Livestock Associated Methicillin Resistant Staphylococcus aureus Strain Infection: comprehensive Review
Received: 26-Aug-2022 / Manuscript No. jvmh-22-74030 / Editor assigned: 29-Aug-2022 / PreQC No. jvmh-22-74030 / Reviewed: 09-Sep-2022 / QC No. jvmh-22-74030 / Revised: 14-Sep-2022 / Manuscript No. jvmh-22-74030 / Accepted Date: 20-Sep-2022 / Published Date: 21-Sep-2022 QI No. / jvmh-22-74030
Abstract
Animal-related methicillin-resistant Staphylococcus aureus is a global threat to the health of livestock and humans and responsible for causing a variety of diseases ranging from superficial skin infections to life-threatening diseases.Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) has gained particular interest since the first findings of LA-MRSA of clonal complex CC398/sequence type (ST) 398 in pigs in 2005 in France and in the Netherlands. This paper emphasizes on livestock-associated methicillin-resistant Staphylococcus aureus infection in different animal species. Clonal complex CC398 was also identified in other animals (including dairy cattle, poultry, dogs,cats, and horses) and it became clear that LA-MRSA should be considered a zoonosis with people with occupational contact with livestock (farmers, veterinarians, and workers at abattoirs) are being most frequently exposed and often colonized. Clonal complex CC398 remains the most commonly identified type of LA-MRSA in most European countries.However, while MRSA CC398 strains have been found in livestock across the globe different strain of LA-MRSA, CC9,appears to be the prominent type in several Asian countries. A verities of secreted and cell surface-associated virulence factors can be promotes adhesion to the host extracellular matrix components, damage host cells, and fight the immune system. Genes encoding the different superantigens, adhesins, proteases, and capsule type are the virulence factors.Livestock-associated methicillin-resistant Staphylococcus aureus (LA- MRSA) has mecA and mecC gene found on a large mobile genetic element called the staphylococcal chromosomal cassette mec (SCCmec) which responsible for complete resistance to nearly all beta-lactam antibiotics including semi-synthetic penicillins such as methicillin, oxacillin,or cloxacillins.
Keywords
Clonal complex 398; Livestock; Livestock-associated methicillin-resistant Staphylococcus aureu; Resistance factor; Virulence factor
Introduction
Staphylococcus aureus is a gram-positive bacterium that colonizes a variety of animal species including humans and regarded as an opportunistic and commensal organism of animals, birds, and humans [1-5]. A wide range of infections can be caused by Staphylococcus aureus, from superficial skin and soft tissue infections to lifethreatening septicemia. Staphylococcus aureus causes a serious public health burden in both hospital and community settings, as well as an economic and animal welfare problem in dairy farming [6-8]. Antibiotic-resistant Staphylococcus aureus in animals are a growing concern because of their possibility transmission of these resistant pathogenic bacteria to humans as food borne pathogens. There is no doubt that the extensive usages of antimicrobials in the livestock industry were enhancing the ability of Staphylococcus aureus to acquire many resistance genes and thus allows the organism to be become more virulent. These resistance genes and the resistant bacteria could be transferred to humans through consumption of animal origin food and in sequence threaten the effectiveness of prevention and control strategies. Dissemination of antimicrobial resistance among Staphylococcus strains is probably due to variations in DNA sequences and horizontal transfer of resistance genes [1]. Methicillin-resistant Staphylococcus aureus (MRSA) strains have been increasingly reported as emerging pathogenic strains that cause great problems in veterinary medicine. The term “MRSA” is related to non-susceptibility to at least one of the antimicrobials in three or more categories and whose resistance to oxacillin or cefoxitin predicts non susceptibility to most categories of β-lactam antimicrobials [1]. About 96% of Staphylococcus aureus strains are resistant to at least one antibiotic, whereas 32% are resistant to multiple antimicrobial classes [8]. The World Health Organization (WHO) has classified MRSA strains as “high priority pathogens,” due to their great threat to the health of both humans and animals. The first identification of MRSA strains was in 1961 in England, after which they have emerged worldwide [3]. Since the 1990s, MRSA has been regarded as a nosocomial or community-associated pathogen and has become a significant public health and worldwide zoonotic problem [2]. Depending on the source of infection there are three types of MRSA categories described, namely hospital-acquired MRSA (HA-MRSA), community-acquired MRSA (CA-MRSA), and livestock-associated MRSA (LA-MRSA). Community-related MRSA infections (CA-MRSA) and hospital-related MRSA (HA-MRSA) have been reported in preliminary studies, whereas MRSA-related livestock (LA-MRSA) infections were first isolated from dairy cows. Lately, LAMRSA has become a major concern as pathogenic bacteria found in livestock, especially in dairy cows, which can transmit methicillin resistance to humans through meat or milk. Livestock-associated methicillin-resistant Staphylococcus aureus belongs to the 398 clonal complex (CC398), known as cattle-related MRSA (LA-MRSA). This type of MRSA isolate can also be found in other animals and can cause of infection in humans [9]. As compare to in developed country especially in Europe, North America and Asia, little is known about the prevalence of LA-MRSA in developing countries, especially in Africa countries only a few studies have been conducted on the epidemiology of livestock-associated methicillin resistant Staphylococcus aureus. Therefore, this review emphasizes livestock-associated Staphylococcus aureus with specially methicillin- resistant Staphylococcus aureus and defines livestock as equine, cattle, goats, pig and poultry which focused on the sources and transmission of livestock-associated methicillinresistant Staphylococcus aureus infection in there different species of livestock, virulence factors, antibiotic resistance, and prevention and control strategies for encouragement of researchers’ to study more about livestock-associated methicillin-resistant Staphylococcus aureus infection for a future.
Objectives of this review
To describe livestock-associated methicillin-resistant Staphylococcus aureus infection in different animal species.
