In-Vitro Application of Synergetic Coalition of Microbial Species Isolated from CETP Sludge and Rhizo-Biome Soil for Escalating Plant Growth
Received: 03-Sep-2021 / Accepted Date: 17-Sep-2021 / Published Date: 24-Sep-2021
Abstract
Application of a microbial consortium having plant growth promoting properties is an economical and environment friendly approach to increase plant growth and crop production yield and this approach can be further incorporated in the utilization of CETP sludge. In this context, this study was conducted to develop an effective consortium from microbial strains isolated from CETP sludge and paddy field soil. 17 bacterial and 8 fungal strains were isolated from both sludge and soil. On the basis of growth study and compatibility test, three bacterial and one fungal strain were selected for the preparation of microbial consortium. Moreover, the prepared consortium was assessed for its efficacy through seed germination method including various parameters. The obtained results confirmed that the seeds treated with microbial consortium showed enhanced seed germination rate and seedling growth hence, confirming the presence of plant growth promoting properties. Therefore, this consortium further can be used as bio-inoculant to increase the plant growth, crop productivity and utilization of sludge generated from CETPs after its treatment with bioinoculant.
Keywords: CETP sludge; Growth study; Compatibilty test; Microbial consortium; Seed germination
Introduction
The word ‘symbiosis’ defines different organisms living together without harming or obstructing each other. A colony enclosed or an amalgamation of multi-species collectively forming an entire unique world. These microbe-microbe interactions are crucial and are associated with environmental recognition followed by transmission of information on molecular and genetic level. Whether it is inter specific or intra specific interaction, they promote the biotransformation of substrates resulting in the increase of biogeochemical cycle rate [1-3]. Moreover, whatever may be the type of interactions, they are indispensable for nourishing and flourishing the microbial communities and their collective activities. Well said ‘someone waste is another one’s treasure’, so these metabolic products secreted by one species become the source of energy for other species. Although these interactions are arbitrated by both physical and molecular mechanisms, but this is the mode of communication between two different species involving plant-microbe, animal-microbe or microbe-microbe interactions [4,5]. In the environment, microorganisms are rarely found as single species populations; rather, there are huge richness and abundance differences within a limited sample, implying that the diverse species have co- evolved, leading to adaptation and specialisation. This leads to the formation of a wide range of cohabitation-facilitating interactions, such as mutualistic and endosymbiotic relationships, as well as antagonistic, competitive, and parasitic relationships.
Plants and microorganisms cohabit or compete for survival via these contact mechanisms, and these cohesive interactions are critical in the treatment of various wastes, whether organic, dry, or hazardous. Bacteria and fungi that act as plant growth promoters can reduce metal phytotoxicity and induce plant growth in two ways: indirectly by stimulating defence mechanisms against phytopathogens, or directly by solubilizing mineral nutrients, producing plant growth promoters, and secreting specific hormones. Therefore, these weapons or defense mechanisms of specific bacteria, actinomycetes or fungi either individually or as a consortium degrade many pollutants and generate byproducts that acts as a catalyst in bio-geochemical cycle. The use of mixed microbial culture: bacteria, fungi and/or algae, for the conversion of pollutants and for increasing soil fertility and plant growth have been used extensively.
Pollution due to inappropriate management of industrial wastewater is one of the major environmental problems. Common Effluent Treatment Plants (CETPs) are considered as one of the viable solution to minimize this pollution [6]. CETPs are treatment systems that are specifically designed to treat wastewater generated by small- scale industrial entities in a cluster [7] and generates a huge amount of sludge which is accumulated in the premises of the CETPs due to lack of further facility for its storage, disposal and treatment in Delhi region. Presently, about 3-4 tons of sludge is generated on daily basis and an average of ~1,400 tons over a year.
Following a two-year period in which CETPs failed to meet the required standards, the National Environmental Engineering Research Institute (NEERI) has proposed short- and long-term plans to improve CETP performance; the former involves the removal of large amounts of sludge, while the latter involves CETP upgradation. As a part of this, a bioremediation process can be included in which sludge treatment with effective measures can be a solution.
By using specific confirmatory methods, interactions between one or two groups/single-single or among different groups can aid in the development of potential consortia/consortium for the treatment of the CETP sludge. In this study, we tried to combine different microbial strain of both bacterial and fungal origins by assembling and selecting the superlative compatible strains isolated from CETP sludge and rhizo-biome of paddy plants. Further, their synergetic effects and their effectiveness on seed germination and plant growth was also examined.
Materials and Methods
Description of sampling site
The CETP from which samples are collected is located at the extreme end (towards eastern side) of the North West district having latitude 28° 42’ 5.8176’’ North and longitude 77° 9’ 51.2568’’ East. It shares a common boundary with North and Central Delhi district. The temperature is about 28-43°C with a population nearly about 3,500. The CETP is managed by the CETP Society as per the Delhi Government CETP Act 2000.
Collection of samples from CETP and rhizo-biome vicinity of paddy field
Collection of sludge samples: Samples of freshly generated and old stored sludge Figure 1 were collected aseptically from CETP premises. Horizontal sampling from the surface (4 corners and middle region) and vertical sampling from a depth of 25 cm was done. The samples were systematically collected, labelled and placed in an ice box and transported to the laboratory for microbiological and physico-chemical analysis.
Collection of samples from rhizo-biome vicinity of paddy field: Soil and rice plants were collected from paddy fields from Bahadurgarh, Haryana. The rice plants (5-6) were uprooted from the irrigated plot along with surrounding soil adherent to the roots from the four corners and one from the center of the plot and were taken to the laboratory in sterile bags for analysis.
