Research Article Open Access
LC-MS Method for the Quantitation of Two Monoclonal Antibodies by Multiple Signature Peptides in Monkey Serum
Rita Mastroianni*, Marina Feroggio, Barbara Marsiglia, Clarissa Porzio Vernino, Simona Riva and Luca Barbero*
QPD - NBE Bioanalytics, RBM-Merck Serono, Via Ribes 1, 10010, Colleretto Giacosa (TO), Italy
- *Corresponding Author:
- Rita Mastroianni
QPD-NBE Bioanalytics
RBM-Merck Serono, Via Ribes 1
10010, Colleretto Giacosa (TO), Italy
Tel: +39-0125222657
Fax: +39-0125222559
E-mail: rita.mastroianni@merckgroup.com
- Luca Barbero
QPD-NBE Bioanalytics
RBM-Merck Serono, Via Ribes 1
10010, Colleretto Giacosa (TO), Italy
Tel: +39-0125222657
Fax: +39-0125222559
E-mail: luca.barbero@merckgroup.com
Received date: June 04, 2015; Accepted date: June 25, 2015; Published date: July 02, 2015
Citation: Mastroianni R, Feroggio M, Marsiglia B, Vernino CP, Riva S et al. (2015) LC-MS Method for the Quantitation of Two Monoclonal Antibodies by Multiple Signature Peptides in Monkey Serum. J Anal Bioanal Tech 6:252. doi: 10.4172/2155-9872.1000252
Copyright: © 2015 Mastroianni R, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Abstract
Evaluation of the in-vivo concentration of monoclonal antibody (mAb) mixtures is a challenging task. Here we report the application of an LC-MS bioanalytical method to quantify in monkey serum the Sym004, an equimolar mixture of two monoclonal antibodies, 992 mAb and 1024 mAb. This method has been assessed accordingly to industry standards and it is based on the determination of two specific signature peptides that report the single mAbs concentrations and on another one peptide, common to the two mAbs, that measures the total concentration of the two target proteins. It is shown that the total concentration is in agreement with the sum of the two measured single concentrations in spiked monkey serum samples. The consistency of the results will allow monitoring of the metabolic fate of different parts of the mAbs, at least in the central body compartment. This can then help to rationalize the design of the protein therapeutics modulating their stability accordingly.
Keywords
Monoclonal antibody; Quantitation; Bioanalysis; Sym004; LC-MS; EGFR; Catabolism
Abbreviations
EGFR: Epidermal Growth Factor Receptor; sEGFR: Extracellular EGFR; mAb: Monoclonal Antibody; ADA: Anti-Drug Antibodies; LC-MS: Liquid Chromatography Mass Spectrometry; IAM: Iodoacetamide; DTT: D-L Dithiothreitol, BIAS%: Percent Relative Error; CoA: Certificate of Analysis; CV%: Percent Coefficient of Variation; IS: Internal Standard; LLOQ: Lower Limit of Quantitation; QC: Quality Control; RS: Reconstitution Solvent; STD: Calibration Standard; ULOQ: Upper Limit of Quantitation; UPLC-MS/MS: Ultra Performance Liquid Chromatography - Tandem Mass Spectrometry; VS(L,M,H): Validation Sample (Low, Medium, High); S/N: Signal-to- Noise ratio; MRM: Multiple Reaction Monitoring; MCX: Mixed Mode Cation Exchange; SD: Standard Deviation
Introduction
Modification of the epidermal growth factor receptor (EGFR; ErbB1) pathway system has been reported to correlate with human malignancies. Increase in ligand production, receptor over-expression, receptor mutations, and/or cross-talk with other receptor systems are the most frequent modifications involved [1-3].
These changes have been linked to the development and maintenance of a malignant phenotype and correlated to poor clinical prognosis [4]. For this reason, the EGFR is an attractive target for anticancer therapy [5].
To date, four EGFR targeting agents (cetuximab, panitumumab, gefitinib and erlotinib) from two distinct drug classes have received FDA approval [6]. These include mAbs directed against the extracellular ligand-binding domain of EGFR and small molecule tyrosine kinase inhibitors (TKIs) directed against the cytosolic catalytic domain of the EGFR.
Although selected patients receive clear benefit from anti-EGFR mAbs, overall single agent response rates are in the order of 10% [5].
When two mAbs against distinct receptor epitopes are combined, rapid and more efficient receptor internalization is observed, followed by EGFR degradation [7]. The mixed antibody treatment is also more effective than single Abs in inhibiting signaling and tumor growth in tissue culture and animal models [8,9].
Sym004 is a recombinant antibody equimolar mixture of a pair of mouse-human chimeric immunoglobulin G1 monoclonal antibodies, 992 mAb and 1024 mAb. Both antibodies have activity against the epidermal growth factor receptor (EGFR) and bind specifically to two distinct non-overlapping epitopes on the extracellular domain III of the EGFR [10,11].
Unlike other anti-EGFR mAbs, Sym004 induces pronounced internalization and degradation of the EGFR, thereby leading to removal of EGFR from the cell surface. This novel mechanism of action is believed to result in superior anti-cancer activity compared to other anti-EGFR mAbs, specifically if resistance or failure to anti-EGFR mAbs is conferred by the presence of high affinity ligands, receptor cross-talk or constitutively activated EGFR. This has been demonstrated both invitro using human cancer cell lines and in-vivo using EGFR-dependent tumor xenografts [11].
Advanced preclinical development of such mAbs mixtures requires the determination of the single PK profiles of each component independently. From this analysis it should be possible to understand how the relative distribution ratio of mAb992 and mAb1024 in the central compartment is modified or maintained, to correlate it to any safety issues, and finally to grasp further pharmacology insights into the mode of action of mixture itself [12]. It is clear that, given mAbs sequence similarities and any binding partners already present in the serum or that may appear (e.g. ADA) during an in-vivo assessment, the bioanalytical method needs more stringent selectivity and sensitivity.
LC-MS methods can fulfill such requirements at different levels of specificity and sensitivity depending on their access to more sophisticated MS technology and to their inherent technological limitations. Three main approaches are used to detect and quantify a protein drug in a complex matrix by LC-MS: top-down, middle-down and bottom-up approaches.
In the top-down approach, the intact molecule is detected and quantified as it is. In this case the highest degree of possible specificity is achieved, when a High Resolution High Accuracy MS device is deployed. On the one hand, the direct determination of the MW can immediately determine whether the molecule itself, or the formulation composition in term of active drugs, has been modified. On the other hand, limitations to this approach are intrinsically linked to ionization techniques (ESI remains the most commonly used ionization mode in LC-MS) that produce many charge states from a single molecule, different glycosylation forms or post-translational modifications that further spread the drug(s) signal over a wide range of m/z units. This impacts the overall MS sensitivity and increases the need for MS accuracy and resolution. Finally, the presence of natural ligands has to be taken into account, since they can increase the complexity of the extraction procedure from the matrix and further reduce the overall sensitivity.
The other two approaches, i.e. middle-down and bottom-up based protocols, involve a progressively extensive cleavage step of the drug(s) to be quantified, i.e. the molecular weight of signature peptides will be above 3 kDa or below 3 kDa respectively. They are less demanding in terms of sample preparation and MS sensitivity, since the number of multiple charge states decreases with the dimension of the molecule to be quantified. This explains why they are extensively used in pharmacokinetic assessments of large protein therapeutics. However, their main drawback stems from the fact that these methods rely on the quantification of a surrogate molecule and not of the whole drug. In fact, any metabolic or elimination effects that impact differently on different parts of the drug or on the drugs present in the formulation administered, could jeopardize the assumption of a direct relation between the concentration of the signature peptide(s) and actual concentration of the drug(s).
To mitigate this problem we are presenting a LC-MS bottom-up method that allows quantitation of two signature peptides specific for each mAb and a third signature peptide that is common to both the mAbs but located on a different part of the mAb molecules, in a single run. Comparison of the concentrations of the three peptides further enhance the LC-MS intrinsic selectivity and provide further insight into the degradation status of the two molecules.
Materials and Methods
In-silico analyses
The identification of the signature peptide candidates consisted of a three-step process. In the first step, peptides derived from enzymatic cleavages by different enzymes were obtained by the ExPASy PeptideCutter tool [13] (http://web.expasy.org/peptide_cutter/). In the second step, the peptides were aligned by using the LALIGN algorithm (http://embnet.vital-it.ch/software/LALIGN_form.html), to obtain paired specific sequences from each mAb.