Literature Review
General Features of Livestock-Associated Methicillin- Resistant Staphylococcus Aureus
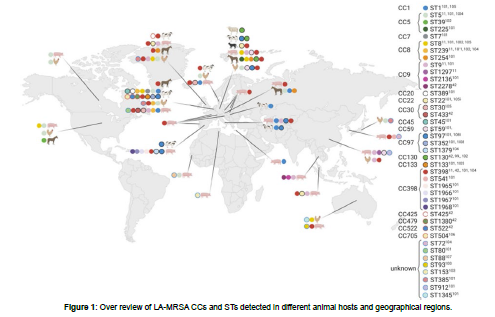
Staphylococcus aureus consists of several lineages, each lineage associated with a specific host group (for example, cattle, poultry, cats, humans, etc.) [2]. Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) has gained particular interest since the first findings of LA-MRSA of clonal complex CC398/sequence type (ST) 398 in pigs in 2005 in France [4] and in the Netherlands [9-15]. This Clonal complex CC398 was also identified in other animals (including dairy cattle, poultry, dogs, cats, and horses) and it became clear that LAMRSA should be considered a zoonosis with people with occupational contact with livestock (farmers, veterinarians, and workers at abattoirs) are being most frequently exposed and often colonized. The LA-MRSA CC398 is also able to cause the same kind of infections in humans as Staphylococcus aureus. In general MRSA cause severe infections in people, this indicates that the animal reservoir of Staphylococcus aureus can have serious consequences for human health [9]. Formerly LA-MRSA, known as nontypeable (NT) MRSA, this non-typeability of the strain isolates was limited to ST398 based on standard pulsed field gel electrophoresis (PFGE) and using of restriction enzyme SmaI [5] and Since then, much evolution has occurred, and more sequence types are included now [2]. Although CC398 is still the most common LA-MRSA worldwide, its prevalence differs geographically; in certain regions, other sequence types are also involved, such as ST9 in Asia and the diversity among sequence types is also greater in some areas than in others. Animal-related methicillin-resistant Staphylococcus aureus (LA-MRSA) is a global threat to the health of livestock and humans. This bacterial strain is responsible for cuasing a variety of diseases ranging from superficial skin infections to life-threatening diseases. Livestock-associated methicillin-resistant Staphylococcus aureus evolved independently from common HA-MRSA or CA- MRSA usually found in humans and mainly belong to Staphylococcus aureus clonal complex CC398 and associated spa types t011, and t034 [7]. Clonal complex 398 shows a broader host range compared to other livestock-associated methicillin-resistant staphylococcus aureus strains and has been detected in cattle, veal calves, poultry, companion animals (dogs and cats), horses, and in humans. However, also other CCs such as CC1, CC97, CC130, and CC5 are found in livestock around the globe [12]. Clonal complex CC398 remains the most commonly identified type of LA-MRSA in most European countries. However, while MRSA CC398 strains have been found in livestock across the globe [8], a different strain of LA-MRSA, CC9, appears to be the prominent type in several Asian countries [15]. The diversity of LA-MRSA appears to be higher in the USA, than in Europe and Asia, with reports of both CC398 and a variety of “human” types of Staphylococcus aureus in livestock [8]. For an overview of the different CCs and STs of LA-MRSA detected in different animal hosts and geographical regions are shown in Figure 1 below. The clonal complex CC398 MRSA usually carries resistance genes to a variety of antimicrobial classes such as tetracycline, macrolides, or aminoglycosides and tends to exhibit increased levels of multiple drug resistances compared with non-CC398 MRSA strains.
The methicillin-resistant gene mecA is found in LA-MRSA strains, but mecC, a gene variant sharing 70% identity at the DNA level with mecA, has been detected in ruminants, pigs, and companion animals, with increasing reports from wild animals [13]. Livestock-associated methicillin-resistant staphylococcus aureus ST398 isolates usually also possess a bunch of different virulence-associated factors such as hemolysins and immune-modulatory factors but commonly lack Panton-Valentine leukocidin (PVL) or staphylococcal enterotoxins (SE), of which a significantly located on mobile genetic elements. In contrast, strains of CC9 commonly harbor enterotoxin genes and tst-1 encoding the toxic shock syndrome toxin [15].
Evolution of MRSA
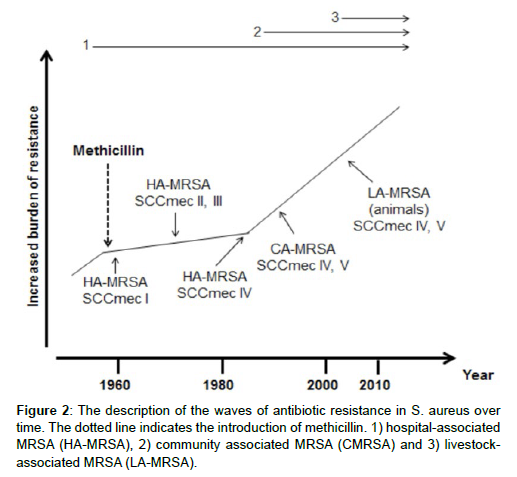
The evolution methicillin-resistant Staphylococcus aureus can be divided into different waves, which correspond to certain time periods. After the first use of methicillin in the clinical environment, MRSA strains were found in the hospitals (end 1950 – 1970) where highly successful lineages emerged around the 1970s until the mid-1980s. Early 1990s, the first MRSA strains emerged in the community independent of the hospital environments. Around 2005, a distinct clone of MRSA, MRSA ST398 or livestock-associated MRSA, was described in livestock animals for the first time. Nowadays, MRSA ST398 can also be found in humans and their environment independent of livestock animals [13].