Isolation, identification, and characterization of microorganisms
CETP sludge: 1 g of each sample (fresh and old sludge sample) was diluted in a ratio of 1:10 with sterile distilled water followed by serial dilution up to 10-7. 0.1 mL of each aliquot was spread over freshly prepared nutrient agar media (Hi-Media, India) for isolating and quantifying bacteria and for fungus, potato dextrose agar medium (Hi- Media, India) was used. All the plates were incubated at 35 ± 2°C for 24 hour (for bacteria) and 27 ± 2°C for 3-5 days (for fungus).
Rhizo-biome soil: 5 g of soil attached from the roots of rice plants was taken in a sterile test tube and mixed with 90 mL of sterile distilled water and agitated at 150 rpm for 30 min and followed by serial dilution upto 10-7 dilution. An aliquot (0.1 mL) of the suspension was spread on the freshly prepared nutrient agar, rhizobium isolation agar (Hi-Media, India) and azotobacter agar (Hi-Media, India). Plates were incubated for 24 hours at 35 ± 2°C. Similarly, potato dextrose agar medium was used to isolate fungal colonies, incubated at 27 ± 2°C for 3-5 days to achieve vigorous growth and then preserved in 20% glycerol vials at -20°C.
For enumeration of bacterial and fungal colonies, pour plate method was adopted with dilutions up to 10-7 in triplicates to minimize the error. All the respective plates were incubated at 35 ± 2°C for 24 hours (for bacteria) and at 27 ± 2°C for 3 days (for fungus), and number of colonies were counted using a digital colony counter (Scientific, SSC- 105, India) and represented as colony forming unit (CFU/g of soil) (Eq 1):
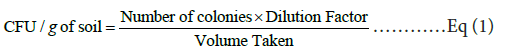
Morphological and biochemical characterization of isolates
Pure culture of individual colony was maintained by sub-culturing them on prepared slants of nutrient agar and potato dextrose agar medium and incubated at 35 ± 2°C for 24 hour (for bacteria) and 27 ± 2°C for 3-5 days (for fungus) respectively. Single colonies were picked up and stained for gram character (bacteria) whereas lactophenol cotton blue staining method was performed for fungus. Microscopic observations were carried out by using bright field/fluorescent microscope (Leica DM3000, Germany). Detailed biochemical characterization was done using Enterobacteriaceae Test Kit (KB001 HiMViC, Hi-Media) and results were interpreted using the chart provided by the manufacturé (for sludge isolates). For rhizo-biome paddy field isolates, certain key biochemical tests (catalase, oxidase, citrate utilization, casein hydrolysis and starch hydrolysis) were performed.
Statistical analysis of bacterial diversity
Simpson index (D) : was used to assess bacterial diversity in all the samples and represented using the given equation (Eq 2):
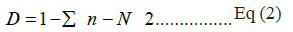
Where, n is the number of specie and N is the total number of species present in a community.
Sorenson’s coefficient (CC):
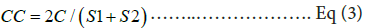
Where, C is the species’ number that is common in any two habitats, S1 is the total number of species found in habitat 1(CETP-fresh sludge/ old sludge/ rhizo-biome soil); S2 is the total number of species found in habitat 2 (CETP-fresh sludge/ old sludge/ rhizo-biome soil) [8].
Compatibility test between selected microbial isolates and preparation of microbial consortium
The sole purpose of conducting compatibility test among microbial strains is to formulate bio-inoculants or consortium that could promote the plant growth. For this purpose, selected bacterial and fungal isolates were tested for compatibility by streaking vertically and horizontally on nutrient agar and potato dextrose agar medium and were then incubated at 35 ± 2°C, 24 hrs (bacteria) and 27 ± 2°C, 3-5 days (fungus) respectively. The plates were observed, and results were recorded for lysis/zone of inhibition (if any) at the juncture of the streaks. Absence of inhibition zone indicates compatible or synergetic relationship between either bacteria-bacteria; bacteria-fungal or fungal-fungal species whereas presence of inhibition zone (if any) indicates incompatibility or antagonistic relationship among the strains. The consortium was composed of three bacterial and one fungal culture. Cells of each isolate were transferred into 100 mL of Luria-Bertani broth (Hi-media) and incubated in shaker incubator (150 rpm) at 30 ± 2°C for 24-48 hours till complete growth was attained.
Growth study experiments of individual and mix consortia
Growth study experiments were performed for both individual species as well as microbial consortia with 24 hour old culture. Growth was studied using UV-VIS spectrophotometer (Thermo fisher Scientific, UV-1900i) and optical density was recorded at 600 nm at different intervals of incubation. Further estimation of biomass concentration and doubling time was also recorded.
Biomass concentration: The density of cell was assessed by measuring the optical density of culture at 600 nm with a UV-VIS spectrophotometer. Concentration of cell was achieved by calculating the weight of dry cell by centrifuging the sample broth at 13,000 g for 5 min (Eppendorf, 5910 R) and then, drying at 80°C to a constant weight [9].
Regeneration/doubling time: Regeneration or doubling time of all individual strains and mix consortium was calculated by using the formula:
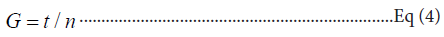
Where, G is the generation time, t is time interval in minutes, and n is number of generations.