In the third and last step, the peptide pairs were checked for matrix interference by the BLAST® (blastp suite) (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&BLAST_PROGRAMS=deltaBlast&PROG_DEFAULTS=on&PAGE_TYPE=BlastSearch&LINK_LOC=BlastHomeAd) with Database set to “Non-redundant protein sequences” and Organism set to “Macaca fascicularis (taxid:9541)”. Only the specific peptide pairs that passed the last two steps were further evaluated by LC-MS to become signature peptides. A further analysis for checking any matrix interference in human serum was performed with the same tool and settings, but with Organism set to “Homo sapiens (taxid: 9606)”.
A similar workflow was used in the setup to select common peptides to the two mAbs, but still specific enough to be quantified in monkey serum. These peptides provide the total concentration of the two mAbs.
Average MW and isoelectric points (pI) were calculated by the ProtParam tool (http://web.expasy.org/protparam/) [13].
Proteins, chemicals and reagents
Monoclonal antibody, 992 mAb and 1024 mAb, were provided by CMC Biologics A/S (Denmark). Labeled Peptide Internal Standards - Stable isotope-labeled amino acids, [13C6 15N] Leucine and [13C5 15N] Valine (> 97% by HPLC assay) were purchased from Bachem (Bubendorf, Switzerland). Acetonitrile (LiChrosolv®, Reag. Ph Eur, gradient grade for liquid chromatography), 2-propanol (LiChrosolv®, gradient grade for liquid chromatography), Methanol (LiChrosolv®, Reag. Ph Eur, gradient grade for liquid chromatography), Formic Acid (Emsure® ACS, Reag. Ph Eur, 98-100% for analysis) were purchased from Merck Millipore (Merck KGaA, Darmstadt, Germany). Ammonia Solution 32% (extra pure), Trypsin from porcine pancreas (Type IXS), Iodoacetamide, IAM (BioUltra, ≥99% NMR), D-L Dithiothreitol, DTT (BioUltra, ≥99.5% RT), Calcium Chloride dihydrate (Reagent Plus® ≥99%), Ammonium bicarbonate (BioUltra, ≥99.5% T), Urea (for electrophoresis gel) and Phosphoric acid (85 wt.% in H2O, 99.99% trace metals basis) were purchased from Sigma–Aldrich (St. Louis, MO). Ultrapure water was from a Millipore Milli-Q system (Merck Millipore, Billerica, MA). Cynomolgus monkey serum was purchased from R.C. Hartelust BV (Tilburg, Nederland).
LC-MS/MS equipment
The UPLC–MS/MS analyses were performed by an Acquity UPLC® (Waters Corporation Milford, MA) system consisting of Binary Solvent Manager, Sample Manager, and Sample Organizer and Column oven. An Acquity UPLC™ CSH (Charged Surface Hybrid) C18 column (2.1 × 150 mm, 1.7 μm particle size; Waters, Milford, MA, USA) was used for the separation. The UPLC system was interfaced with an AB SCIEX TripleQuad™ 5500 mass spectrometer (AB SCIEX, Toronto, Canada) used as the detector. Analyst software v.1.5.1 was used for data acquisition and processing.
Monoclonal antibodies-spiked serum samples
Stock solutions of monoclonal antibodies 992 mAb and 1024 mAb were prepared in ammonium bicarbonate buffer 100 mM at a concentration of 1 mg/mL, separately. Protein-spiked serum samples were prepared by diluting protein stock solutions into blank monkey serum followed by further serial dilution in blank monkey serum to obtain the final concentrations desired. 992 mAb and 1024 mAb calibration standard concentrations were 150.0, 250.0, 500.0, 1000.0, 2500.0, 5000.0, 7500.0, 10000.0 ng/ml for the method performance evaluation runs (for the total signature peptide the concentration levels are twice those listed); VS concentrations for all mAb analytes were 150.0, 450.0, 1500.0, 8500.0 and 10000.0 ng/ml. The m992 IS, 1024 mAb IS and total mAbs IS working solutions were prepared at a concentration of 5000.0 ng/ml, 2500.0 ng/ml and 2500.0 ng/ml, respectively.
Serum sample digestion
50 μL of serum samples (STD, VS) were transferred to 96-well polypropylene microplates. 992 mAb, 1024 mAb and total mAbs Internal standards (5 μL) (Table 1), Urea 2M solution (700 μL), DTT 250 mM solution (70 μL) were added to each well followed by vortex mixing at 750 rpm for 30 min. at 60°C. IAM 0.5M solution (45 μL) was added to each well followed by vortex mixing at 750 rpm for 45 min. at RT. Calcium chloride 87.5 mM (10 μL) solution and trypsin solution at 15 mg/mL concentration (85 μL) were added to each well followed by vortex mixing at 750 rpm for 40 min. at 60°C.
| Labeled peptide | Sequence | #of aa | MW (Av.), Da |
|---|---|---|---|
| HC-3-1024 mAb IS | Acetyl-NH-VKQRPGQG-[L(13C6;15N)]-EWIGEINPSSGR-COOH | 21 | 2357.6 |
| HC-1-992 mAb IS | NH2-EVQLQPGSE-[L(13C6;15N)]-VRPGASVKLS-CONH2 | 21 | 2228.5 |
| HC-3-total IS | Acetyl-NH-YRVVSVLT-[V(13C5;15N)]-LHQDWLNGK-COOH | 18 | 2175.5 |
Table 1: Primary sequence of the stable isotopes (internal standards).
Sample clean-up
SPE clean-up: The digests of the serum samples were mixed with 15 μl of phosphoric acid (85%, %W/V) and then loaded onto an Oasis® MCX SPE plate. The samples were washed sequentially with 2 mL each of: 2% formic acid in water; 1 mL of 10% methanol; 1 mL of 5% ammonium hydroxide in water. The analytes were eluted with 1 mL of 5% ammonium hydroxide in 60/40 methanol/water and then dried down. The dried samples were reconstituted into 150 μl of 1% formic acid in water and analyzed by LC-MS/MS.
Chromatographic and mass spectrometric conditions
A gradient solvent system consisting of mobile phase A (0.1% formic acid in water), and mobile phase B (0.1% formic acid in acetonitrile) was used. The column temperature was set at 45°C. The gradient was as follows: 0-0.5 min 10% B; 0.5-5.0 min 10-18% B; 5.0-8.5 min 30% B; 8.6-10.6 min 95%B; 10.7-16.5 min 10%B. The flow rate was 0.3 ml/ min, and the injection volume was 20 μl. The mass spectrometer was operated in ESI positive mode. The following parameters were used: curtain gas 30 psi; ion source gas one, 45 psi; ion source gas two, 50 psi; temperature 450°C; ion-spray voltage 5200 V. The MRM channels monitored for the surrogate peptides and their IS are listed in Table 2. The dwell time for each MRM channel was 50 ms.
Results
| Peptide | Precursor ion (m/z) | Product ion (m/z) |
|---|---|---|
| HC-3-1024 mAb | 694.4 [M+3H]3+ | 916.5 y9+1 ion |
| HC-3-1024 mAb | 694.4 [M+3H]3+ | 866.5 b8+1 ion |
| HC-1-992 mAb | 674.7 [M+3H]3+ | 648.8 y13+2 ion |
| HC-1-992 mAb | 674.7 [M+3H]3+ | 712.9 y14+2 ion |
| HC-1-992 mAb | 674.7 [M+3H]3+ | 777.0 y15+2 ion |
| HC-3-total | 603.3 [M+3H]3+ | 805.4 y14+2 ion |
| HC-3-1024 mAb IS | 696.7 [M+3H]3+ | 859.4 y8+2 ion |
| HC-1-992 mAb IS | 677.3 [M+3H]3+ | 714.4 y14+2 ion |
| HC-3-total IS | 605.3[M+3H]3+ | 807.4 y14+2 ion |
Table 2: MRM transitions and charge states for surrogate peptides and the stable isotopes (internal standards).
