Research Article Open Access
Isolation and Evaluation of PAH Degrading Bacteria
| Raden Darmawan1,3*, Haruhiko Nakata1, Hiroto Ohta1, Takuro Niidome1, Kiyoshi Takikawa2 and Shigeru Morimura1 | ||
| 1Graduate School of Science and Technology, Kumamoto University, Kumamoto, Japan | ||
| 2Center for Marine Environment Studies, Kumamoto University, Kumamoto, Japan | ||
| 3Department of Chemical Engineering, Institut Teknologi Sepuluh Nopember (ITS), Surabaya, Indonesia | ||
| Corresponding Author : | Raden Darmawan Graduate School of Science and Technology Kumamoto University Kumamoto, Japan Tel: +81-96-342-3669 Fax:+81-96-342-3669 E-mail: rdarmawan09@gmail.com |
|
| Received February 23, 2015; Accepted March 24, 2015; Published March 27, 2015 | ||
| Citation: Darmawan R, Nakata H, Ohta H, Niidome T, Takikawa K, et al. (2015) Isolation and Evaluation of PAH Degrading Bacteria. J Bioremed Biodeg 6:283. doi:10.4172/2155-6199.1000283 | ||
| Copyright: © 2015 Darmawan R, et al. This is an open-a ccess article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | ||
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
||
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
Polycyclic aromatic hydrocarbons (PAHs) degrading bacteria can be isolated from polluted coastal environments. They were close to Burkholderia fungorum and Mycobacterium gilvum, and had nidA, nidA3, pdoA2, and pcaH genes. In the pyrene and fluoranthene degradation experiment using a minimum medium, B. fungorum isolate no. 1 and M. gilvum isolate no. 13 were able to degrade 98.6% + 1.9 of pyrene after 20 days, and 99.9% ± 0.1 after a 16-day incubation period, respectively. Moreover, fluoranthene could be consumed by B. fungorum isolate no. 1 and M. gilvum isolate no. 13 at the level of 99.6% ± 0.7 after 24 days, and 100% after a 28-day incubation period, respectively.
| Keywords |
| Isolation; Evaluation; PAH; Minimum medium; HPLC |
| Introduction |
| Polycyclic aromatic hydrocarbons (PAHs) are one class of organic pollutants that are considered the most hazardous due to their toxic, mutagenic, and carcinogenic properties [1]. Generally, PAHs are generated from pyrogenic, biogenic, and petrogenic sources [2]. PAHs may be classified on the basis of their aromatic benzene rings number; low-molecular-weight (LMW) compounds consist of two or three rings, and high-molecular-weight (HMW) compounds, more than three rings [3]. |
| In recent years, there has been an increasing amount of literature on the removal of PAHs. One of the most widely used and promising technology to break down the pollutants is bioremediation, which uses selected microorganisms such as fungi, yeast, and bacteria [4]. For the destruction of PAH contaminants, bioremediation is considered a better process than chemical or physical remediation processes because the technique is economical and efficient [5]. Bacteria have been studied extensively, and there is information regarding their ability to degrade xenobitics, including PAHs [6]. PAHs could be degraded by bacteria under aerobic conditions through the initial oxidation of the aromatic ring, which is catalyzed by the dioxygenase enzyme. nidA and nidB genes are encoding genes for large and small subunits of napthalene-inducible dioxygenase [7,8]. The pcaH was significant in the β-ketoadipate pathway [9] where the pathway as it conserves biochemically, and is a major class of non-heme-iron containing dioxygenase [10]. Moreover, a dioxygenase gene, nidA3B3, was detected in Mycobacterium vanbaalenii PYR-1 as an alternate degradation pathway which could catalyze both the initial dihydroxylation of pyrene [11] to be pyrene cis-4,5-dihydrodiol, and an alternate detoxification pyrene pathway to be pyrene cis-1,2-dihydrodiol [12]. The pdoA2 gene had role in the oxidation of PAHs and the detoxification of PAH catechols into more hydrosoluble compounds [12,13]. |