In-vitro seed germination by agar plate and test tube methods
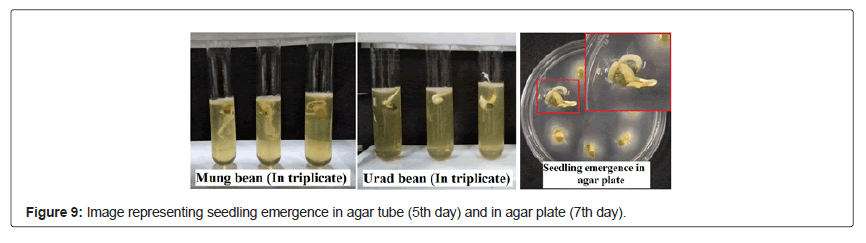
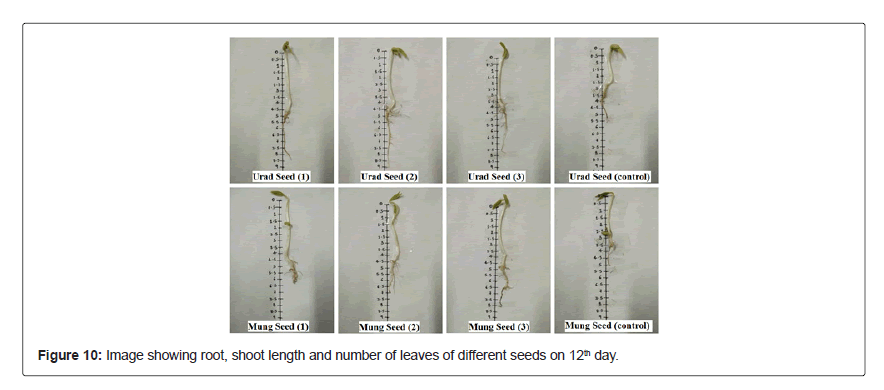
20 seeds of each urad bean (Vigna mungo) and mung bean (Vigna radiata) of fabaceae family were used in this germination test. Seeds were procured from IARI, Pusa, New Delhi. All the seeds were surface sterilized with 70% ethanol followed by 7% sodium hypochlorite solution for 5 min and washed repeatedly with sterile distilled water [10]. Seed treatment was done by submerging the seeds in the microbial consortium containing 108 cells/mL overnight. Parallel set of experiments were performed under similar conditions with sterile distilled water for control set. For experimentation, agar plate and test tube method was used to test the seed germination in in-vitro condition. 0.5% semi-solid agar media was used and pre-treated seed were placed in semi-solid agar plates and test tube (in triplicates) and were incubated at 23 ± 2°C for 12 days. Observations were noted from 4th days onwards for number of seedlings germinated till 12th day to evaluate the standard germination of the seedlings. However, measurement of root and shoot length (in cm) of the seedlings were recorded on 12th day for each replica and mean of root and shoot length was calculated.
Germination parameters
After the end of 12th day, few germination and emergence parameters for growth rate were recorded which are as follows:
FGP% or final germination : percentage represents the total number of seedlings at the end of the experiment:
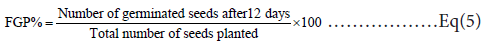
Speed germination (S.G): was calculated by the formula [11]:
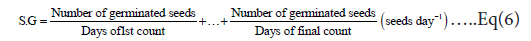
MGT or mean germination time: was calculated by the formula [12]:
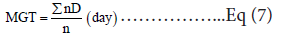
Where n is the number of seeds that germinated on the day D observation, and D denotes the number of days since the start of germination.
CVG or coefficient of velocity of germination: was calculated by the formula [13]:
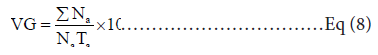
Where N denotes the seeds that germinated on day a, and T denotes the number of days from sowing.
GRI or germination rate index: was calculated by the formula [14]:
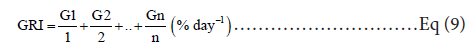
Where G1, G2.Gn are the germination percentages from the first to the 12th day after sowing; 1, 2, and n are the days of the first, second, and final counts, respectively.
Seedling emergence test: The seedling percent emergence was calculated by the formula [15]:

Where Ta denotes the number of days since sowing, Na denotes the number of seeds that have emerged, and S is the total number of seeds used.
Results And Discussion
Physiochemical characterization of CETP sludge samples
Table 1 represents the physico-chemical characteristics and metal contents of fresh and old sludge. It can be observed that the fresh sludge was somehow sandy in consistency while old sludge was silty. Both the samples showed neutral pH but, organic carbon content was found to be high in fresh sludge (5.12%), compared to old sludge (2%). Bulk density of fresh as well as old sludge samples was analogous, whereas porosity of fresh sludge was 80% compared to old sludge (70%). Both the samples exhibited good water holding capacity. The metal content was also found to be remarkably high in fresh sludge compared to old sludge samples.
| Parameter | Fresh sludge | Old sludge |
|---|---|---|
| pH | 7.3 | 7 |
| Electrical conductivity Ms/Cm | 4.16 | 4.5 |
| Organic carbon, (%) | 5.12 | 2.0 |
| Sand, (%) | 60 | 40 |
| Silt, (%) | 32 | 47 |
| Clay, (%) | 3 | 6 |
| Textural class | Sandy loam | Silty loam |
| Bulk density (g/cm3) | 0.7 | 0.73 |
| Porosity, (%) | 80 | 70 |
| Water holding capacity, (%) | 125 | 110 |
| Fe | 10440 | 7562 |
| Cu | 1910 | 960 |
| Pb | 2050 | 1082 |
| Mn | 7530 | 4073 |
| Ni | 1270 | 530 |
| Zn | 110 | 38 |
| Cr | 11770 | 9121 |
Table 1: Physico-chemical characteristics and metal content (mg/kg) of fresh and old sludge samples.