In-silico analysis
The primary sequences of the light and heavy chain of the two mAbs (Table 3) were analyzed as described in the Materials and Methods section. The enzyme selected for the cleavage was trypsin since it was able to provide peptides with well distributed length and charge, and at least 3 to 4 peptide pairs for heavy as well as light chains specific for each mAb Fab part. From these, the selectivity analysis conducted by the BLAST suite revealed the candidate signature peptides. These findings are reported in Tables 4 and 5 for the mAb Fab parts.
 |
Table 3: Primary sequences of 992 mAb and 1024 mAb. The signature peptides are reported in bold.
| mAb | Sequence | #of aa | MW Da | pI | "Macaca fascicularis BLASTP” | "Homo sapiens BLASTP" | Identity% vs. HC of the other mAb |
|---|---|---|---|---|---|---|---|
| 992 | EVQLQQPGSELVRPGASVK | 19 | 2022.2 | 6.24 | OK | OK | 78.9 |
| 992 | ASGYTFTSYWMHWVK | 15 | 1864.1 | 8.55 | OK | "Not OK” Chain B, Mature Metal Chelatase Catalytic Antibody With Hapten pdb|3FCT|B | 93.3 |
| 992 | QRPGQGLEWIGNIYPGSR | 18 | 2028.2 | 8.75 | OK | OK | 82.4 |
| 992 | ATLTVDTSSSTAYMQLSSLTSEDSAVYYCTR | 31 | 3352.6 | 4.03 | OK | OK | 90.3 |
| 992 | NGDYYVSSGDAMDYWGQGTSVTVSSASTK | 29 | 3034.1 | 3.93 | OK | OK | 80.0 |
| 1024 | QVQLQQPGAELVEPGGSVK | 19 | 1964.2 | 4.53 | OK | "Not OK” Chain B, Anti-blood Group A Fv pdb|1JV5|B" | 78.9 |
| 1024 | ASGYTFTSHWMHWVK | 15 | 1838.0 | 8.65 | OK | OK | 93.3 |
| 1024 | QRPGQGLEWIGEINPSSGR | 19 | 2081.2 | 6.14 | OK | OK | 82.4 |
| 1024 | SSSTAYMQFSSLTSEDSAVYYCVR | 24 | 2682.9 | 4.37 | OK | OK | 91.7 |
| 1024 | YYGYDEAMDYWGQGTSVTVSSASTK | 25 | 2766.9 | 4.03 | OK | OK | 80.0 |
Table 4: Result summary of the in-silico selection process for Fab Heavy Chains (HC). “OK” and “Not OK” mean “no match” or “match”, respectively, for known protein sequences present in monkey or human proteome. When a match is present, some examples of proteins containing that exact sequence are listed. The level of identity with the corresponding peptide in the same region on the other mAb is reported (I to L not considered) as Identity% score. The average MW, and calculated isoelectric point (pI) are also reported for each peptide.
| mAb | Sequence | #of aa | MW Da | pI | "Macaca fascicularis BLASTP" | "Homo sapiens BLASTP" | Identity % vs. LC of the other mAb |
|---|---|---|---|---|---|---|---|
| 992 | DIQMTQTTSSLSASLGDR | 18 | 1911.0 | 4.21 | OK | "Not OK Chain A, Anti-Blood Group A Fv, pdb|1JV5|A, pdb|1IKF|L, pdb|3U0W|L" | 50.0 |
| 992 | TSQDIGNYLNWYQQKPDGTVK | 21 | 2455.6 | 5.63 | OK | OK | NA |
| 992 | LLIYYTSR | 8 | 1028.2 | 8.59 | OK | "Not OK”, immunoglobulin VL region=humanized bispecific antibody [human, Peptide Recombinant, 107 aa, gb|AAB24132.1|" | NA |
| 992 | LHSGVPSR | 8 | 851.9 | 9.76 | Not OK | Not OK | NA |
| 1024 | DIVMTQAAFSNPVTLGTSASISCR | 24 | 2469.8 | 5.83 | OK | "Not OK, anti-GlcNAc antibody variable region:SUBUNIT=light chain prf||1911357B" | 50.0 |
| 1024 | FSSSGSGTDFTLR | 13 | 1361.4 | 5.84 | OK | OK | NA |
| 1024 | VEAEDVGVYYCAQNLELPYTFGGGTK | 26 | 2824.1 | 4.00 | OK | OK | NA |
Table 5: Result summary of the in-silico selection process for Fab Light Chains (LC). “OK” and “Not OK” mean no match or match, respectively, for known protein sequences present in monkey or human proteome. When a match is present, some examples of proteins containing that exact sequence are listed. The level of identity with the corresponding peptide in the same region on the other mAb is reported (I to L not considered) as Identity% score. The average MW, and calculated isoelectric point (pI) are also reported for each peptide.
The in-silico analysis regarding the shared mAb Fc parts are reported in Table 6. As described by Furlong et al. [14], these tryptic peptides can be used to obtain the concentration of a humanized mAb in non-human matrices.
| Peptide | Sequence | Human chain subclass | # of aa | MW (Av.), Da | pI |
|---|---|---|---|---|---|
| LC-1 | TVAAPSVFIFPPSDEQLK | NA | 18 | 1946.2 | 4.37 |
| LC-2 | SGTASVVCLLNNFYPR | NA | 16 | 1740.9 | 7.94 |
| LC-3 | VDNALQSGNSQESVTEQDSK | NA | 20 | 2136.1 | 3.92 |
| LC-4 | DSTYSLSSTLTLSK | NA | 14 | 1502.6 | 5.83 |
| HC-1 | TPEVTCVVVDVSHEDPEVK | IgG1 | 19 | 2082.3 | 4.17 |
| HC-2 | FNWYVDGVEVHNAK | IgG1 | 14 | 1677.8 | 5.32 |
| HC-3 | VVSVLTVLHQDWLNGK | IgG1, IgG4 | 16 | 1808.1 | 6.71 |
| HC-4 | GFYPSDIAVEWESNGQPENNYK | IgG1, IgG2, IgG4 | 22 | 2544.6 | 4.00 |
| HC-5 | TTPPVLDSDGSFFLYSK | IgG1 | 17 | 1874.0 | 4.21 |
Table 6: List of common tryptic Fc peptides from 992 and 1024 mAbs [14]. None of them is present in monkey serum.
Optimization of experimental conditions and selection of signature peptides
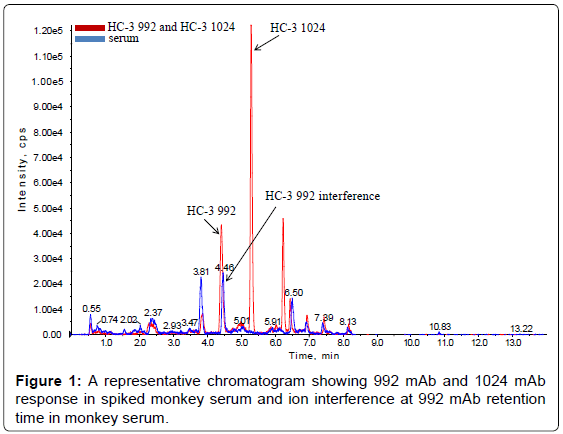
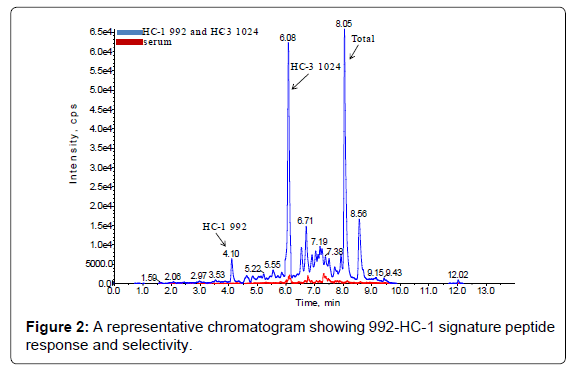
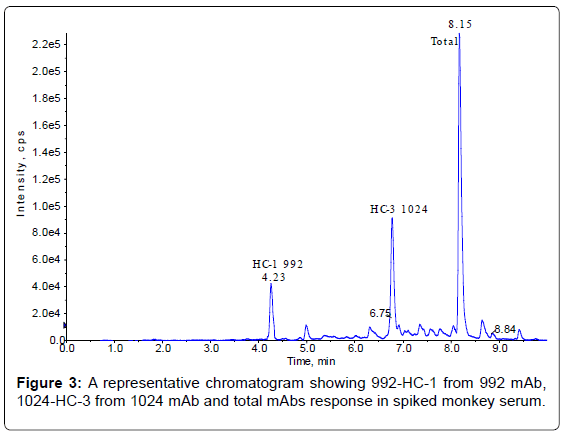