Molecular characterization of livestock associated methicillin resistant stphylococcus aureus
At present, livestock -associated MRSA is found in different animal species and especially livestock animals, resulting in the name change to livestock-associated MRSA (LA-MRSA) or MRSA ST398. Livestock -associated MRSA has some specific features that distinguish this type from the other two MRSA types. First, LA-MRSA is non-typeable with Pulsed Field Gel Electrophoresis (PFGE) using SmaI restriction. Second, antibiotic resistance for antibiotics used in livestock farming has been observed. Third, in general, LA-MRSA does not carry human virulence genes. Last, more variation was observed in the SCCmec carriage: MRSA ST398 mainly carries SCCmec type IVa and V although at present others (e.g. SCCmec type III, V (5C2&5), IX and X) have been described [14, 7]. In general, LA-MRSA is PVL negative, but PVL positive strains have been reported [11]. Livestock -associated MRSA has the ability to acquire additional antibiotic resistance genes (such as dfr (trimethoprim) and erm (macrolides)) and mobile elements with for example human virulence genes [7, 6].
Isolation and characterization of MRSA
Various matrices can be sampled such as humans, animals, environment, air, carcasses and meat samples. In humans, three locations are sampled for clinical screenings: nose, throat and perineum [15]. In most pig studies, nasal swabs are taken. Locations such as on the skin behind the ear, the perineum, organs and faeces were also sampled. Pletinckx and colleagues [4]. Reported that the best sampling location for retrieving MRSA was the skin behind the ears. However, the authors suggested that environmental contamination and not colonization of this site should be considered. For environmental sampling, dust swabs, environmental wipes or sponges were described. Moreover, manure, water and feed samples were also taken. Three sampling methodologies exist for air sampling: impingement, filtration or open plates. For carcasses, the best sampling method is usage of an abrasive sponge compared to swabbing and cutting off samples. No study reported on the best sampling location for MRSA ST398 on a carcass. Different sampling methodologies for meat samples are available. In general, “hard” meat samples (such as ribs, ears, forelimbs) are swabbed or soaked into broth and manually mixed. Ten to twenty-five grams of “soft” meat samples (such as steak, bacon and minced meat) are homogenized into broth. It has been reported that the use of enrichment improved the MRSA isolation in clinical studies. Various studies investigated the effect of enrichment on the isolation of MRSA ST398. In general, addition of salt (6.5-7.5%) to the enrichment medium increased the MRSA ST398 detection. Staphylococci are salt-tolerant and salt inhibits the contaminating or commensal flora in the samples. In Dutch studies, after the salt enrichment step, an antibiotic enrichment step followed. Similar percentages of MRSA positive samples were observed when comparing the Belgian study (enrichment in salt) and the Dutch studies (enrichment in salt and afterwards enrichment in antibiotics). For environmental samples, the same salt-enrichment is used. Few studies have been performed on pig carcasses and in most studies; the same enrichment as described above was used. For meat samples, enrichment also occurs in salt-enriched and/or antibiotic-enriched medium. After enrichment, the samples are inoculated onto selective media. At first, agars supplemented with antibiotics were used. Throughout the years, specific chromogenic media for MRSA have been developed, such as chromIDTM MRSA agar (BioMerieux), Brilliance MRSA agar (Oxoid) and others on which MRSA colonies have a specific color. Good performances for MRSA ST398 in human and pig nasal swabs have been reported for chromIDTM MRSA, MRSASelectTM , Brilliance MRSA agar and MRSA screen, but not for Oxacillin-screening agar.
Confirmation of MRSA ST398
Once suspect colonies are obtained, biochemical tests or Polymerase Chain Reaction (PCR) protocols can be used for the confirmation of MRSA. At present, a lot of (real-time) PCRs exist for the confirmation of MRSA. However, only three MRSA ST398 specific PCRs have been described: one based on the sau1hsd1 gene (encodes a restriction modification system) and mecA gene, one based on four ST398 specific genes and another real-time PCR based on a 124bp ST398 specific sequence.
Multilocus Sequence Typing
During Multilocus Sequence Typing (MLST), single nucleotide variations in internal fragments of seven housekeeping genes are detected. Housekeeping genes belong to the core genes of the genome and are essential for the survival of a bacterium. Changes in these genes occur slower and equal to long periods of evolution. For S. aureus, the seven genes are: Carbamate kinase (arcC), Shikimate dehydrogenase (aroE), Glycerol kinase (glpF), Guanylate kinase (gmk), Phosphate acetyltransferase (pta), Triosephosphate isomerase (tpi) and Acetyl coenzyme A acetyltransferase (yqiL). MLST is a PCR based method during which the internal fragments of the genes (500bp) are amplified and subsequently sequenced. Each allele receives a number and a numeric code is obtained per strain. Based on this numeric code, each strain can be classified into a sequence type (ST). All MRSA ST398 strains have 3-35-19-2-20-26-39 as numeric code. At present, a database is available with all obtained alleles for each housekeeping gene, which makes comparisons much easier. Closely related STs (one allele difference = single locus variant or two alleles difference = double locus variants) can be classified in a clonal complex (CC). The CC receives the name of the most ancestral sequence type. For example, CC398 consists of sequence types ST398, ST541, ST572, ST753, ST1965, ST1966, ST1967, ST1968, ST1969 and others of which ST398 is considered as the ancestral type. MLST is a standardized, objective and highly reproducible method. Moreover, a standard nomenclature of CCs and STs is used and comparison between laboratories is possible.
It is a good technique for population studies (bacterial population structure and evolution) However, MLST encounters a high cost and a low throughput, is time consuming and has only a moderate discriminatory power.
Spa typing
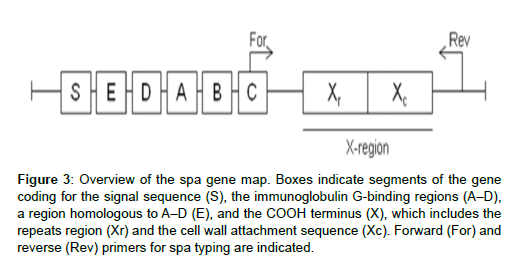
Another PCR-based and sequence based method is spa typing . This technique involves amplification and sequence analysis of the repeats (24 bp in length) in the X region of the protein a gene (Figure 2).