| A high concentration of PAH compounds was detected in sediment from Tanoura Bay, Yatsushiro Sea, Japan, with a mean concentration of 30,200 ng/g (dry weight), in which the two highest concentration of PAHs were fluoranthene and pyrene [14]. The PAHs are two toxic pollutants of the 16 PAHs considered a priority by the U.S.EPA, and are good aromatic ring models for studying the biodegradation of HMW PAHs [15]. The mean concentration of PAHs in Tanoura Bay was approximately 3 times higher than the heavily polluted concentration in Tokyo Bay, with a mean concentration of 9,546 ng/g [16]. |
| Due to their spacious distribution causing ubiquitous pollutants and environmental problems, therefore, there is urgent requisition for cleaning up the pollutants. The recent study was designed to isolate and evaluate PAHs (pyrene and fluoranthene) degrading ability for the future bioremediation in the polluted coastal site. nidA, nidA3, pdoA2, and pcaH genes were targeted in the investigation of the PAHs degrading ability [17]. |
| Materials and Methods |
| Chemicals and preparation of stock solution |
| Fluoranthene and pyrene were purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan) and Nacalai tesque, Inc. (Kyoto, Japan), respectively. Each PAH stock solution was prepared in acetone with a concentration of 5 mg/mL. All chemicals used in this research were laboratory grade, with>98.0% analytical standards. |
| Preparation of sample |
| Sediment samples were collected with a sterile spatula on November 26, 2011 and October 3, 2013 from surface sediment during low tide in Tanoura Bay, Yatsushiro Sea. The samples were then entered into a 50-mL polypropylene conical tube and placed within sterile polythene bags containing ice to be brought to the laboratory. |
| Enrichment culture and isolation of PAH-degrading bacteria |
| per litre. The culture medium was adjusted to a final pH of 7.00 by using 0.5 mol/L of sodium hydroxide solution. The MM-pyrene plates were prepared with MM and 1.5% of powder agar, followed by the spraying of 2000 μg of pyrene on the surface of the solid medium. A portion of 0.5 g soil samples were added into the 200 mL-Erlenmeyer flask containing 50 mL of MM and 100 mg/L of pyrene. It was shaken in an orbital shaker at a speed of 180 rpm at 30oC in the dark. The enrichments were incubated under aerobic conditions for four weeks. One mL aliquots of enrichment cultures were transferred into another sterile Erlenmeyer flask containing fresh MM and pyrene. One mL of aliquots was also serially diluted subsequently, and 0.1 mL of culture broth was spread onto the MM-Pyrene plates. They were incubated at 30oC and routinely checked for colony growth. After about two weeks, the bacterial colonies were growing and showing zones of pyrene clearing. The colonies were picked and serially diluted, and then streaked onto new MM-Pyrene agar plates. A single colony was picked and streaked again on other new MM-Pyrene plates. |
| Polymerase chain reaction (PCR) |
| Thirteen isolates were examined for amplification of the target genes. All PCR amplifications were carried out using the TAKARA TP600 thermal cycler (Takara Bio, Inc, Kyoto, Japan). The Go-Taq Master Mix (Promega, Madison, Wisconsin) was used with 49 μL of total volume. The PCR reagent mix consisted of colony picked up as a template, 2 μL of forward and reverse primer, of which the concentration was 25 μM each, 25 μL of Go-Taq Green, and 20 μL of nuclease-free water. Sequences of used primer sets were summarized in Table 1. After preheating at 95°C for 5 minutes, 30 cycles of heating at 95°C for 30 s, annealing, and extension at 72°C for 2 minutes were performed. Annealing temperatures were 62°C for 30 s, 63°C for 45 s, 54°C for 45 s, 54°C for 45 s, and 62°C for 60 s applied with primer sets including nidA 508F/508R, CD34F/CD34R, pdoA2F/ pdoA2R, nidA3F/ nidA3R, and 27F/518R, respectively. Five μL of PCR products and 1 μL of loading buffer were mixed and quantified by electrophoresis in 1.5% (w/v) agarose gel (Takara L03). Subsequently, the electrophoresis results were visualized by staining the gels in 5 mg/L of ethidium bromide solution for 20 minutes. UV transillumintor (Toyobo Fas III, Tokyo, Japan) was used to view the targeted DNA band. After the targeted band appeared, the remaining PCR products were purified by a PCR clean-up kit (Mo Bio Laboratories, Inc, Canada) according to its protocol. |