Isolation, characterization and enumeration of microorganisms from CETP sludge and rhizo-biome of paddy field
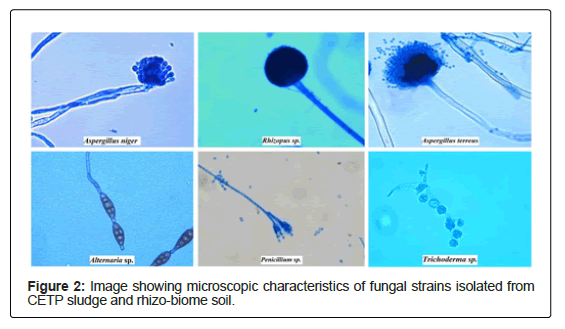
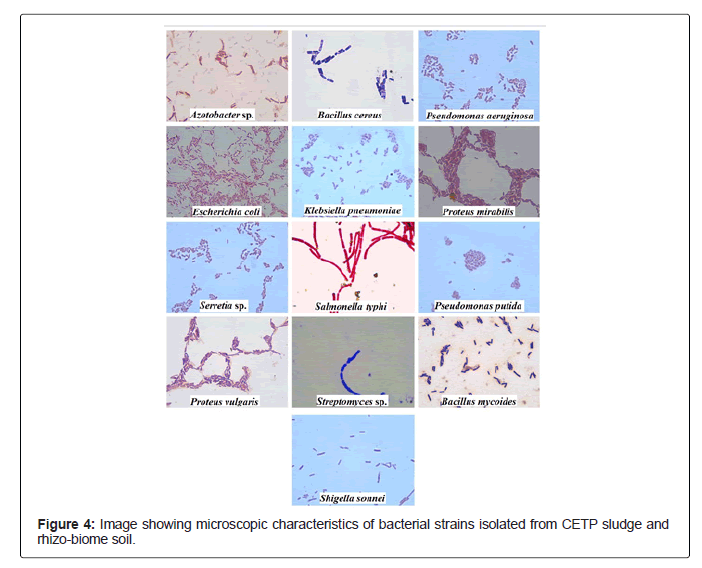
A total of 17 bacterial and 8 fungal strains were isolated from CETP sludge and rhizo-biome soil samples. Morphological characterization of the bacterial isolates based on Gram staining revealed that 9 of CETP isolates were gram negative rods while 1 was gram positive rod whereas isolates from rhizo-biome soil of paddy field showed 4 were gram negative rods and 3 were gram positive rods as observed in Figure 2.
A detailed observation on cell morphology such as shape, texture, margin, size, and count (CFU/g soil/sludge) are presented in the Table 2. A total 14 biochemical tests (as instructed in the manufactures interpretation chart) as illustrated in Table 3 were performed for 10 bacterial strains isolated from fresh and old sludge which were identified as E.coli, Pseudomonas aeruginosa, Bacillus cereus, Klebsiella pnemoniae, Proteus mirabilis, Pseudomonas putida, Salmonella typhi and Shigella sonnei respectively. Whereas for rhizo-biome bacterial strains, key biochemical tests revealed their identity as Azotobacter sp., Bacillus cereus, Serretia sp., Streptomyces sp., Pseudomonas putida, Proteus vulgaris and Bacillus mycoides respectively. Results were further confirmed through Bergey’s manual of determinative bacteriology for preliminary identification of the isolates [16].
| Sample | Isolates | Mean CFU/gm | Colony morphology | Microscopic character | |||
|---|---|---|---|---|---|---|---|
| Size | Shape and margin | Colour | Texture | ||||
| Fresh sludge | FS1 | 27 x10-6 | Large | Circular, Entire | White | Smooth | Gram - , Rod shaped |
| FS2 | 36 x10-6 | Small | Circular, Undulate | White | Matte | Gram - , Rod shaped | |
| FS3 | 18 x10-6 | Medium | Circular, Undulate | White | Shiny | Gram + , Rod shaped | |
| FS4 | 22 x10-6 | Medium | Circular, Entire | Cream | Mucoid | Gram - , Rod shaped | |
| Old sludge | OS1 | 16 x10-6 | Small | Irregular, Undulate | Transparent | Mucoid | Gram - , Rod shaped |
| OS2 | 30 x10-6 | Small | Circular, Undulate | White | Matte | Gram - , Rod shaped | |
| OS3 | 21 x10-6 | Medium | Umbonate, Entire | Translucent | Mucoid | Gram - , Rod shaped | |
| OS4 | 18 x10-6 | Large | Circular, Entire | White | Smooth | Gram - , Rod shaped | |
| OS5 | 38 x10-6 | Small | Circular, Entire | Cream | Shiny | Gram - , Rod shaped | |
| OS6 | 42 x10-6 | Small | Circular, Entire | Transparent | Smooth | Gram - , Rod shaped | |
| Rhizo-biome soil | RS1 | 29 x10-6 | Medium | Circular, Entire | Cream | Slimy | Gram - , Rod shaped |
| RS2 | 17 x10-6 | Medium | Circular, Undulate | White | Shiny | Gram + , Rod shaped | |
| RS3 | 23 x10-6 | Small | Circular, Entire | Pinkish red | Shiny | Gram - , Rod shaped | |
| RS4 | 29 x10-6 | Medium | Irregular, Undulate | White | Rough | Gram + , Rod shaped | |
| RS5 | 27 x10-6 | Medium | Umbonate, Entire | Translucent | Mucoid | Gram - , Rod shaped | |
| RS6 | 35 x10-6 | Small | Irregular, Undulate | Cream | Rough | Gram - , Rod shaped | |
| RS7 | 28 x10-6 | Small | Rhizoid, Curled | White | Matte | Gram + , Rod shaped | |
Table 2: Morphological and microscopic characteristics of bacterial strains isolated from CETP sludge and rhizo-biome soil sample.