Multilocus variable-number tandem-repeat analysis
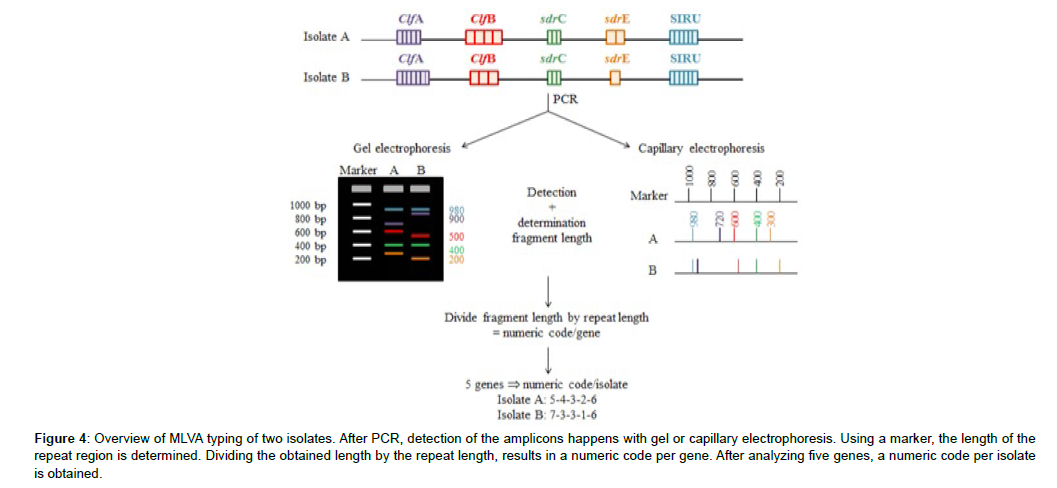
A third PCR-based method is Multilocus Variable-number tandem-repeat Analysis (MLVA). MLVA detects the number of repeats in the repeat region of different selected genes. After amplification, two detection methods are available: gel electrophoresis and capillary electrophoresis. Subsequent to detection, the length of the repeat region is determined. This length is divided by the repeat length. In such way, the total number of the repeats of which a repeat region consists, is determined. Based on the number of repeats, a number is assigned to each repeat region and as a result a digit code per strain is obtained (Figure 3).
Figure 3: Overview of the spa gene map. Boxes indicate segments of the gene coding for the signal sequence (S), the immunoglobulin G-binding regions (A–D), a region homologous to A–D (E), and the COOH terminus (X), which includes the repeats region (Xr) and the cell wall attachment sequence (Xc). Forward (For) and reverse (Rev) primers for spa typing are indicated.
Figure 4: Overview of MLVA typing of two isolates. After PCR, detection of the amplicons happens with gel or capillary electrophoresis. Using a marker, the length of the repeat region is determined. Dividing the obtained length by the repeat length, results in a numeric code per gene. After analyzing five genes, a numeric code per isolate is obtained.
SCCmec typing
As mentioned before, acquisition of the SCCmec cassette results in methicillin resistance. Staphylococcal Chromosomal Cassette mec consists of a mecA gene complex, a ccr gene complex and three joining regions, which can be used to determine the SCCmec cassette type, present in an MRSA isolate. Due to the variation in SCCmec types, different PCR-based methods have been described throughout the years. However, seen the diversity and subtypes, none of the described methods is able to identify all cassettes and non-typeable cassettes have often been reported. In general, two approaches for SCCmec typing have been used. At first, the methods detected loci, specific for SCCmec types I, II, III and IV. More recently, the ccr (1 to 5) and mec (A, B, C2) complexes are identified. Afterwards, the results are combined to determine the SCCmec type. In addition, detection of loci in junkyard regions can help to determine the subtype. An international working group provided a standard nomenclature for the SCCmec types (www.sccmec.org). However, no standardized protocol was proposed and available for SCCmec typing. The use of diverse PCRs results in a relatively high cost and low throughput of samples. Moreover, this method is not as discriminatory as the others.
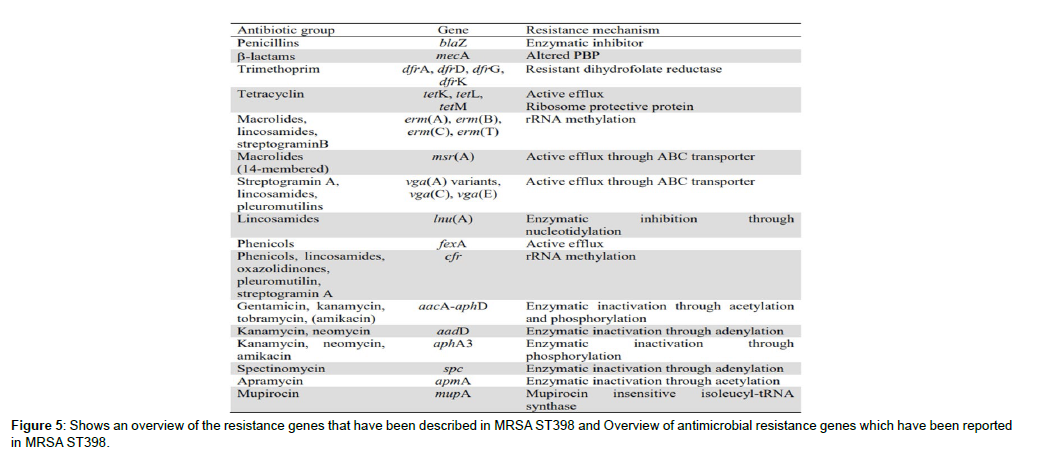
Pulsed Field Gel ElectrophoresisPulsed Field Gel Electrophoresis (PFGE) is not a PCR based method. During PFGE, bacterial cells are immobilized after embedding in agarose and after cell lysis, the whole genome becomes available. Restriction of the whole genome occurs with a “rare cutter” endonuclease, which had only few cutting sites in the genome. Different enzymes are available and each enzyme has a specific cutting site, such as CCCGGG for the enzyme SmaI. As a result, restriction generates large DNA fragments between 10 and 1000 Kb. Detection of these large fragments occurs in an electric field that is periodically changed, allowing efficient separation of the DNA fragments. In general, the obtained patterns are (Figure 5) analyzed using an Unweighted Pair Group Method using Averages (UPGMA) clustering algorithm. At present, international PFGE databases and a standardized protocol (HARMONY) are available where profiles can be submitted and compared. PFGE with SmaI restriction has long been considered as the gold standard for S. aureus and MRSA. But, MRSA ST398 appeared nontypeable using this method. PFGE is able to detect more recent evolution in strains and genetic changes can lead to differences in fragments. However, the detected differences remain uncharacterized. PFGE is highly discriminatory, commonly used, reproducible and standardized. Nevertheless, as the analysis of the patterns is still subjective, variation in the results might occur. In addition, PFGE is a technically demanding and time consuming technique.