| Homology analysis of isolated strains |
| The obtained partial 16S rRNA gene sequences were compared to those in the National Center for Biotechnology Information (NCBI) database using the Basic Local Alignment Search Tool (BLAST) [18]. |
| PAH-degradation experiments |
| PAH-degradation experiments were carried out using test tubes containing 2 mL of MM, and added PAHs with a final concentration of 100 mg/L. The PAHs used in this study were pyrene and fluoranthene. The inoculum was prepared by culturing concentrated strains in 0.05× Luria Bertani (LB) medium at 30°C for 24 hours. Subsequently, after centrifugation at 10,000 rpm for 10 minutes, the cells were harvested and washed twice with a 0.85% NaCl solution. The optical density of the cell was adjusted by using a UV-visible spectrophotometer (UV- 1700 Model TCC-240A, Shimadzu, Japan) at 600 nm to OD of 0.1 [19], The initial colony forming unit (CFUs) was calculated through the growth on the LB agar plates, where it was 7 × 106 CFU/mL and 6 × 106 CFU/mL for B. fungorum isolate no. 1 and M. gilvum isolate no. 13, respectively. The cell inoculum of 1 mL was added to 50 mL of MM and divided to each test tube of 2 mL by shaking at 180 rpm for 1 hour previously. The culture was shaken and incubated under aerobic conditions in the dark in a shaker at 30°C and at a speed of 180 rpm. Abiotic and growth control were performed by adding PAHs into MM without inoculum, and inoculum into MM without PAHs, respectively. Every 4 days, the samples were taken and examined for the remaining amount of PAHs and optical density (OD). The remaining PAH concentration was analyzed using the HPLC method, and the growing cells were measured by spectrophotometer. |
| Measurement of PAHs |
| The residual concentration of PAHs was extracted using a C18 cartridge before they were injected to the HPLC. The solid-phase extraction (SPE) method was chosen for the PAHs extraction because the SPE recovery values for all compounds of PAHs were better than the liquid-liquid extraction (LLE) [20]. Dichloromethane (CH2Cl2) was used as a solvent for the elution step of the analytes because of its low boiling temperature (39.6oC). Acetonitrile with 0.1% of trifluoroacetic acid (TFA) was used as a mobile phase. TFA was used to enhance the mass of the cluster ions to acquire the exact measurements of mass without increasing it [21]. The remaining concentration of pyrene and fluoranthene were analyzed using Hitachi HPLC equipped with FL Detector L-2485 and Diode Array Detector L-2455. The software used was EZChrom Elite version 3.1.7, assembled with an L-column ODS (4.6 mm I.D. × 150 mm, Chemicals Evaluation and Research Institute, Japan). The ODS column was used for the reverse-phase chromatography analysis. The absorption wavelength was adjusted at 250 nm and at 350 nm. |
| Results and Discussion |
| Homology analysis of isolated strains |
| Thirteen PAH-degrading bacteria were isolated from enrichment cultures containing MM, contaminated soil, and pyrene. They grew rapidly on MM-pyrene agar plates and showed clear zone around their colonies, and had nidA gene. Therefore, they were identified through the analysis of the partial 16S rRNA gene sequence. As presented in Table 2, the isolated bacteria clasified four classes, namely β-Proteobacteria, γ-Proteobacteria, α-Proteobacteria and Actinobacteria with 98-99% of similarity. |