| S.No | Biochemical test | Fresh sludge | Old sludge | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| FS1 | FS2 | FS3 | FS4 | OS5 | OS6 | OS7 | OS8 | OS9 | OS10 | ||
| E.coli | P. aeruginosa | B.cereus | K.pneumoniae | Proteus mirabilis | P. aeruginosa | P. putida | E.coli | S. typhi | Shigella sonnei | ||
| 1 | Lysine | + | - | - | + | - | - | + | + | + | - |
| 2 | Ornithine | V | - | - | - | + | - | V | V | - | + |
| 3 | Urease | - | V | - | + | + | V | - | - | - | - |
| 4 | TDA | - | - | - | - | + | - | - | - | - | - |
| 5 | Nitrate | + | + | - | + | + | + | + | + | + | + |
| 6 | H2S | - | - | - | - | + | - | - | - | + | - |
| 7 | Citrate utilization | - | + | + | + | V | + | + | - | - | - |
| 8 | Voges proskauer’s | - | - | + | + | V | - | - | - | - | - |
| 9 | Methyl red | + | - | - | V | + | - | - | + | + | + |
| 10 | Indole | + | - | - | - | - | - | + | + | - | - |
| 11 | Malonate | - | - | - | + | - | - | - | - | - | - |
| 12 | Saccharose | V | - | - | + | V | - | - | V | - | - |
| 13 | Glucose | + | + | + | + | + | + | + | + | + | + |
| 14 | Lactose | + | - | - | + | - | - | - | + | - | - |
“+”means Positive (more than 90%), “-”means Negative (more than 90%), “V”means 11-89% positive
Table 3: Biochemical characteristics of bacterial strains isolated from fresh and old sludge sample.
The LPCB stained fungal isolates were examined under bright field/fluorescent microscope Figure 3. Genus selection identification was done based on their morphological and microscopic examination using taxonomic keys in Table 4 available in the literature [17,18]. List of the identified fungal isolates include Aspergillus niger, Alterneria sp., Rhizopus sp., Penicillium sp., Aspergillus terreus and Trichoderma sp. from fresh, old sludge and rhizo-biome of paddy field.
| Sample | Isolate | Mean CFU/gm | Colonial characteristics | Microscopical Characteristics | Identified fungal strain | |
|---|---|---|---|---|---|---|
| Nature of hyphae | Type of spore | |||||
| Fresh sludge | F1 | 3 x 10-5 | Yellow coloured, cottony colony | Septate | Glubose shaped conidiospore | Aspergillus niger |
| F2 | 6 x 10-5 | Greyish, dense wooly colony | Septate | Glubose shaped conidiospore | Alterneria sp. | |
| Old sludge | F3 | 4 x 10-5 | Yellowish brown, dense cottony colony | Aseptate | Glubose shaped sporangiospores | Rhizopus sp. |
| F4 | 5 x 10-5 | Greyish, dense wooly colony | Septate | Glubose shaped conidiospore | Alterneria sp. | |
| F5 | 3 x 10-5 | Dark green colony with fast spreading mycelium | Aseptate | Globose shaped conidiospores | Penicillium sp. | |
| Rhizo-spheric soil | F6 | 7 x 10-5 | Brown coloured, hairy colony | Septate | Globose shaped conidiospores | Aspergillus terreus |
| F7 | 3 x 10-5 | Cream colored cottony colony | Septate | Oblong shaped conidiospores | Trichoderma sp. | |
| F8 | 6 x 10-5 | Light green coloured, cottony colony | Septate | Round shaped conidiospores | Penicillium sp. | |
Table 4: Morphological and microscopic characteristics of fungal strains isolated from CETP sludge and rhizobiome soil sample.
Enumeration of bacterial and fungal isolates were carried out using colony counting method, represented as mean value, CFU/gm of soil or sludge. Highest bacterial count in fresh sludge was observed to be 36 x 10-6 CFUg-1 whereas in old CETP sludge it was found to be with mean value of 42 x 10-6 CFUg-1 and fungal count with mean value of 6 x 10-5 CFUg-1 and 5 x 10-5 CFUg-1 in fresh and old sludge respectively. Comparing the results with the rhizo-biome soil of paddy field, highest bacterial count with a mean value of 35 x 10-6 CFUg-1 and fungal count of 7 x 10-5 CFUg-1 was recorded.
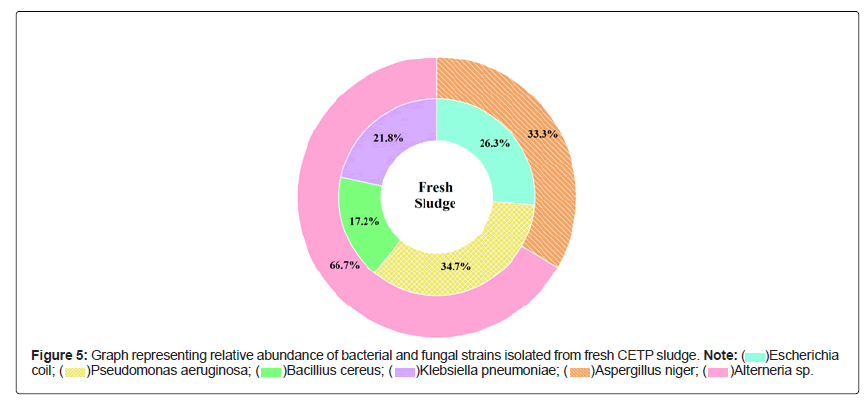
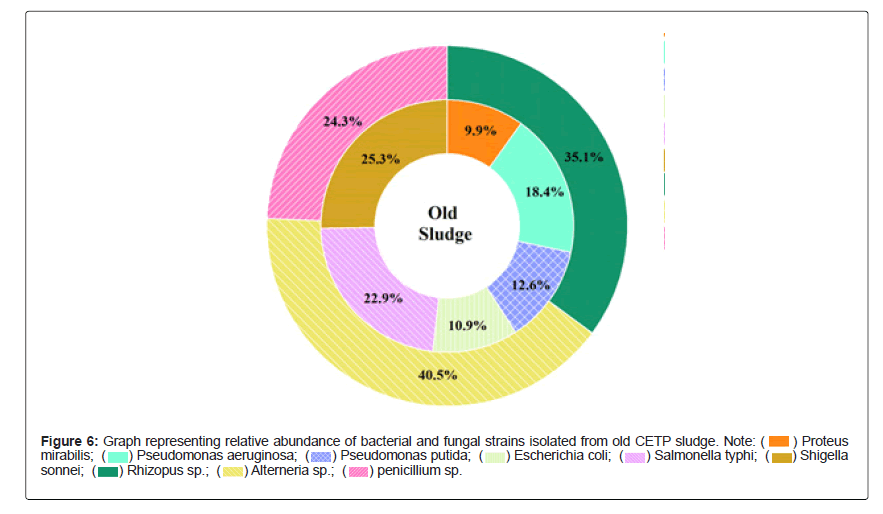
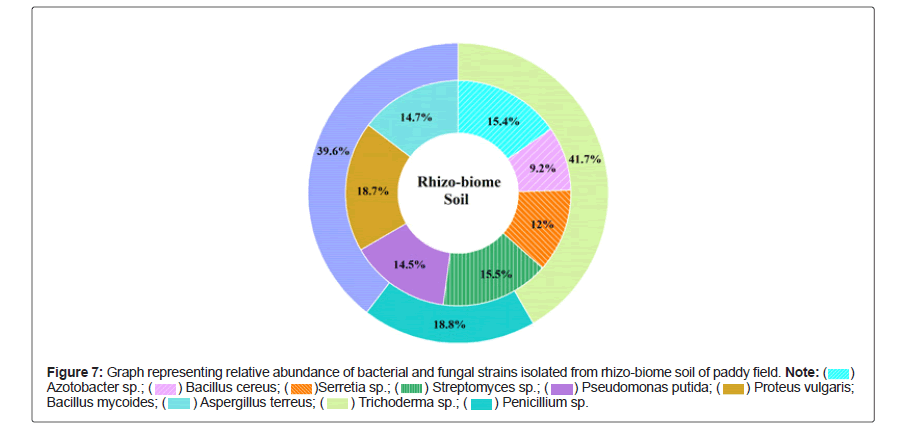
Graphs showing the relative abundance (%) of bacterial and fungal strains isolated from fresh, old sludge are presented in Figures 4 and 5 suggesting that the samples were dominated by Pseudomonas aeruginosa (34.7%), Shigella sonnei (25.3%) and Alterneria sp. (66.7% and 40.5%) respectively. In contrast, to rhizo-biome paddy field sample Figure 6 was dominated by Proteus vulgaris (18.7%), Bacillus mycoides (15.4%) and Aspergillus terreus (41.7%)