Antimicrobial susceptibility typing
Phenotypic analysis
To gain more insights into the characteristics of bacterial strains, their resistance to antimicrobial agents may be determined. At present, two standardized methods are available: disk diffusion tests and Minimal Inhibitory Concentration (MIC) determination. When performing disk diffusion, a bacterial suspension is applied onto a universal growth medium (for example Mueller Hinton Agar) and disks containing a fixed concentration of an antimicrobial agent are applied. After incubation, the halo around the disk is measured and according to interpretation tables, a strain is considered sensitive, intermediate or resistant. When determining the MIC, a bacterium is exposed to different, mostly clinical relevant, concentrations of antimicrobial agent. The lowest concentration that inhibits the bacterium is considered the MIC. For the detection of MRSA, the antibiotics oxacillin and cefoxitin are often use. In MRSA ST398 strains, acquired resistance to tetracyclin, trimethoprim, macrolides, lincosamides, aminoglycosides, chloramphenicol and fluoroquinolones was observed. A large variety in antibiotypes has been reported. After antimicrobial susceptibility testing, an adjusted treatment for bacterial infections can be used. In addition, the evolution of antimicrobial resistance in strains can be studied. Still, the method is time-consuming but at present, automated systems such as Vitek and DiversiLab® are available.
Genotypic analysis
When a bacterium becomes resistant to antimicrobial agents, this is due to mutations in the bacterial genome or the acquisition of resistance genes. To date, different molecular techniques have been described to determine these genes. However, presence of a resistance gene does not necessarily mean that the bacterium is resistant to that antibiotic.
DNA microarray analysis
DNA microarray analysis consists of different steps. First, DNA is isolated, amplified and labeled (for example by incorporation of biotin- 16-dUTP). Subsequently, this labeled DNA is hybridized with a chip on which target genes or probes are fixed. When a certain gene is located on the strain DNA, hybridization occurs and a signal is observed. Possible target genes are antimicrobial resistance genes, entero- and exotoxin genes and others. To date, this method was used to compare MRSA ST398 strains originating from pigs and humans and pigs alone. The rapid microarray technology has the potential to detect an almost unlimited number of genes within a single reaction. Moreover, commercial and automated platforms are available. However, this technology is expensive and requires specific equipment.
Whole genome sequencing
A new approach in many studies is whole genome sequencing (WGS). In the 1970s, the first method for genome sequencing has been developed. Throughout the years, better DNA preparation protocols and automated platforms have been developed which increased the use of WGS. To date, the genome of 49 MRSA CC398 strains has been sequenced. WGS is the ultimate tool to discriminate between closely related strains and eases high resolution phylogenetic reconstruction. Nevertheless, equipment for WGS is expensive and WGS is considered impractical for routine.
Molecular epidemiology
Upon the first description of MRSA ST398, methods such as MLST and PFGE with SmaI restriction were used to characterize this new MRSA type. Later, other methods such as spa or SCCmec typing were also used for identification. Epidemiology is the study of determinants of health, disease, and productivity in populations of humans, plants or animals. Molecular epidemiology involves the use of molecular methods to study possible transmission routes of a bacterium, relationships between isolates and others. The definition of molecular epidemiology does not include identification of isolates. When studying the molecular epidemiology of isolates, a combination of two or more typing methods can be useful. It has been reported that combining methods increases the discriminatory power. For example, the discriminatory power, reported by Rasschaert and colleagues was 0.74 for spa typing and 0.81 for MLVA typing. Combining spa and MLVA typing resulted in a discriminatory power of 0.87.To date, in many studies, molecular tools were used for MRSA ST398 identification (MLST, spa typing, PFGE). However, few studies on the molecular epidemiology of MRSA ST398 in pigs have been performed. The early studies on MRSA ST398 involved molecular typing of limited numbers of isolates and in most cases the use of MLST, spa typing and PFGE with SmaI restriction. When it was reported that other restriction enzymes were able to generate a fingerprint in PFGE, some research groups used these other enzymes. Besides MLST, spa typing and PFGE, methods such as SCCmec typing, determination of antibiotic resistance or virulence genes, micro-array or whole genome sequencing have been used for molecular epidemiology. MLVA typing has scarcely been used in the molecular epidemiology of MRSA ST398.
Sources and Transmission of Livestock Associated Methicillin-Resistant Staphylococcus Aureus Infection
Livestock associated methicillin-resistant Staphylococcus aureus has a characteristics distributed rapidly among animals, humans, and the environment, through direct contact between animals and humans and spread through water, air, food, and contaminated surfaces. Livestockassociated methicillin-resistant Staphylococcus aureus poses a zoonotic risk to humans, especially those workers close-contact with animals were more exposed group. The horizontal route of transmission among animals or humans may occur. Livestock associated methicillinresistant Staphylococcus aureus transmission between humans, an animal production and companion animal is common and also it can be transmitted from vertebrate animals to humans. Staphylococcus aureus infections can be present in both humans and animals and transmitted in both directions, such infections are defined as “amphixenoses”. Although most of the isolates having been similar to common bovine strains, a human-to-animal transmission of a new MRSA strain acquired by the farmer seems more likely due to the observed antimicrobial characteristics. Since then, MRSA colonization has been reported in dogs, cats, horses, cattle, pigs, rabbits and poultry. It has even been described in wild birds such as magpies and vultures. The first animalto- human transmission of Staphylococcus aureus was reported in dairy sheep. Shortly after, the first case of MRSA transmission from animal to human, including human to-human transmission of the same strain was reported.