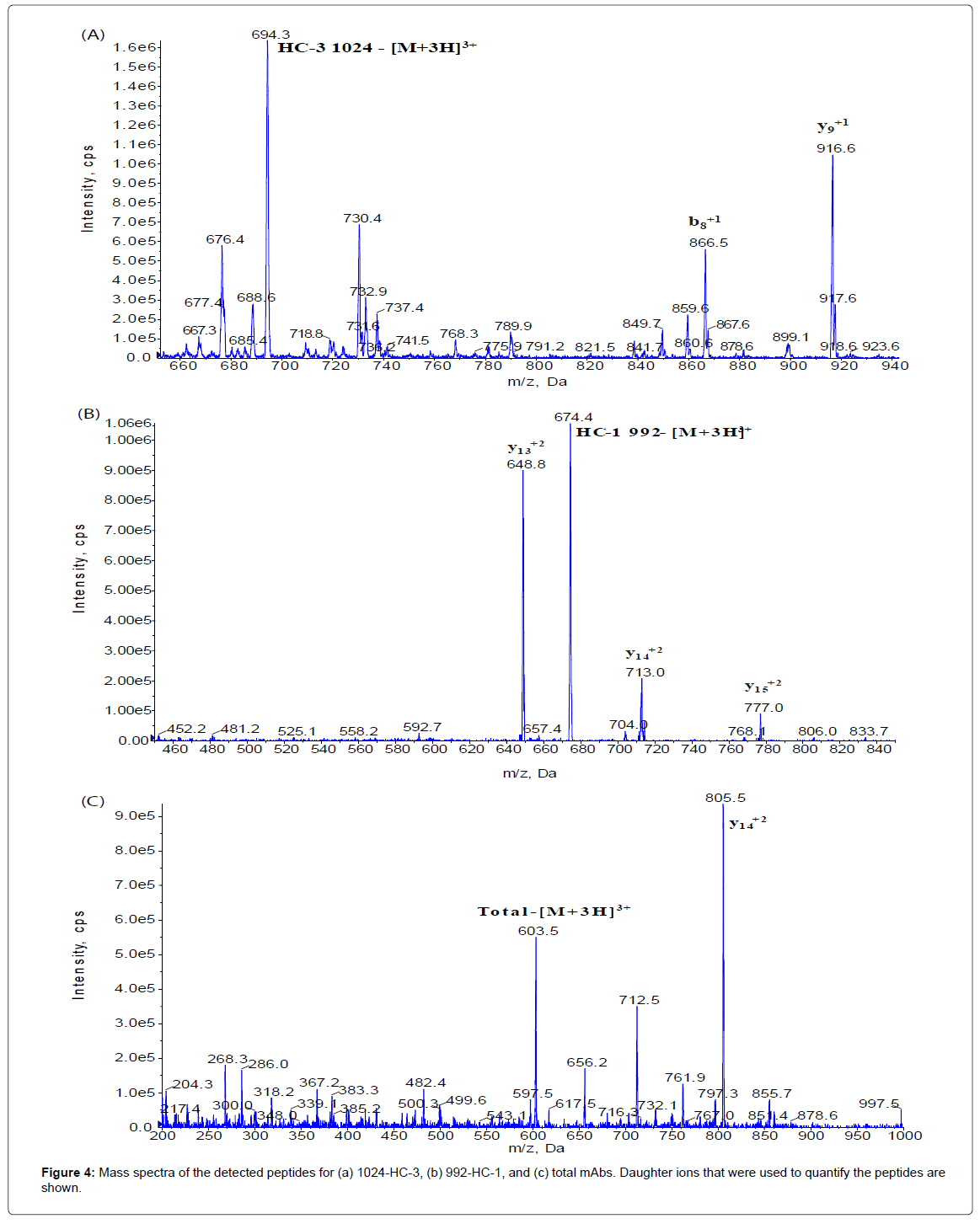
The method set-up was thoroughly investigated in order to obtain the most selective peptides, the best SPE conditions and the appropriate chromatographic conditions for peptide separation. During this phase, different SPE cartridges and different solvent mixtures were tested to obtain acceptable results in term of sensitivity, robustness, reproducibility and selectivity. Initially two signature peptides, 992-HC-3 and 1024-HC-3, coming from paired regions were monitored and analyzed using a BEH C18 Column (1 × 100 mm, 1.7 μm, Waters), given their very intense MS signal. Unfortunately the 992-HC-3 peptide from 992 mAb, did not show sufficient selectivity and sensitivity on different MRM transitions when monitored in a biological matrix, while the 1024-HC-3 peptide from 1024 mAb showed acceptable selectivity and sensitivity in a biological matrix (Figure 1). Modifications in chromatographic conditions (including column length) and SPE purification were not able to solve the issue with 992-HC-3 peptide. Of the other 992 mAbs peptides, 992-HC-1 was found to be the second choice in term of sensitivity. Therefore its selectivity was investigated and considered to be sufficient in matrix samples (Figure 2). In order to improve HC-1-992 peptide sensitivity, a CSH C18 column and some gradient modifications were introduced (see LC-MS/MS equipment and Chromatographic and mass spectrometric conditions section, respectively). With these modifications, 992-HC-1 could be selected as mAb signature peptide. Additionally, quantitation of a signature peptide coming from the common region of the two monoclonal antibodies was introduced. This strategy was applied in order to confirm the analytical response from HC-1-992 (992 mAb) and HC-3-1024 (1024 mAb) (Figures 3 and 4).
Method performances
The method was assessed by evaluating linearity, sensitivity, selectivity, accuracy and precision and matrix effect according to the recent industry guidelines and adopting white papers suggestions [15-18] for NBE LC/MS analysis. For both the mAb analytes, a single calibration curve range and a single QC/VS concentration set were analyzed using the same LC method that incorporated all surrogate peptides.
Selectivity, specificity and sensitivity
Selectivity and specificity assessments were performed by evaluating potential analytes traces in different types of serum sample: a) zero sample; b) five different blank matrix sources; c) LLOQ prepared in five different matrix sources in six replicates each; d) 1024 mAb at ULOQ concentration level spiked in the 992 mAb calibration standard curve from STD1 to STD 4 (the most critical concentrations to be determined) and vice-versa. No analyte ions were found in the blank serum samples or in the zero samples, which indicated that serum matrices and the IS do not interfere with the determination of either monoclonal antibodies or total mAbs. The lower limit of detection was 150 ng/mL for both the surrogate peptides (from 992 mAb and 1024 mAb) and 300 ng/mL for the total surrogate peptide (common region of the two mAbs) with a S/N ratio above 5, which represented a concentration of about 1.00 nM, 1.00 nM and 2.00 nM of analytes (about 20 fmol, 20 fmol and 40 fmol) injected on the column for QRPGQGLEWIGEINPSSGR, EVQLQQPGSELVRPGASVK and VVSVLTVLHQDWLNGK peptides, respectively.
Both monoclonal antibodies at LLOQ level met the required accuracy (%BIAS ± 25) and precision (%CV ≤25) criteria in all the sources of matrices tested (Table 7) and in all the signature peptides considered. The total mAb determination was performed at LLOQ+25% only, during preliminary experiments. It was also demonstrated that a high concentration (ULOQ) of one monoclonal antibody does not affect the response of the second monoclonal antibody at low concentrations, and vice-versa (specificity). Moreover, the accuracy (%BIAS ± 25) of the total mAbs quantified (sum of one mAb spiked at ULOQ level and the second one spiked at low levels, and vice-versa) gave confirmation of the method consistency (Table 8).
| 1024 Monoclonal antibody | |||||
|---|---|---|---|---|---|
| Parameters | Matrix 1 | Matrix 2 | Matrix 3 | Matrix 4 | Matrix 4 |
| Mean Conc. (ng/mL) | 157.4 | 161.1 | 163.6 | 142.0 | 167.8 |
| BIAS% | 4.9 | 7.4 | 9.1 | -5.3 | 11.9 |
| SD | 11.7 | 21.0 | 27.4 | 18.7 | 28.4 |
| CV% | 7.4 | 13.0 | 16.7 | 13.2 | 16.9 |
| 992 Monoclonal antibody | |||||
| Parameters | Matrix 1 | Matrix 2 | Matrix 3 | Matrix 4 | Matrix 4 |
| Mean Conc. (ng/mL) | 169.9 | 150.0 | 169.2 | 135.8 | 172.9 |
| BIAS% | 13.3 | 0.0 | 12.8 | -9.5 | 15.3 |
| SD | 33.6 | 27.7 | 26.4 | 9.7 | 10.5 |
| CV% | 19.8 | 18.5 | 15.6 | 7.1 | 6.1 |
Table 7: Selectivity of the LC-MS/MS analysis of 1024 mAb and 992 mAb spiked at LLOQ in five different monkey serum lots.