Natural pH and high content of organic carbon in the sludge, favors the growth of copious and frequently occurring environmental bacteria namely Bacillus, Azotobacter, Pseudomonas, Actinobacteria [19,20]. On the other hand, some major enteric group of water-borne pathogens such as Escherichia, Shigella, Salmonella and Proteus were predominately found out. Most of these microorganisms pose high health risk potential for humans and animals [21-23] therefore; selective choice was made based on their non-pathogenic ability to induce or stimulate growth promoting substances either directly or with co-existence and competence to build synergetic association.
Statistical analysis of microbial community
The microbial diversity was obtained by using Simpson’s Index and Sorenson’s Coefficient and represented in Table 5. Simpson’s Index showed variation in bacterial diversity from 0.735 to 0.856 whereas for fungal diversity, variation goes from 0.444 to 0.674 suggesting the higher microbial diversity in the rhizo-biome of paddy soil compared to the CETP fresh and old sludge samples. Sorenson’s coefficient indicates the similarity of microbial phyla between two samples, the higher the coefficient the more the similarity between two habitats [8]. Table 5 illustrates that fresh and old sludge had higher microbial similarity i.e. 0.40 for both bacterial and fungal community respectively compared to rhizo-biome of paddy soil.
| Habitat | Bacterial diversity | Fungal diversity | |
|---|---|---|---|
| Simpson’s index | Fresh Sludge | 0.735 | 0.444 |
| Old Sludge | 0.817 | 0.652 | |
| Rhizo-biome Soil | 0.856 | 0.674 | |
| Soreson’s coefficient | Fresh Sludge; Old Sludge | 0.40 | 0.40 |
| Old Sludge; Soil | 0.15 | 0.33 | |
| Fresh Sludge; Soil | 0.18 | 0.00 | |
Table 5: Microbial diversity in sludge and soil habitat.
Observations on compatible/incompatible mixed culture
terial and fungal strains in different combination of two bacteria and one fungal strain, respectively as indicated in Table 6. The results interpret that Trichoderma was compatible with all three of the selected bacterial strains i.e. P. aeruginosa, B. mycoides and B. cereus. T. viride/T. harzianum and P.fluorescens have synergetic compatibility thus inducing plant growth [24]. While the combination of P. aeruginosa, B. mycoides and B.cereus with Aspergillus and Penicillium were incompatible and partially compatible respectively, showing antagonistic activities towards each other. It was observed that the due to profuse growth and sporulation by Penicillium, and Aspergillus sp., the growth of bacterial isolates was restricted. Penicillium sp. which is a common soil inhabitant, and a filamentous fungus is well known for producing diversified range of bioactive secondary metabolites that are anti-bacterial in origin with well-proven biological activities [25,26] whereas most of the group of ubiquity ascomycetes such as Aspergillus sp. secretes secondary by-products and anti-oxidants that inhibit or subside the growth of bacteria when grown on same plate [27,28]. Growth of the bacteria, sporulation, and mycelia growth [29- 31] were some of the significant parameters observed and recorded to be developed into a potential consortium.
| Combination of different isolates | Mycelial growth | Sporulation | Bacterial growth | Compatibility |
|---|---|---|---|---|
| Bacillus mycoides: Pseudomonas aeruginosa: Trichoderma sp. | +++ | ++ | ++ | Compatible |
| Pseudomonas aeruginosa: Bacillus cereus: Trichoderma sp. | +++ | +++ | +++ | Compatible |
| Bacillus mycoides: Pseudomonas aeruginosa: Aspergillus sp. | +++ | +++ | - | Non-compatible |
| Pseudomonas aeruginosa: Bacillus cereus: Aspergillus sp. | +++ | +++ | + | Non-compatible |
| Bacillus mycoides: Pseudomonas aeruginosa: Penicillium sp. | +++ | ++ | + | Partial compatible |
| Pseudomonas aeruginosa: Bacillus cereus: Penicillium sp. | ++ | ++ | + | Partial compatible |
+++ - Good, ++ - Average, + - Poor, -- - No growth
Table 6: Compatibility among different isolates from CETP sludge and paddy soil.