Methicillin-resistant Staphylococcus aureus in Different Livestock Animals
Methicillin-resistant Staphylococcus aureus in Dairy Cattle
Methicillin-resistant Staphylococcus aureus has become a major concern in society because these pathogenic bacteria are often found in farm animals and can infect humans. In livestock MRSA were first reported in the early 1970s, when the bacterium Staphylococcus aureus was isolated from milk of mastitis cows in Belgium and categorized in the CC398 group. Community-related MRSA infections (CAMRSA) and hospital-related MRSA (HA-MRSA) have been reported in preliminary studies, whereas MRSA-related livestock (LA-MRSA) infections were first isolated from dairy cows. Lately, LA-MRSA has become a major concern as pathogenic bacteria found in livestock, especially in dairy cows, which can transmit methicillin resistance to humans through meat or milk .
Livestock-associated methicillin-resistant Staphylococcus aureus can colonize the udder and cause intramammary infection in dairy cattle, sometimes leading to clinical mastitis and relevant economic losses. However, the epidemiology of MRSA in dairy cattle is yet to be fully studied and the rate of the infection is not clearly understood. A high variability of inter-herd and intra-herd prevalence has been reported so far. Moreover, on the base of the genetic relatedness observed among isolates detected from different sources, different patterns of transmission between and within investigated farms have been described. Like in pigs, clonal complex 398 LA-MRSA is the predominant MRSA type in dairy herds in Europe. Also, in studies from Brazil, China, and Israel, LA-MRSA CC398 was found in mastitis milk samples. Very recently, LA-MRSA exclusively belonging to CC398 was found on 20 German dairy farms. In contrast, ST9 MRSA was found in milk samples in Southeast Asia. In most studies, transmissions from cow to cow were indicates that as there is usually a predominant MRSA strain within herds. However, the environment may also act as a reservoir of MRSA strains found in dairy herds, too.
2.7.1. Methicillin-resistant Staphylococcus aureus in Chickens and other Poultry
In poultry, MRSA strains were isolated for the 1st time from arthritis cases in South Korea; then, different strains have been isolated from live birds and retail poultry products. The presence of MRSA in live poultry has been reported and has highlighted the increase in the antibiotic resistance rate, especially in recently isolated strains.
Although the prevalence of MRSA in poultry is lower compared to the prevalence in pigs and cattle, MRSA have been detected along the whole poultry production chain from farm to fork. The prevalence rates of MRSA in poultry and poultry meat varied between geographical regions, with highest MRSA prevalence being observed in South America (27%) and lowest MRSA prevalence being observed in North America (1%). In poultry various clonal lineages MRSA was isolated. In Europe, the main lineage detected in broiler chicken and chicken retail meat products was CC398, but CC5, CC8, CC9, and CC80 were also isolated from poultry. A Canadian study investigating LA-MRSA isolates from chicken meat and broiler flock samples (2013-2014) reported a prevalence of 1.3% in chicken meat samples and no positive samples from broiler chickens and all isolates were assigned to ST398 and ST8.
Methicillin-resistant Staphylococcus aureus in Pigs
Livestock associated Methicillin-resistant Staphylococcus aureus is frequently detected along the entire pork production chain with predominant lineages changing over time and differing between geographical regions. Livestock associated methicillin-resistant Staphylococcus aureus CC398 first detected in pig was in 2005 in France and in the Netherlands. Livestock associated methicillin-resistant Staphylococcus aureus CC398 has since become widespread across pig breeding and production farms all over Europe, with prevalence rates varying substantially among different European countries. The movements Pig between farms are considered an important means spreading of LA-MRSA CC398. A recent study in Danish showed that the prevalence in herds raised conventionally (89%) substantially exceeded the prevalence in free range herds (20%). Similar to Europe, the prevalence of LA-MRSA in pigs in Asian countries varies widely among different geographical regions. Different studies report a prevalence of 1% in Japan and Malaysia, 3% in South Korea, 11% in China, 10-40% in Thailand, 16-39% in Hong Kong, and 4- 43% in Taiwan.
Methicillin resistant Staphylococcus aureus in Other Animals
Livestock associated methicillin-resistant Staphylococcus aureus has been detected not only in livestock, but also found in other companion animal’s cats, dogs, mice, and rat. Livestock associated methicillinresistant Staphylococcus aureus, mainly CC398, plays an important role, Particularly in horses, CC398 MRSA are massively prevalent worldwide and represent nearly 90% of the MRSA isolated from equine wound infections in Germany. Methicillin resistant Staphylococcus aureus belonging to livestock-associated clonal lineages were exclusively found in companion animals in regions with high livestock density. (Kaspar and colleagues further concluded that the presence of LA-MRSA among pets and probable dissemination in clinical settings supports the necessity of a One Health approach to address the potential threats due to MDRO-carrying companion animals.