| Sample | Nominal Conc. (ng/mL) | 992 mAb Calculated Conc. (ng/mL) | 992 mAb %BIAS | 1024 mAb Calculated Conc. (ng/mL) | 1024 mAb %BIAS | 1024 mAb at ULOQ | 992 mAb at ULOQ | Sum of 992 mAb and 1024 mAb at ULOQ | Sum of 1024 mAb and 992 mAb at ULOQ | Total calculated conc. (ng/mL) | %BIAS | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Std 1 | 150.0 | 158.0 | 105.3 | - | - | 11051.6 | - | 11209.6 | - | 11120.5 | 99.2 | ||||
| Set 1 | Std 2 | 250.0 | 263.0 | 105.2 | - | - | 10426.9 | - | 10689.9 | - | 11502.6 | 107.6 | |||
| Std 3 | 500.0 | 590.0 | 118.0 | - | - | 10491.9 | - | 11081.9 | - | 12180.1 | 109.9 | ||||
| Std 4 | 1000.0 | 1146.6 | 114.7 | - | - | 10154.0 | - | 11300.6 | - | 12687.6 | 112.3 | ||||
| Std 1 | 150.0 | - | - | 161.6 | 107.8 | - | 11730.0 | - | 11891.6 | 12320.1 | 105.0 | ||||
| Set 2 | Std 2 | 250.0 | - | - | 239.0 | 95.6 | - | 13011.3 | - | 13250.3 | 12249.6 | 94.1 | |||
| Std 3 | 500.0 | - | - | 434.3 | 86.9 | - | 14181.3 | - | 14615.6 | 9934.21 | 70.1 | ||||
| Std 4 | 1000.0 | - | - | 997.4 | 99.7 | - | 10596.9 | - | 11594.3 | 12312.7 | 116.2 | ||||
| Sample | 992 and 1024 mAb nominal conc. (ng/mL) | 1024 mAb calculated conc. (ng/mL) | 992 mAb calculated conc. (ng/mL) | Sum of 1024 mAb and 992 mAb calculated conc. (ng/mL) | Total mAbs calculated conc. (ng/mL) | %Bias | |||||||||
| VS-LLOQ_1 | 150.0 | 165.8 | 141.5 | 307.2 | 301.6 | 98.2 | |||||||||
| VS-LLOQ_2 | 150.0 | 167.1 | 144.6 | 311.7 | 288.6 | 92.6 | |||||||||
| VS-LLOQ_3 | 150.0 | 173.1 | 162.1 | 335.3 | 290.1 | 86.5 | |||||||||
| VS-LLOQ_4 | 150.0 | 164.6 | 117.2 | 281.8 | 288.8 | 102.5 | |||||||||
| VS-LLOQ_5 | 150.0 | 152.4 | 152.2 | 304.6 | 297.7 | 97.7 | |||||||||
| VS-LLOQ_6 | 150.0 | 125.5 | 160.0 | 285.5 | 310.9 | 108.9 | |||||||||
| VS-LLOQ_7 | 150.0 | 141.8 | 148.9 | 290.7 | 263.1 | 90.5 | |||||||||
| VS-LLOQ_8 | 150.0 | 162.0 | 153.6 | 315.6 | 264.0 | 83.6 | |||||||||
| VS-LLOQ_9 | 150.0 | 162.5 | 156.7 | 319.2 | 313.8 | 98.3 | |||||||||
| VS-LLOQ_10 | 150.0 | 150.7 | 164.1 | 314.7 | 266.1 | 84.5 | |||||||||
| VS-LLOQ_11 | 150.0 | 152.6 | 158.2 | 310.7 | 267.0 | 85.9 | |||||||||
| VS-Low_1 | 450.0 | 498.5 | 460.1 | 958.5 | 942.4 | 98.3 | |||||||||
| VS-Low_2 | 450.0 | 509.0 | 490.1 | 999.1 | 1074.1 | 107.5 | |||||||||
| VS-Low_3 | 450.0 | 479.4 | 469.8 | 949.2 | 1016.4 | 107.1 | |||||||||
| VS-Low_4 | 450.0 | 530.8 | 477.9 | 1008.7 | 956.2 | 94.8 | |||||||||
| VS-Low_5 | 450.0 | 511.5 | 513.8 | 1025.3 | 1120.3 | 109.3 | |||||||||
| VS-Low_6 | 450.0 | 447.1 | 479.5 | 926.7 | 1007.3 | 108.7 | |||||||||
| VS-Low_7 | 450.0 | 414.8 | 450.2 | 865.0 | 783.9 | 90.6 | |||||||||
| VS-Low_8 | 450.0 | 462.6 | 458.4 | 921.0 | 873.7 | 94.9 | |||||||||
| VS-Low_9 | 450.0 | 393.5 | 424.1 | 817.6 | 1019.4 | 124.7 | |||||||||
| VS-Low_10 | 450.0 | 446.2 | 523.4 | 969.6 | 1124.5 | 116.0 | |||||||||
| VS-Low_11 | 450.0 | 461.5 | 377.6 | 839.1 | 1072.3 | 127.8 | |||||||||
| VS-Medium_1 | 1500.0 | DEV | DEV | DEV | DEV | DEV | |||||||||
| VS-Medium_2 | 1500.0 | 1701.1 | 1837.1 | 3538.3 | 3181.9 | 89.9 | |||||||||
| VS-Medium_3 | 1500.0 | 1634.9 | 1750.2 | 3385.1 | 3569.9 | 105.5 | |||||||||
| VS-Medium_4 | 1500.0 | 1717.1 | 1782.8 | 3499.9 | 3593.0 | 102.7 | |||||||||
| VS-Medium_5 | 1500.0 | 1706.7 | 1614.5 | 3321.1 | 3235.2 | 97.4 | |||||||||
| VS-Medium_6 | 1500.0 | 1475.4 | 1632.8 | 3108.2 | 2586.1 | 83.2 | |||||||||
| VS-Medium_7 | 1500.0 | 1524.1 | 1389.1 | 2913.1 | 2789.6 | 95.8 | |||||||||
| VS-Medium_8 | 1500.0 | 1443.1 | 1424.3 | 2867.4 | 2832.3 | 98.8 | |||||||||
| VS-Medium_9 | 1500.0 | 1697.3 | 1497.9 | 3195.1 | 3387.2 | 106.0 | |||||||||
| VS-Medium_10 | 1500.0 | 1698.5 | 1722.1 | 3420.6 | 3285.3 | 96.0 | |||||||||
| VS-Medium_11 | 1500.0 | 1875.6 | 1792.7 | 3668.3 | 3591.1 | 97.9 | |||||||||
| VS-High_1 | 8500.0 | 8401.0 | 8187.5 | 16588.5 | 15863.2 | 95.6 | |||||||||
| VS-High_2 | 8500.0 | 8948.2 | 9003.0 | 17951.2 | 16840.3 | 93.8 | |||||||||
| Sample | 992 and 1024 mAb nominal conc. (ng/mL) | 1024 mAb calculated conc. (ng/mL) | 992 mAb calculated conc. (ng/mL) | Sum of 1024 mAb and 992 mAb calculated conc. (ng/mL) | Total mAbs calculated conc. (ng/mL) | %Bias | |||||||||
| VS-High_3 | 8500.0 | 8501.8 | 8986.3 | 17488.1 | 17506.2 | 100.1 | |||||||||
| VS-High_4 | 8500.0 | 8797.9 | 8114.8 | 16912.7 | 19210.4 | 113.6 | |||||||||
| VS-High_5 | 8500.0 | 8881.8 | 8691.8 | 17573.6 | 18023.8 | 102.6 | |||||||||
| VS-High_6 | 8500.0 | 8191.7 | 8791.02 | 16982.8 | 14244.0 | 83.9 | |||||||||
| VS-High_7 | 8500.0 | 8800.2 | 8909.84 | 17710.0 | 15840.6 | 89.4 | |||||||||
| VS-High_8 | 8500.0 | 8421.5 | 9575.83 | 17997.4 | 14445.5 | 80.3 | |||||||||
| VS-High_9 | 8500.0 | 8429.3 | 8714.6 | 17143.8 | 17328.8 | 101.1 | |||||||||
| VS-High_10 | 8500.0 | 8183.6 | 9253.3 | 17436.9 | 16197.1 | 92.9 | |||||||||
| VS-High_11 | 8500.0 | 8406.3 | 8774.3 | 17180.6 | 16795.2 | 97.8 | |||||||||
| VS-ULOQ_1 | 10000.0 | 11091.2 | 13600.9 | 24692.1 | 22622.1 | 91.6 | |||||||||
| VS-ULOQ_2 | 10000.0 | 11414.5 | 11984.6 | 23399.1 | 24885.9 | 106.4 | |||||||||
| VS-ULOQ_3 | 10000.0 | 10760.8 | 11311.2 | 22072.0 | 21742.9 | 98.5 | |||||||||
| VS-ULOQ_4 | 10000.0 | 11190.5 | 12435.0 | 23625.5 | 22692.7 | 96.1 | |||||||||
| VS-ULOQ_5 | 10000.0 | 11018.2 | 14001.5 | 25019.7 | 24498.1 | 97.9 | |||||||||
| VS-ULOQ_6 | 10000.0 | 10099.1 | 11867.0 | 21966.1 | 16253.0 | 74.0 | |||||||||
| VS-ULOQ_7 | 10000.0 | 11463.1 | 11306.4 | 22769.5 | 20383.4 | 89.5 | |||||||||
| VS-ULOQ_8 | 10000.0 | 11299.2 | 10712.1 | 22011.3 | 17992.2 | 81.7 | |||||||||
| VS-ULOQ_9 | 10000.0 | 11412.3 | 11301.9 | 22714.2 | 23017.8 | 101.3 | |||||||||
| VS-ULOQ_10 | 10000.0 | 11870.9 | 11608.8 | 23479.7 | 24067.1 | 102.5 | |||||||||
| VS-ULOQ_11 | 10000.0 | 10427.4 | 11056.1 | 21483.5 | 21557.2 | 100.3 | |||||||||
Table 8: Evaluation of total mAbs accuracies when compared to the sum of individual mAbs measured concentrations. DEV stands for deviation from the sample preparation procedure. In Set1 and Set2 are also reported the specificity experiment results where one mAb at ULOQ is spiked over a concentration curve of the other mAb. The last column reports the accuracy of the calculated total mAb concentrations (%BIAS) where the nominal concentration was assumed to be the sum of the individual mAbs calculated concentrations. Out of accuracy acceptance criteria, i.e. ±20% BIAS% (±25%BIAS % at LLOQ) are shown in bold.