| Pyrene degradation ability of isolates |
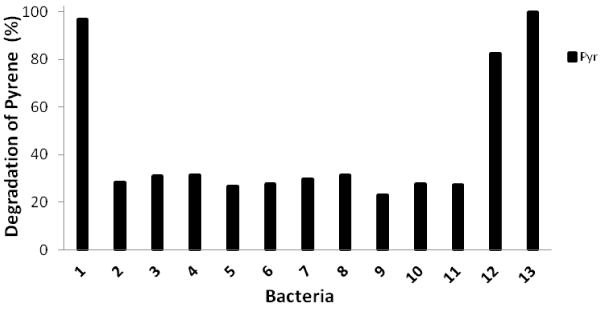
| Biodegradation tests of pyrene with thirteen identified bacteria within 16 days of incubation were carried out and their degradation abilities were shown in Figure 1. The percentage of pyrene degradation was from 26.3%-31.5% for 10 isolates. However, isolate no. 1 (close to Burkholderia fungorum), isolate no. 12 (close to Mycobacterium vanbaleeni) and isolate no. 13 (close to Mycobacterium gilvum) showed high degradation percentage such as 96.2%, 82.2% and 100%, respectively. |
| The PCR amplifications targeting nidA, nidA3, pdoA2, and pcaH genes were carried out for all isolates. Only three isolates, namely isolate no. 1, isolate no. 12 and isolate no.13 which showed high pyrene degradation ability had all four genes. Isolate no. 9, which was close to Pseudomonas vancouverensis, had nidA and pcaH genes. Other 9 isolates had only nidA gene. It seemed that the whole pathway was necessary for the complete degradation of pyrene. |
| The nidA, nidA3, pdoA2, and pcaH genes were identified for the degradation of PAHs in other reports [7,9,11,13]. The naphthalene inducible pyrene dioxygenase gene (nidA), with the α-subunit pyrene dioxygenase, has a critical role in the initial hydroxylation of PAHs, which was identified to be a transmembrane protein; PAHs that are hydrophobic tend to split into the cell membranes [22]. Furthermore, particularly for pyrene, M. vanbaalenii PYR-1 yielded pyrene cis-4,5- dihydrodiol using this enzyme during the initial degradation pathway [12]. The presence of the pcaH gene-encoding enzyme of protocatechuate 3,4-dioxygenase has an important part in the β-ketoadipate pathway [9]. The existence of the nidA3 and pdoA2 genes suggested that these genes had key roles for the degradation of pyrene into a more hydrosoluble compound in the 16 days of cultivation. The pdoA2 gene encoding of the α-subunit of phenanthrene ring-hydroxylating oxygenase was involved in the initial PAH-degradation pathway [12]. This gene has a role in the detoxification of PAH catechols into more hydrosoluble compounds by delivering less reactive methoxy-derivatives compounds. Furthermore, the nidA3 gene encoding of the α-subunit of fluoranthene/pyrene ringhydroxylating oxygenase (RHO) functions as a terminal oxygenase of RHO for degrading fluoranthene and pyrene. The gene played dual roles in the initial and alternative PAH-degradation pathway [12]. It was broken down to be pyrene cis-4,5-dihydrodiol and pyrene cis-1,2- dihydrodiol, respectively. So, the appearance of four genes, including nidA, nidA3, pdoA2, and pcaH, in the bacteria will have a substantial role on PAH degradation completely. |
| Degradation of pyrene and fluoranthene by two isolates |
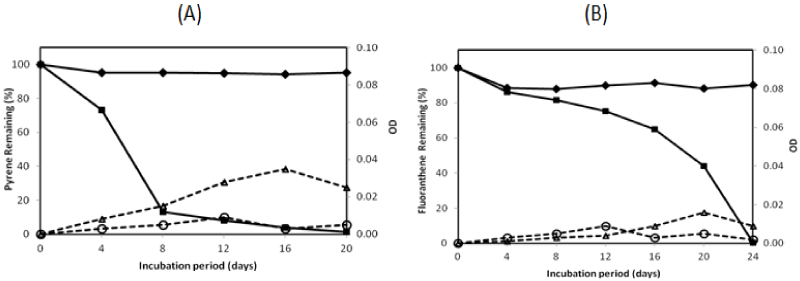
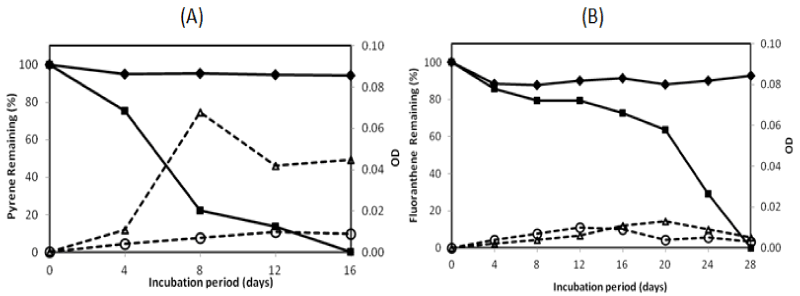
| There were two isolates which could degrade pyrene more than 95% in the 16-day incubation period, namely isolate no. 1 and no. 13. They were investigated on their ability to degrade two PAHs completely by extending the incubation period. The profile of pyrene and fluoranthene in the biodegradation experiments for these bacteria is exhibited in Figures 2 and 3. The experiment on PAHs’ biodegradation showed that B.fungorumisolate no. 1 and M.gilvum isolate no. 13 were able to degrade 98.6% ± 1.9 of pyrene after 20 days and 99.9% ± 0.1 after 16 days, respectively. Furthermore, fluoranthene could be consumed by B.fungorumisolate no. 1 and M. gilvum isolate no. 13 at the level of 99.6% ± 0.7 after the 24-day incubation period, and 100% after 28 days, respectively. The removal of fluoranthene were preceded by an extended degradation lag period, whose results indicated that the lack of fluoranthene removal over the incubation period may be a product of the poor degradation rates, rather than a lack of fluoranthene degradative capacity [23]. |