Virulence and Resistance Factors
Virulence Factors of livestock associated methicillinresistant Staphylococcus aureus
Successful infection produced by Staphylococcus aureus both humans and animals depend on virulence factors of the organism. A verities of secreted and cell surface-associated virulence factors can be promotes adhesion to the host extracellular matrix components, damage host cells, and fight the immune system. Genes encoding the different superantigens, adhesins, proteases, and capsule type are the virulence factors. For the adhesins, the genes for fibronectin-binding protein A, elastin-binding protein and extracellular fibrinogen-binding protein were almost always present. The whole genome sequence of an MRSA ST398 isolate obtained from a human contain a novel staphylococcal pathogenicity island (SaPI) that appeared to be a composite of sequences from SaPibov, SaPI5, which is found in strain USA300 and the novel part encoded a staphylococcal complement inhibitor variant and a variant of von Willebrand factor-binding protein. The ability of acquire foreign DNA may be one of most dangerous features of MRSA CC398, this important for capable of acquiring virulence genes, and its acquisition of the Panton– Valentine leukocidin (PVL) gene. Regarding other virulence factors, staphylococcal enterotoxins occasionally have been reported in LA-MRSA CC398 in pigs and turkeys. In contrast, genes encoding adhesion factors, proteases, hemolysins, other leukocidins, and superantigen-like proteins have been detected frequently in LAMRSA CC398 strain isolates. Formations of biofilms were an additional level of complexity for many infections. Biofilms consists a combinations of bacteria embedded in polysaccharides, proteins, extracellular DNA. Biofilm formation is a potent immune evasion strategy, both through physical blocking of access and through active immune evasion because of secreted immune evasion components. Formation biofilm in bacteria also play an important role in resistance to antimicrobial agents and these helps to maintain chronic infections. Several livestock methicillinresistant Staphylococcus aureus contain the ica (intercellular adhesion) genes, which encode products involved in biofilm formation. Free DNA also helps to build biofilms in combination with b-toxin encoded by the hlb (β-Hemolysin) gene, which acts not only as a toxin, but also as a DNA binding protein in biofilm formation.
Resistance factors for livestock associated methicillinresistant Staphylococcus aureus
Majority of MRSA strain carry the mecA gene encode penicillin binding protein 2a (PBP2a), which has a low tropism to all β-lactam antibiotics, is the corner stone responsible for producing MRSA phenomenon. This mecA gene responsible for complete resistance to nearly all beta-lactam antibiotics including semi-synthetic penicillins such as methicillin, oxacillin, or cloxacillin. The mecA gene is found on the MRSA chromosome (SCCmec) have seven types of SCCmec (I–VII) of SCCmec were identified up to date. Methicillin resistance (MecC) gene is another a beta lactam resistance gene that was first recognized in 2011, and is less well understood than mecA. Like mecA, mecC is located on SCCmec and it codes for a different version of PBP2a, which is also interfere with the effects of beta-lactam antibiotics on cell walls. However, a recent paper suggests that mecC-encoded PBP2a may mediate resistance to some beta-lactam drugs, but not others. This could raise the possibility of treatment with some drugs that are ineffective against mecA-bearing MRSA. Many mecC-bearing organisms seem to belong to lineages of staphylococci associated with animals. Some of these lineages appear to have a wide host range.
Public Health Importance of Livestock Associated Methicillin-Resistant Staphylococcus Aureus
Livestock associated Methicillin-resistant Staphylococcus aureus can occur in people who have direct contact with livestock like farmers, veterinarians, or slaughterhouse employees. In the livestock production system, slaughterhouse staff in contact with live animals exhibited particularly high prevalence rates of MRSA carriage.
In general prevalence of MRSA human population has been reported to be in the range of 0.8– 1.3%. But in case of veterinary personnel, the prevalence reported ranged between 0 and 50% with an overall mean prevalence of 8% across studies. Most of the studies conducted in Europe clonal complex CC398 strain were the most commonly reported dominant LA-MRSA strain as zoonotic implication. In Asia some other strains such as CC5 (usually spa type t002) were reported as more dominant. Although CC398 being were considered the classical LA-MRSA zoonotic strain, it originated from humans methicillinsusceptible S. aureus . In a study executed in Spain in 2016, 58% of pig farm workers were carriers of LA-MRSA CC398, compared to an MRSA prevalence rate of <0.5 % in the general population. Another recent systematic review also stresses the role of occupational livestock contact and in particular pig contact enhancing the risk of LA-MRSA colonization. A study repeatedly screening Dutch veterinarians for LA-MRSA colonization over a period of 2 years found that 44% of veterinarians were LA-MRSA carriers at one or more measurement time points, with 13% being persistently colonized with the same strain. Livestock-associated methicillin-resistant staphylococcus aureus strains represent a small proportion (3.9%) of the isolated MRSA in humans in the European Union, but their proportion was higher (≥10%) in five countries (Belgium, Denmark, Netherlands, Slovenia and Spain). Several studies have concluded that humans in close contact with animals have greater risk of being colonized by LA-MRSA than the rest of the population.
Prevention and Control Strategies
A modern farming system characterized by densely populated livestock, intensive livestock farming, and excessive use of antibiotics can encourage the emergence of new LA-MRSA cases in the future. To avoid transmission from animal to human and animal to animal adjusting the distance between farm animals, isolation of diseased animals in separate area, brushing dung, and bathing dairy cows regularly will be an effective method to reduce transmission LAMRSA transmission. Improvement of biosecurity and environmental cleanliness of dairy cattle pens, as well as regular animal and human health care arrangements are important to prevent the spread of LAMRSA strains.
Prevention and control of MRSA are based on the application of hygienic measures in farms as well as proper processing, handling, and cooking of retail animal origin food products. To avoid the possibility of zoonotic transmission, cooperation between veterinary and human practitioners is very important. Regular monitoring of MRSA strains from livestock is necessary to understand the changes in the genetic selection and zoonotic potential of these resistant strains. To prevent MRSA infections, awareness, public health education, and good hygiene are critical, especially in veterinary medicine. Sanitary education of food handlers has great value to decrease their potential role as reservoirs and shedders of MRSA. In addition, good hygienic practice should be taken to ensure the safety of food products, and a proper risk assessment should be applied for further clarification of the possible health hazards for consumers. Recommended the proper application of good manufacturing and hygiene practices and well-designed hazard analysis of a critical control point program in slaughterhouses and processing units to avoid contamination of meat with MRSA.