Assessment of matrix effects
Matrix effect assessment was conducted using the post-extraction spike method. It quantitatively assesses matrix effects by comparing the response of the surrogate peptides in neat solvent to the response of the surrogate peptide spiked into a blank matrix sample that has gone through the sample preparation process. The monoclonal antibodies, at QC-Low and QC-High level concentrations, were spiked in five different monkey serum sources in order to investigate also the matrix effect values among different lots of serum. The results reported in Table 9 show that there is no significant matrix effect on either analyte.
| 1024 Monoclonal antibody | ||||
|---|---|---|---|---|
| AN/IS Area ratio in samples spiked after extraction | AN/IS Area ratio in neat solvent | |||
| Parameters | VS Low 450.0 ng/mL | VS High 8500.0 ng/mL | VS Low 450.0 ng/mL | VS High 8500.0 ng/mL |
| Mean area ratio | 0.658 | 9.66 | 0.501 | 9.39 |
| Normalized matrix effect | 1.3 | 1.0 | ||
| n | 6 | 6 | 6 | 6 |
| 992 Monoclonal antibody | ||||
| AN/IS Area ratio in samples spiked after extraction | AN/IS Area ratio in neat solvent | |||
| Parameters | VS Low 450.0 ng/mL | VS High 8500.0 ng/mL | VS Low 450.0 ng/mL | VS High 8500.0 ng/mL |
| Mean area ratio | 0.334 | 5.13 | 0.275 | 5.92 |
| Normalized matrix effect | 1.2 | 0.9 | ||
| n | 6 | 6 | 6 | 6 |
Table 9: Matrix effect of the LC-MS/MS analysis of 1024 mAb and 992 mAb in monkey serum.
Accuracy and precision
Accuracy and precision were determined in one run of intra-batch and three runs of inter-batch serum samples containing 1024 mAb, 992 mAb and the total mAbs at five concentration levels (LLOQ, VSLow, VS-Medium, VS-High and ULOQ). The intra-batch relative mean accuracy of back calculated concentrations of the VS compared with theoretical ones ranged from 2.4% to 12.7%, from -6.3% to 16.4% and from -2.2% to 16.4% for QRPGQGLEWIGEINPSSGR peptide (1024 mAb), EVQLQQPGSELVRPGASVK peptide (992 mAb) and for VVSVLTVLHQDWLNGK (total mAbs), respectively. It should be noted that one QC at the medium concentration level was out of the acceptable limit of the assay for both the analytes but was excluded from the calculation since it was an outlier (known sample processing error). The intra-assay precision (%CV) ranged from 2.2 to 4.6, from 4.3 to 11.7 and from 2.0 to 7.4 for QRPGQGLEWIGEINPSSGR peptide (1024 mAb), EVQLQQPGSELVRPGASVK peptide (992 mAb) and VVSVLTVLHQDWLNGK (total), respectively (Table 10).
| 1024 Monoclonal antibody | |||||
|---|---|---|---|---|---|
| Parameters | VS LLOQ 150.0 ng/mL | VS Low 450.0 ng/mL | VS Medium 1500.0 ng/mL | VS High 8500.0 ng/mL | VS ULOQ 10000.0 ng/mL |
| Mean Conc. (ng/mL) | 164.6 | 505.8 | 1690.0 | 8706.2 | 11095.0 |
| Accuracy (%BIAS) | 9.7 | 12.4 | 12.7 | 2.4 | 10.9 |
| SD | 7.6 | 18.8 | 37.3 | 241.2 | 239.2 |
| Precision (%CV) | 4.6 | 3.7 | 2.2 | 2.8 | 2.2 |
| n | 5 | 5 | 4* | 5 | 5 |
| 992 Monoclonal antibody | |||||
| Parameters | VS LLOQ 150.0 ng/mL | VS Low 450.0 ng/mL | VS Medium 1500.0 ng/mL | VS High 8500.0 ng/mL | VS ULOQ 10000.0 ng/mL |
| Mean Conc. (ng/mL) | 143.5 | 482.3 | 1746.1 | 8596.7 | 9372.9 |
| Accuracy (%BIAS) | -4.3 | 7.2 | 16.4 | 1.1 | -6.3 |
| SD | 16.7 | 20.7 | 94.8 | 425.9 | 771.4 |
| Precision (%CV) | 11.7 | 4.3 | 5.4 | 5.0 | 8.2 |
| n | 5 | 5 | 4* | 5 | 5 |
| Total | |||||
| Parameters | VS LLOQ 300.0 ng/mL | VS Low 900.0 ng/mL | VS Medium 3000.0 ng/mL | VS High 17000.0 ng/mL | VS ULOQ 20000.0 ng/mL |
| Mean Conc. (ng/mL) | 293.4 | 1021.9 | 3395.0 | 17488.8 | 23288.3 |
| Accuracy (%BIAS) | -2.2 | 13.5 | 13.2 | 2.9 | 16.4 |
| SD | 5.9 | 76.0 | 216.6 | 1256.3 | 1341.9 |
| Precision (%CV) | 2.0 | 7.4 | 6.4 | 7.2 | 5.8 |
| n | 5 | 5 | 4* | 5 | 5 |
| *One outlier of a set of five values, excluded from the statistic calculation | |||||
Table 10: Intra-assay accuracy and precision of the LC-MS/MS analysis of 1024 mAb, 992 mAb and total mAbs in monkey serum.
As shown in Table 11, for QRPGQGLEWIGEINPSSGR peptide (1024 mAb), inter-assay accuracy (% Bias) of less than 10.9% and inter-assay precision (%CV) of less than 9% was achieved. For peptide EVQLQQPGSELVRPGASVK (992 mAb), inter-assay accuracy (%Bias) of less than 9.6% and inter-assay precision (%CV) of less than 11.1% was achieved, while for VVSVLTVLHQDWLNGK (total mAbs) inter-assay accuracy (%Bias) of less than 11.0% and inter-assay precision (%CV) of less than 12.3% was achieved. The accuracy and precision of all the peptides are well below the 20% (25% for VS at LLOQ level) acceptance criteria typically used in LC–MS/MS applied to large molecule bioanalysis.