Growth study of selected microbial isolates
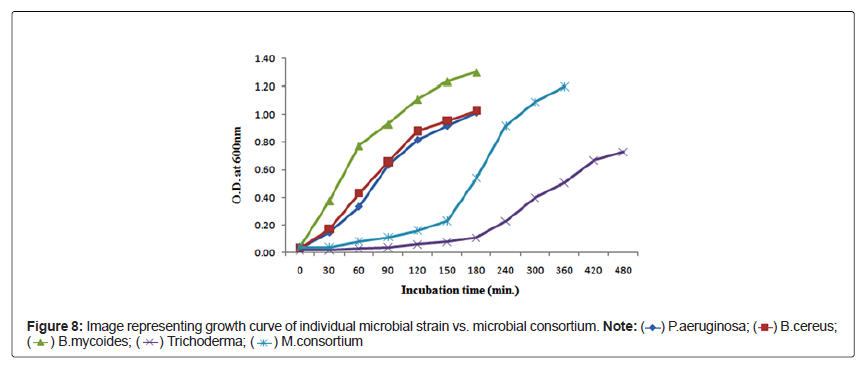
To understand and accelerate the growth of microbial consortium, growth study experiments were performed using individual as well as mixed cultures. Figure 7 represents the growth curve of individual strains versus mixed consortium as function of time. Biomass /cellular concentration and doubling time of the respective isolates and mixed consortium are shown in the Table 7.
| Organism | Biomass concentration | Doubling time |
|---|---|---|
| (mg/ml) | (In minutes) | |
| Pseudomonas aeruginosa | 0.61 | 36.5 |
| Bacillus cereus | 0.73 | 40.8 |
| Bacillus mycoides | 0.7 | 46.2 |
| Trichoderma sp. | 0.56 | 77 |
| Microbial Consortium | 2.42 | 43.3 |
Table 7: Doubling time of individual microbial strains and prepared microbial consortium.
| Seed treatment | Type of seed | Germination parameters | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Emergence index | FGP% | S.G | MGT | CVG | GRI | Mean root length (cm) | Mean shoot length (cm) | Mean number of leaves | ||
| Microbial consortium | Urad bean | 12 | 95 | 8 | 3.4 | 6.2 | 40 | 4.2 | 4.5 | 2 |
| Mung bean | 12 | 100 | 8 | 2.5 | 4.3 | 39.8 | 3 | 4.6 | 2 | |
| Normal distilled water | Urad bean | 10.8 | 85 | 5.8 | 4 | 2.9 | 28.8 | 1.9 | 3.5 | 1 |
| Mung bean | 11.4 | 90 | 5.9 | 2.8 | 2.6 | 29.3 | 1.4 | 3.2 | 2 | |
Table 8: Seed germination parameters after treatment with microbial consortium..
Seed germination by agar plate and tube method
The seeds, urad bean and mung bean treated with microbial consortium showed high final germination percentage i.e. 95 and 100 resp., compared to control (normal distilled water) i.e. 85 for urad seeds and 90 for mung seeds. Moreover, highest germination index and coefficient of velocity of germination was recorded for the seeds treated with microbial consortium which gives an indication of rapidity of seed germination [32]. Speed of germination was also observed to be more than control set. Whereas, the mean germination time was low for consortium treated seeds i.e 3.4 and 2.5 for urad bean and mung bean seed respectively as lower the MGT, the faster a seed population has germinated [33].
Figure 8 represents the emergence of seedling in consortium treated seeds, whereas the root, shoot length and number of leaves of both seeds Figures 9 and 10 treated with microbial consortium and normal distilled water (control) on 12th day is illustrated in Table 8, which shows that consortium treated seeds has increased root and shoot length in compare to the control set, suggesting that the prepared consortium consisting B.cereus, B.mycoides, P.aeruginosa and Trichoderma sp. had plant growth promoting properties and was effective enough to accelerate the seed germination rate and growth of seedling. These results are in favor with the study of Roshni et al. [34] who reported the synergestic effect of microbial consortia application on seed germination rate of wheat seeds, which in our study are mung bean seeds and urad bean seeds.
Conclusion
This research study summarized the isolation, identification and characterization of bacterial and fungal isolates from CETP sludge and rhizobiome soil of paddy field and preparation of a microbial consortium to check its efficacy on seed germination of mung bean (Vigna radiata) and urad bean (Vigna mungo). On basis of all the findings, this study suggests that the application of microbial consortium significantly increased the seed germination rate and seedling growth which indicates the presence of plant growth promoting properties in used microbes i.e. B.cereus, B.mycoides, P.aeruginosa and Trichoderma sp. The results of this study concluded that the prepared microbial consortium have necessary potential to encourage the plant growth and further can be used as bioinoculant to treat the generated CETP sludge to increase its fertility and promotes its utilization.
Acknowledgment
The authors would like to express their gratitude to their Director, CSIR-National Environmental Engineering Research, Institute, Nagpur, for allowing them to carry out this work at Delhi Zonal Center.
Data Availability Statement
All data generated or analyzed during this study are included in this article.
References
- Li J, Gu J (2007) Complete degradation of dimethyl isophthalate requires the biochemical cooperation between Klebsiella oxytoca Sc and Methylobacteriummesophilicum Sr isolated from wetland sediment. Sci Total Environ 380: 181-187.