Conclusion and Recommendations
Although little known about the prevalence of LA-MRSA in developing countries, especially in Africa countries, several studies have been performed in developed countries especially in European countries. Methicillin-resistant Staphylococcus aureus (MRSA) strains have been increasingly reported as emerging pathogenic strains that cause great problems in veterinary medicine. Depending on the source of infection there are three types of MRSA categories described, namely hospital-acquired MRSA (HA-MRSA), community-acquired MRSA (CA-MRSA), and livestock-associated MRSA (LA-MRSA). Clonal complex CC398 remains the most commonly identified type of LAMRSA in most European countries. However, while MRSA CC398 strains have been found in livestock across the globe different strain of LA-MRSA, CC9, appears to be the prominent type in several Asian countries. The diversity of LA-MRSA appears to be higher in the USA, than in Europe and Asia, with reports of both CC398 and a variety of “human” types of Staphylococcus aureus in livestock. Majority of MRSA strain carry the mecA gene encode penicillin binding protein 2a (PBP2a), which has a low tropism to all β-lactam antibiotics, is the corner stone responsible for producing MRSA phenomenon, This mecA gene found on a large mobile genetic element called the staphylococcal chromosomal cassette mec (SCCmec) It responsible for complete resistance to nearly all beta-lactam antibiotics including semi-synthetic penicillins such as methicillin, oxacillin, or cloxacillin. MecC is another a beta lactam resistance gene that was first recognized in 2011, and is less well understood than mecA. Like mecA, mecC is located on SCCmec and it codes for a different version of PBP2a, which is also interfere with the effects of beta-lactam antibiotics on cell walls. Livestock associated methicillin-resistant Staphylococcus aureus can occur in people who have direct contact with livestock like farmers, veterinarians, or slaughterhouse employees. In the livestock production system, slaughterhouse staff in contact with live animals exhibited particularly high prevalence rates of MRSA carriage. To prevent MRSA infections, awareness, public health education, and good hygiene are critical, especially in veterinary medicine. Sanitary education of food handlers has great value to decrease their potential role as reservoirs and shedders of MRSA. In addition, good hygienic practice should be taken to ensure the safety of food products, and a proper risk assessment should be applied for further clarification of the possible health hazards for consumers. Based on the above conclusion the following recommendations are forwarded:
• Veterinarian Researcher should be study more about the occurrence and economic impact of livestock associated methicillin resistance staphylococcus areus in livestock production system.
• Integrated approach between human health provider with veterinarian should applied for better prevention and control of livestock associated methicillin resistance staphylococcus
• The animal owners and animal health service providers should give more emphasis on intensive livestock production system
• Animal health professionals should create public awareness with regarding about public health concerns and participate in disease control and prevention practices.
References
- Abd El-Ghany WA (2021) Staphylococcus aureus in poultry, with special emphasis on methicillin-resistant strain infection: A comprehensive review from one health perspective. Int J One Health 7: 257-267.
- Algammal AM, Hetta HF, Elkelish A, Alkhalifah DH, Hozzein WN, et al. (2021) Methicillin-Resistant Staphylococcus aureus (MRSA): One Health Perspective Approach to the Bacterium Epidemiology Virulence Factors Antibiotic-Resistance and Zoonotic Impact. Infection and Drug Resistance13: 3255-3265.
- Argudin MA, Fetsch A, Tenhagen BA, Hammerl JA, Hertwig S, et al. (2010) High heterogeneity within methicillin-resistant Staphylococcus aureus ST398 isolates, defined by Cfr9I Macrorestriction-Pulsed-Field Gel Electrophoresis profiles and spa and SCCmec types. Applied and Environmental Microbiology 76: 652-658.
- Armand-Lefevre L, Ruimy R, Andremont A (2005) Clonal comparison of Staphylococcus aureus isolates from healthy pig farmers, human controls and pigs. Emerg Infect Dis 11: 4-2.
- Basset P, Feil EJ, Zanetti G, Blanc DS (2011) The evolution and dynamics of methicillin-resistant Staphylococcus aureus. In: Tibayrenc M editor Genetics and Evolution of Infectious Diseases. Elsevier, London 669-676.
- Becker A, Forster DH, Kniehl E (2002) Oxacillin resistance screening agar base for detection of methicillin-resistant Staphylococcus aureus. Journal of Clinical Microbiology 40: 4400-4401.
- Bernier-Lachance J, Arsenault J, Usongo V, Parent É, Labrie J, et al. (2020) Prevalence and characteristics of livestock associated methicillin-resistant Staphylococcus aureus (LAMRSA) isolated from chicken meat in the province of Quebec Canada. PLoS One 15: 0227183.
- Romazanvoc F (2001) Cestode zoonosis: Echinococcosis and Cysticercosis: An emergent and Global problem. 34-57.
- Sazmand A, Joachim A (2017) Parasitic diseases of camels in Iran (1931 – 2017): A literature review. Parasite 24:21.
- Singh BB, Sharma R, Sharma JK, Juyal PD (2010) Parasitic zoonoses in India: An overview. Rev Sci Tech Off Int Epiz 29: 629–637.
- Ahmadi NA, Meshkehkar M (2011) An abattoir-based study on the prevalence and economic losses due to cystic echinococcosis in slaughtered herbivores in Ahwaz, south-western Iran. J Helminthol 85: 33-39.
- Jenkinsa DJ, Romig T, Thompson RCA (2005) Emergence or re-emergence of Echinococcus spps. A global update. Int J parasitol 35: 1205- 1219.
- Santivanez S, Garcia HH (2010) Pulmonary cystic echinococcosis. Curr Opin Pulm Med 16: 257-261.
- Bosch T, de Neeling AJ, Schouls LM, van der Zwaluw KW, Kluytmans JAJW, et al. (2010) PFGE diversity within the methicillin-resistant Staphylococcus aureus clonal lineage ST398. BMC Microbiology 10: 1-7.
- Boye K, Westh H (2011) Variations in spa types found in consecutive MRSA isolates from the same patients. Fems Microbiology Letters 314: 101-105.
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Citation: Debeb D, Tsehayneh B (2022) Livestock Associated Methicillin Resistant Staphylococcus aureus Strain Infection: comprehensive Review. J Vet Med Health 6: 160.
Copyright: © 2022 Debeb D. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Usage
- Total views: 2420
- [From(publication date): 0-2022 - Nov 17, 2025]
- Breakdown by view type
- HTML page views: 1972
- PDF downloads: 448