the hsa-miR-145-5p/KLF4 axis. J Gene Med 23.
- Chen L, Zou W, Zhang L, Shi H, Li Z, et al. (2021) ceRNA network development and tumor-infiltrating immune cell analysis in hepatocellular carcinoma. Med Oncol 38.
- Wu X, Sui Z, Zhang H, Wang Y and Yu Z (2020) Integrated Analysis of lncRNA-Mediated ceRNA Network in Lung Adenocarcinoma. Front Oncol 10.
- Guo K, Qian K, Shi Y, Sun T and Wang Z (2021) LncRNA-MIAT promotes thyroid cancer progression and function as ceRNA to target EZH2 by sponging miR-150-5p. Cell Death Dis 12.
- Qi X, Zhang D-H, Wu N, Xiao J-H, Wang X, et al. (2015) ceRNA in cancer: possible functions and clinical implications. J Med Genet 52: 710-718.
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Citation: Hao S, Wu B, Cheng H, Li X, Zhang W, et al. (2024) LncRNA MicroarrayAnalysis of Non-small Cell Lung Cancer A549 Cell Lines Treated with Phycocyanin.Cell Mol Biol, 69: 302.
Copyright: © 2024 Hao S, et al. This is an open-access article distributed underthe terms of the Creative Commons Attribution License, which permits unrestricteduse, distribution, and reproduction in any medium, provided the original author andsource are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Usage
- Total views: 628
- [From(publication date): 0-2024 - Apr 27, 2025]
- Breakdown by view type
- HTML page views: 427
- PDF downloads: 201