- Kimura Zi, Okabe S (2013) Acetate oxidation by syntrophic association between Geobacter sulfurreducens and a hydrogen-utilizing exoelectrogen. ISME J 7: 1472-1482.
- Liang J, Bai Y, Hu C, Qu J (2016) Cooperative Mn(II) oxidation between two bacterial strains in an aquatic environment. Water Res 1(89): 252-60.
- Wolin MJ, Miller TL, Stewart CS (1997) Microbe-microbe interactions. Springer 467-491.
- Sharma A, Verma RK (2018) Root–Microbe Interactions: Understanding and Exploitation of Microbiome. Springer 52: 323-339.
- Vyas M, Modhera BK, Vyas V, Sharma A (2011) Performance forecasting of common effluent treatment plant parameters by artificial neural network. ARPN J Eng Appl Sci 6(1): 38-42.
- Mhete M, Eze PN, Rahube TO, Akinyemi FO (2020) Soil properties influence bacterial abundance and diversity under different land-use regimes in semi-arid environments. Scientific African 7: e00246.
- Shim H, Hwang B, Lee S, Kong S (2005) Kinetics of BTEX biodegradation by a coculture of Pseudomonas putida and Pseudomonas fluorescens under hypoxic conditions. Biodegradation 16(4): 319-327.
- Singh AV, Chandra R, Goel R (2013) Phosphate solubilization by Chryseobacterium sp. and their combined effect with N and P fertilizers on plant growth promotion. Arch Agro Soil Sci. 59 (5): 641-651.
- Ellis RA, Roberts EH (1981) The quantification of ageing and survival in orthodox seeds. Seed Sci Technol 9: 373-409.
- Scott S, Jones R, Williams W (1984) Review of data analysis methods for seed germination. Crop Science 24: 1192-1199.
- Esechie H (1994) Interaction of salinity and temperature on the germination of sorghum. J Agron Crop Sci 172: 194-199.
- Copeland LO (1976) Principals of Seed Science and Technology. Burgess publishing company 164-165.
- Garrity G, Brenner DJ, Krieg NR, Staley JR (2005) Bergey’s Manual of Systematic Bacteriology.
- St-Germain G, Summerbell R (2003) Identifying Filamentous Fungi: A Clinical Laboratory Handbook. Rev Inst Med Trop S Paulo 45(3): 152.
- Bonomo RA, Szabo D (2006) Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin Infect Dis 43(2): S49-56.
- Kaskhedikar M, Chhabra D (2010) Multiple drug resistance in Aeromonas hydrophila isolates of fish. Food Microbiol 28: 157-168.
- Hunter PR, Colford JM, LeChevallier MW, Binder S, Berger PS (2001) Waterborne diseases. Emerg Infect Dis 7(3): 544.
- Colwell RR (2004) Infectious disease and environment: cholera as a paradigm for waterborne disease. Int Microbiol 7(4): 285-289.
- Arnone RD, Walling JP (2007) Waterborne pathogens in urban watersheds. J Water Health 5(1): 149-162.
- Rini CR, Sulochana KK (2006) Management ofseedling rot of chilli (Capsicum annuum L.) using Trichoderma spp. and fluorescent pseudomonads (Pseudomonas fluorescens). J Trop Agric 44(1-2): 79-82.
- RanÄić A, Soković M, Karioti A, Vukojević J, Skaltsa H (2006) Isolation and structural elucidation of two secondary metabolites from the filamentous fungus Penicillium ochrochloron with antimicrobial activity. Environ Toxicol Pharmacol 22(1): 80-84.
- Scott N, Sang L, Yukihiro A, Jong A, Hyuncheol O, et al. (2012) Aflaquinolones A-G: secondary metabolites from marine and fungicolous isolates of Aspergillus spp. J Nat Prod 75(3): 464-472.
- Hamed AA, Abdel-Aziz MS, Abd El Hady FK (2018) Antimicrobial and antioxidant activities of different extracts from Aspergillus unguis SPMD-EGY grown on different media. Bull Natl Res Cent 42(29).
- Stahl PD, Christensen M (1992) In vitro mycelial interactions among members of a soil microfungal community. Soil Biol Biochem 24: 309-316.
- Alam M, Fakhru’l-Razi A, Molla A, Abd-Aziz S (2003) Optimization of Compatible Mixed Cultures for Liquid State Bioconversion of Municipal wastewater Sludge. Water, Air, Soil, Pollut 149: 113-126.
- Molla AH, Fakhru'l-Razi A, Abd-Aziz S, Hanafi MM, Alam Mz (2001) In-vitro compatibility evaluation of fungal mixed culture for bioconversion of domestic wastewater sludge. World World J Microbiol Biotechnol 17: 849-856.
- Jones K, Sanders D (1987) The influence of soaking pepper seed in water or potassium salt solutions on germination at three temperatures. J Seed Technol 11(1): 97-102.
- Orchard T (1977) Estimating the parameters of plant seedling emergence. Seed Sci Technol 5(1): 61–69.
- Roshani Khan A, Singh AV, Upadhayay VK, Prasad B (2020) Development of Potential Microbial Consortia and their Assessment on Wheat (Triticum aestivum ) Seed Germination. 38: 6-16.
Citation: Singh S, Sharma S, Yadav N, Goyal SK (2021) In-vitro Application of Synergetic Coalition of Microbial Species Isolated from Cetp Sludge and Rhizo- Biome Soil for Escalating Plant Growth. J Bioremediat Biodegrad 12: 010.
Copyright: © 2021 Singh S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Usage
- Total views: 2973
- [From(publication date): 0-2021 - Apr 21, 2025]
- Breakdown by view type
- HTML page views: 2321
- PDF downloads: 652