| Previous studies have reported that there are a very limited number of bacteria taxonomic groups, including the dominant PAH degraders, namely Sphingomonas, Pseudomonas, Burkholderia, and Mycobacterium [24-26]. According to some other reports [27-29], bacteria belonging to genus Burkholderia and Mycobacterium could degrade PAHs significantly. Burkholderia sp. was isolated to utilize several PAHs, such as naphtalene, penantherene, and pyrene [27,30]. The first report on Mycobacterium sp. showed that the strain could degrade 94.9% in 0.5 mg/L, with pyrene as the sole energy source in a minimal basal salt medium after 4 days of incubation [29,30]. Mycobacterium sp. could be the major member genus of the indigenous PAH-degrading bacteria, wherein the genus had been isolated from many polluted environments [25,31]. In this study, B. fungorum isolate no. 1 and M.gilvum isolate no. 13 were isolated from Tanoura Bay, Yatsushiro Sea, as promising bacteria to degrade pyrene and fluoranthene for future bioremediation. These strains could degrade both pyrene and fluoranthene properly into more hydrosoluble compounds. During the PAH-degrading experiment, the medium coloring was observed. There was no switch in color on pyrene degradation. In contrast, in the fluoranthene degradation experiment, both isolates showed the color change of medium to a reddish hue after 10-days of incubation. The medium color probably came from metabolite accumulation [13]. |
| The PAHs’ concentration of abiotic control was decreased from 100% to 94.19%, and from 100% to 88.09% for pyrene and fluoranthene, respectively. The decreasing concentration of abiotic controls was a possibility due to volatilization [23]. The growth controls increased to 0.009 OD and 0.014 OD for B. fungorum isolate no. 1 and M. gilvum isolate no. 13, respectively. The OD of growth controls was lower than with pyrene, in which the OD containing pyrene increased to 0.035 and 0.068 for B.fungorum isolate no. 1 and M.gilvum isolate no. 13, respectively. The higher OD containing pyrene compared with those of growth control in the experiments indicate that both B.fungorum |
| isolate no. 1 and M. gilvum isolate no. 13 could utilize pyrene as a sole carbon and energy source, and there was no significant pyrene toxicity to bacteria. Meanwhile, for fluoranthene, the OD of growth control was higher than the OD of bacteria in the initial incubation. This was followed by an OD of bacteria that was higher than the OD of growth control after 16 days, in which the OD of fluoranthene increased to 0.016 and 0.013 for B.fungorum isolate no. 1 and M. gilvum isolate no. 13, respectively.Some isolates in liquid culture with fluoranthene grew very slowly, although they consistently yielded clearing zones with PYGT plates containing fluoranthene [26]. Furthermore, the differences in cometabolism and the lower degradation pathway may lead to the inefficiency in fluoranthene degradation [31]. Therefore, fluoranthene degradation took longer than the pyrene degradation. |
| The pyrene degradation needed 16-20 days. Meanwhile, the entire fluoranthene degradation required 24-28 days incubation. This result indicated that the process of pyrene degradation was easier than the fluoranthene degradation. It might be caused by the enzymatic specificity from two isolates. Furthermore, an extended degradation lag period occurred between pyrene and fluoranthene degradation was a probability due to the enzymatic induction. |
| PAHs biodegradation spectrums |
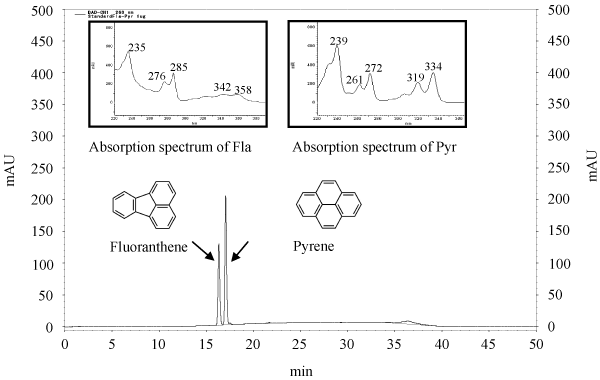