| 1024 Monoclonal antibody | |||||
|---|---|---|---|---|---|
| Parameters | VS LLOQ 150.0 ng/mL | VS Low 450.0 ng/mL | VS Medium 1500.0 ng/mL | VS High 8500.0 ng/mL | VS ULOQ 10000.0 ng/mL |
| Mean Conc. (ng/mL) | 156.2 | 468.6 | 1647.4 | 8542.1 | 11095.2 |
| Accuracy (%BIAS) | 4.1 | 4.2 | 9.8 | 0.5 | 10.9 |
| SD | 13.6 | 42.4 | 131.3 | 270.3 | 504.6 |
| Precision (%CV) | 8.7 | 9.0 | 8.0 | 3.2 | 4.5 |
| n | 11 | 11 | 10* | 11 | 11 |
| 992 Monoclonal antibody | |||||
| Parameters | VS LLOQ 150.0 ng/mL | VS Low 450.0 ng/mL | VS Medium 1500.0 ng/mL | VS High 8500.0 ng/mL | VS ULOQ 10000.0 ng/mL |
| Mean Conc. (ng/mL) | 152.8 | 465.9 | 1644.3 | 8818.4 | 10428.8 |
| Accuracy (%BIAS) | 0.6 | 3.5 | 9.6 | 3.7 | 4.3 |
| SD | 13.2 | 40.5 | 160.4 | 419.1 | 1158.5 |
| Precision (%CV) | 8.8 | 8.7 | 9.8 | 4.8 | 11.1 |
| n | 11 | 11 | 10* | 11 | 11 |
| Total | |||||
| Parameters | VS LLOQ 300.0 ng/mL | VS Low 900.0 ng/mL | VS Medium 3000.0 ng/mL | VS High 17000.0 ng/mL | VS ULOQ 20000.0 ng/mL |
| Mean Conc. (ng/mL) | 286.5 | 999.1 | 3205.2 | 16572.3 | 21792.0 |
| Accuracy (%BIAS) | -4.5 | 11.0 | 6.8 | -2.5 | 9.0 |
| SD | 18.9 | 104.5 | 360.3 | 1475.0 | 2687.7 |
| Precision (%CV) | 6.6 | 10.85 | 11.2 | 8.9 | 12.3 |
| n | 11 | 11 | 10* | 11 | 11 |
| *One outlier of a set of five values, excluded from the statistic calculation | |||||
Table 11: Inter-assay accuracy and precision of the LC-MS/MS analysis of 1024 mAb, 992 mAb and total mAbs in monkey serum.
Standard curve
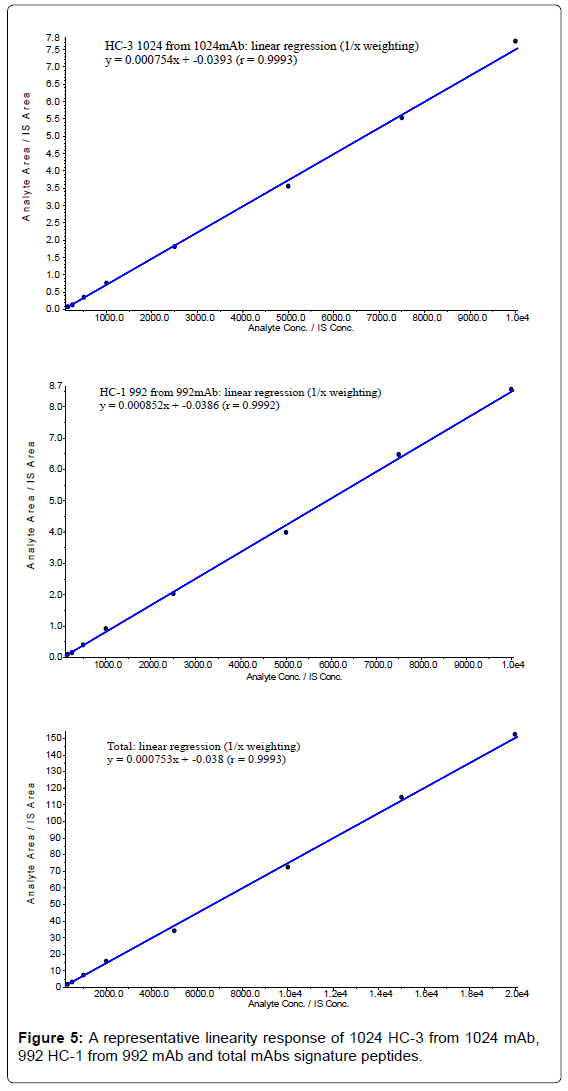
The calibration curves of 1024 mAb, 992 mAb and total mAbs in monkey serum (Figure 5) were generated automatically by using the algorithm classic of Analyst software. The linear dynamic range evaluated was between 150 ng/mL and 10000 ng/mL for 1024 mAb and 992 mAb, and from 300 ng/mL to 20000 ng/mL for total mAbs. Individual standard curve concentration data in monkey serum are shown in Table 12.
| 1024 Monoclonal antibody | ||||||
|---|---|---|---|---|---|---|
| Nominal conc. (ng/mL) | Calculated conc. (ng/mL) | Accuracy (%Bias) | Calculated conc. (ng/mL) | Accuracy (%Bias) | Calculated conc. (ng/mL) | Accuracy (%Bias) |
| 150.0 | 136.4 | -9.0 | 155.4 | 3.6 | 146.8 | -2.1 |
| 250.0 | 271.0 | 8.4 | 228.9 | -8.5 | 151.4 | *Dev |
| 500.0 | 506.5 | 1.3 | 516.7 | 3.3 | 549.2 | 9.8 |
| 1000.0 | 1064.2 | 6.4 | 1060.6 | 6.1 | 980.0 | -2.0 |
| 2500.0 | 2302.6 | -7.9 | 2457.1 | -1.7 | 2421.1 | -3.2 |
| 5000.0 | 5098.9 | 2.0 | 4769.3 | -4.6 | 4582.0 | -8.4 |
| 7500.0 | 7092.6 | -5.4 | 7390.1 | -1.5 | 7835.3 | 4.5 |
| 10000.0 | 10427.8 | 4.3 | 10321.9 | 3.2 | 10135.6 | 1.4 |
| 992 Monoclonal antibody | ||||||
| Nominal conc. (ng/mL) | Calculated conc. (ng/mL) | Accuracy (%Bias) | Calculated conc. (ng/mL) | Accuracy (%Bias) | Calculated conc .(ng/mL) | Accuracy (%Bias) |
| 150.0 | 141.1 | -5.9 | 152.6 | 1.7 | 128.8 | -14.1 |
| 250.0 | 265.5 | 6.2 | 222.7 | -10.9 | 222.4 | -11.0 |
| 500.0 | 436.1 | -12.8 | 513.3 | 2.7 | 584.6 | 16.9 |
| 1000.0 | 1147.0 | 14.7 | 1119.4 | 11.9 | 1122.2 | 12.2 |
| 2500.0 | 2721.4 | 8.9 | 2428.7 | -2.9 | 2466.2 | -1.4 |
| 5000.0 | 4489.4 | -10.2 | 4730.4 | -5.4 | 4855.1 | -2.9 |
| 7500.0 | 6643.5 | -11.4 | 7646.6 | 2.0 | 7514.5 | 0.2 |
| 10000.0 | 11056.0 | 10.6 | 10086.2 | 0.9 | 10006.2 | 0.1 |
| Total MABS | ||||||
| Nominal conc. (ng/mL) | Calculated conc. (ng/mL) | Accuracy (%Bias) | Calculated conc. (ng/mL) | Accuracy (%Bias) | Calculated conc. (ng/mL) | Accuracy (%Bias) |
| 300.0 | 290.1 | -3.3 | 297.9 | -0.7 | 240.1 | -20.0 |
| 500.0 | 527.1 | 5.4 | 486.3 | -2.7 | 349.4 | *Dev |
| 1000.0 | 935.2 | -6.5 | 1039.0 | 3.9 | 1101.4 | 10.1 |
| 2000.0 | 2140.5 | 7.0 | 2156.1 | 7.8 | 2250.6 | 12.5 |
| 5000.0 | 4707.9 | -5.8 | 4589.8 | -8.2 | 4977.7 | -0.4 |
| 10000.0 | 10526.8 | 5.3 | 9673.2 | -3.3 | 9922.9 | -0.8 |
| 15000.0 | 14727.5 | -1.8 | 15267.2 | 1.8 | 14693.3 | -2.0 |
| 20000.0 | 19944.9 | -0.3 | 20290.7 | 1.5 | 20114.1 | 0.6 |
| *Dev: Deviation from nominal concentration | ||||||
Table 12: Individual standard curve concentration data of the LC-MS/MS analysis of 1024 mAb, 992 mAb and total mAbs in monkey serum.
With the exception of a single 1024 mAb calibration point, the deviations of the back-calculated concentrations from their nominal values were all within ±20.0% (within ±25.0% at LLOQ) for all the surrogate peptides. The correlation coefficients (r) from three batches of calibration samples were 0.999, between 0.993 and 0.999 and 0.999 for QRPGQGLEWIGEINPSSGR peptide (1024 mAb), EVQLQQPGSELVRPGASVK (992 mAb) and VVSVLTVLHQDWLNGK (total mAbs), respectively.
These results indicated that this LC-MS method can provide sensitive, specific, selective, precise and accurate analysis of 1024 mAb, 992 mAb and total mAbs in monkey serum.
Therefore it met the validation criteria set by Health Authorities’ guidelines, industry best practice and opinion leaders’ white papers in bioanalytical method validation.
Discussion
In-silico analysis led to the selection of 3-4 candidate signature peptides specific for each mAb, that were then tested using LC-MS to assess sensitivity and selectivity in the real matrix. An additional criterion that guided the selection was to select, as far as possible, tryptic peptide pairs deriving from homologous mAb regions. This was taken to allow for the possibility of differential in-vivo degradation on different parts of the mAbs. Selecting peptides from paired regions would normalize the measured concentration by leveling off possible differential catabolic effects.
This was not entirely possible, since the sensitivity and selectivity for the N-terminus 1024 mAbs candidate were not ideal. Nevertheless, the two selected specific peptides derive from Heavy Chain N-terminus regions of the mAbs.
Since the quantitative method had to be developed in a non-human matrix and the mAbs belong to the h-IgG1 class, it was hypothesized that a further improvement could be the quantitation of peptide(s) not present in monkey matrix but common to the two mAbs. This had already been reported by Furlong et al. [14], but in our work the investigation was extended. We used them for the total quantitation of the mAbs, but also attempted a comparison between the concentrations from the mAbs specific signature peptides and the concentration obtained from non-specific peptides. The added value lies not only in the consistency of the concentrations obtained, but in the help it can provide in understanding if and how the two mAbs degrades in-vivo.
To this end, first of all we had to prove that the method (for each of the three signature peptides independently) was robust, sensitive and specific enough to meet design requirements. This was successfully demonstrated by assessing linearity, accuracy and precision, sensitivity, selectivity, specificity, and matrix effect according to GLP industry guidelines for each of the three signature peptides.
Upon this solid basis, we were then able to confirm, that the total mAbs concentration in spiked monkey serum samples was consistent with the sum of the specific single concentrations from the two mAbs within the same accuracy criteria that were applied for method performance assessment, i.e. 20% BIAS% (25% for LLOQ) (Table 12): 94% of the results met the acceptance criteria. Of course this should be further verified doing incurred samples reanalysis, and in spiked incurred samples to have a more complete picture of the applicability of the principle.
As a future perspective, this method represents a seed of moving the LC-MS bottom-up approach to more reliable and informative quantification procedure.
In real samples, due to catabolic processing, we could hypothesize four different situations (Table 13). In the first case the sum of 992 mAb and 1024 mAb is identical to the concentration determined for the total mAbs: we could therefore infer that there is not any difference in the catabolic fate of the different HC regions. In the second situation, the HC N-terminus degrades differently with respect to the HC C-terminus, but identical degradation has occurred on the two HC N-termini of the two mAbs. Situation 3 is the opposite case from situation 2: there is a differential degradation between the two HC N-termini of the two mAbs, but no differences on the HC C-termini. Finally, as in case 4, the two HC chains are subject to different degradation processes. For the first time, these kinds of measurements will allow identification of which part of the molecule is impacted by degradation and connect this parameter directly to their relative body exposure. This information is of paramount importance in molecule design, and can be used to finetune the molecular structure in order to maximize its efficacy.
| Measured signature peptide concentrations | ||||
|---|---|---|---|---|
| Condition | 992 mAb | 1024 mAb | Tot mAbs | Degradation inference |
| 1 | C992 = C | C1024 = C | =2*C | = at HC N-term and at C-term Identical degradation |
| 2 | C992 = C | C1024 = C | ≠2*C | = at HC N-term ≠ C-term. Different degradation at C-term |
| 3 | C992 | C1024 | = C992+C1024 | ≠at HC N-term = at C-term, but the same on individual mAb at HC N-term and C-term. Different catabolism of one mAb |
| 4 | C992 | C1024 | ≠C992+C1024 | Different catabolism, ≠ at HC N-term ≠ at C-term |
Table 13: The four possible situations that can be reported from the measurement of in-vivo samples with the multi-analyte method, with some potential implications described. C: concentration, C992 specific 992mAb concentration, C1024 specific 1024mAb concentration, HC heavy chain. Two values are considered as identical if the differences are within 20% of BIAS% (25% at LLOQ).
References
- Peghini PL, Iwamoto M, Raffeld M, Chen YJ, Goebel SU, et al. (2002) Overexpression of epidermal growth factor and hepatocyte growth factor receptors in a proportion of gastrinomas correlates with aggressive growth and lower curability. Clin Cancer Res 8: 2273-2285
- Damstrup L, Kuwada SK, Dempsey PJ, Brown CL, Hawkey CJ, et al. (1999) Amphiregulin acts as an autocrine growth factor in two human polarizing colon cancer lines that exhibit domain selective EGF receptor mitogenesis. Br J Cancer 80: 1012-1019.
- Wong AJ, Ruppert JM, Bigner SH, Grzeschik CH, Humphrey PA, et al. (1992) Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci U S A 89: 2965-2969.
- Arteaga CL (2001) The epidermal growth factor receptor: from mutant oncogene in nonhuman cancers to therapeutic target in human neoplasia. J Clin Oncol 19: 32S-40S.
- Mendelsohn J, Baselga J (2006) Epidermal growth factor receptor targeting in cancer. Semin Oncol 33: 369-385.
- Wheeler DL, Dunn EF, Harari PM (2010) Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nat Rev Clin Oncol 7: 493-507.
- Friedman LM, Rinon A, Schechter B, Lyass L, Lavi S, et al. (2005) Synergistic down-regulation of receptor tyrosine kinases by combinations of mAbs: implications for cancer immunotherapy. Proc Natl Acad Sci U S A 102: 1915-1920.
- Ben-Kasus T, Schechter B, Lavi S, Yarden Y, Sela M (2009) Persistent elimination of ErbB-2/HER2-overexpressing tumors using combinations of monoclonal antibodies: relevance of receptor endocytosis. Proc Natl Acad Sci U S A 106: 3294-3299.
- Spangler JB, Neil JR, Abramovitch S, Yarden Y, White FM, et al. (2010) Combination antibody treatment down-regulates epidermal growth factor receptor by inhibiting endosomal recycling. Proc Natl Acad Sci U S A 107: 13252-13257.
- Pedersen MW, Jacobsen HJ, Koefoed K, Hey A, Pyke C, et al. (2010) Sym004: a novel synergistic anti-epidermal growth factor receptor antibody mixture with superior anticancer efficacy. Cancer Res 70: 588-597.
- Koefoed K, Steinaa L, Søderberg JN, Kjær I, Jacobsen HJ, et al. (2011) Rational identification of an optimal antibody mixture for targeting the epidermal growth factor receptor. MAbs 3: 584-595.
- Skartved NJ, Jacobsen HJ, Pedersen MW, Jensen PF, Sen JW, et al. (2011) Preclinical pharmacokinetics and safety of Sym004: a synergistic antibody mixture directed against epidermal growth factor receptor. Clin Cancer Res 17: 5962-5972.
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, et al. (2005) Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM (ed) The Proteomics Protocols Handbook. Humana Press, pp. 571-607.
- Furlong MT, Zhao S, Mylott W, Jenkins R, Gao M, et al. (2013) Dual universal peptide approach to bioanalysis of human monoclonal antibody protein drug candidates in animal studies. Bioanalysis 5: 1363-1376.
- DeSilva B, Garofolo F, Rocci M, Martinez S, Dumont I, et al. (2012) 2012 white paper on recent issues in bioanalysis and alignment of multiple guidelines. Bioanalysis 4: 2213-2226.
- Stevenson L, Garofolo F, DeSilva B, Dumont I, Martinez S, et al. (2013) 2013 White Paper on recent issues in bioanalysis: 'hybrid'--the best of LBA and LCMS. Bioanalysis 5: 2903-2918.
- European Medicines Agency (2011) Guideline on bioanalytical method validation, pp. 1-23.
- Food and Drug Administration (2001) Guidance for Industry: Bioanalytical Method Validation. U.S. Department of Health and Human Services, pp. 1-25.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 18158
- [From(publication date):
August-2015 - Jul 06, 2025] - Breakdown by view type
- HTML page views : 13453
- PDF downloads : 4705