| The standard curve, retention time (RT), and PAHs’ spectrum for pyrene and fluoranthene are shown in Figure 4. The peaks were released at 17.09 minutes and 16.36 minutes for pyrene and fluoranthene, respectively. As reported by some researchers, the pyrene peak showed greater retention than the fluoranthene peak [32,33]. We have also detected the absorption spectrum with maxima at 239, 261, 272, 319, and 334 μm for pyrene, and at 235, 276, 285, 342 and 358 μm for fluoranthene. The absorption spectrum of the HPLC analysis obtained from the diode array detector. The analysis of the pyrene and fluoranthene extracts indicated that the presence of metabolite products appeared more readily. |
| Comparison of PAHs’ degradation with other studies |
| There were other studies using different strains within same genus where a comparison of PAHs’ degradation can be seen in Table 3. It was a positive corelation between used substrate and the PAHs degradation ability, except for fluoranthene degradation with Burkholderia VUN 10001. The capability of fluoranthene degradation from Burkholderia VUN 10001 was lower than B. fungorum isolate no. 1. Pyrene and fluoranthene degradation using some rich substrates, such as basal salts medium (BSM) and basal salts medium with yeast extract (BSMY), have been reported [30,34]. The recent study showed that pyrene degradation ability of Burkholderia VUN 10001 (100%) using BSM liquid medium was higher than B. fungorum isolate no. 1 (87%) using MM medium. Moreover, using BSMY medium, the capability of pyrene and fluoranthene degradation of Mycobacterium 1B can be attained completely after 6 day and 20 day, respectively, although it was supplemented with higher initial concentration of pyrene 250 mg/L. The PAH degradation of Mycobacterium 1B was better than M. gilvum isolate no. 13 utilizing MM medium, where 77.7% of pyrene and 70.9% of fluoranthene can be consumed for 8 and 24 day of incubation period, respectively. The results indicate that both BSM and BSMY mediums could increase the degradation ability of PAHs. The BSM medium added with different additives, such as peptone, glucose, sucrose, ethanol, methanol and yeast have been investigated where the study indicating that the supplement of yeast extract could increase specific growth and phenantherene degradation rate significantly [27]. |
| The research results have shown that all of these genera can utilize PAHs, particularly pyrene and fluoranthene, though they used different media. Four genes detected in these bacteria had important role in PAH degradation by encoding terminal oxygenase of the RHO enzyme. Nevertheless, this study showed that the both isolated bacteria, B. fungorum isolate no. 1 and M. gilvum isolate no. 13 can degrade the PAHs well, despite the use of a limited media, namely minimum medium (MM). The recent study suggests that minimum medium can be used as an alternate medium in PAHs degradation experiments. |
| Conclusion |
| This study has shown that PAH-degrading bacteria can be isolated from a polluted coastal environment and they have a good ability for PAHs degradation. There is a positive correlation between the four genes considered and the ability for HMW PAH degradation. B. fungorum isolate no. 1 and M. gilvum isolate no. 13 can degrade pyrene and fluoranthene almost completely, although the experiment used a finite medium, namely minimum medium (MM). The isolates might be applied as candidate bacteria for future bioremediation in polluted coastal sites. These results may be applicable to hydrosoluble compounds as intermediate metabolite products after PAHs degradation. Further studies need to be carried out in order to elucidate the degradation products. |
| Acknowledgements |
| We would like to thank the Directorate General of Higher Education (DIKTI) - Ministry of Education and Culture of the Republic of Indonesia for the scholarship given to Darmawan in order to pursue his doctoral program. |
References
|
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 |
Figures at a glance
 |
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 22965
- [From(publication date):
May-2015 - Apr 04, 2025] - Breakdown by view type
- HTML page views : 18088
- PDF downloads : 4877
