Research Article Open Access
Investigating Splicing Variants Uncovered by Next-Generation Sequencing the Alzheimer's Disease Candidate Genes, CLU, PICALM, CR1, ABCA7, BIN1, the MS4A Locus, CD2AP, EPHA1 and CD33
Naomi Clement1#, Anne Braae1#, James Turton1, Jenny Lord1, Tamar Guetta-Baranes1, Christopher Medway1, Keeley J Brookes1, Imelda Barber1, Tulsi Patel1, Lucy Milla1, Maria Azzopardi1, James Lowe2, David Mann3, Stuart Pickering-Brown3, Noor Kalsheker1, Peter Passmore4, Sally Chappell1# and Kevin Morgan1†*; Alzheimer’s Research UK (ARUK) Consortium†1Human Genetics Group, School of Life Sciences, Queens Medical Centre, University of Nottingham, Nottingham, UK
2Neuropathology, School of Medicine, Queens Medical Centre, University of Nottingham, Nottingham, UK
3Clinical Neuroscience Research Group, Greater Manchester Neurosciences Centre, University of Manchester, Salford, UK
4Centre for Public Health, School of Medicine, Dentistry, and Biomedical Sciences, Queen’s University Belfast, Belfast, Northern Ireland, UK
5†The Alzheimer’s Research UK (ARUK) Consortium comprises Peter Passmore, David Craig, Janet Johnston, Bernadette McGuinness, Stephen Todd, Queen’s University Belfast, UK; Reinhard Heun, Royal Derby Hospital, UK; Heike Kölsch, University of Bonn, Germany; Patrick G. Kehoe, University of Bristol, UK; Emma R.L.C. Vardy, University of Salford, UK; Nigel M. Hooper, Stuart Pickering-Brown, University of Manchester, UK; Julie Snowden, Anna Richardson, Matt Jones, David Neary, Jenny Harris, Salford Royal NHS Foundation Trust, UK & University of Manchester, UK; Keeley Brookes, Christopher Medway, James Lowe, Kevin Morgan, University of Nottingham, UK; A. David Smith, Gordon Wilcock, Donald Warden, University of Oxford (OPTIMA), UK; Clive Holmes, University of Southampton, UK
- *Corresponding Author:
- Kevin Morgan
Human Genetics, School of Life Sciences, A Floor
West Block, Room 1306, Queens Medical Centre
University of Nottingham, Nottingham, NG7 2UH
United Kingdom
Tel: +44 115 8230724
E-mail: Kevin.Morgan@nottingham.ac.uk
Received date: October 03, 2016; Accepted date: October 22, 2016; Published date: October 29, 2016
Citation: Clement N, Braae A, Turton J, Lord J, Guetta-Baranes T, et al. (2016) Investigating Splicing Variants Uncovered by Next-Generation Sequencing the Alzheimer’s Disease Candidate Genes, CLU, PICALM, CR1, ABCA7, BIN1, the MS4A Locus, CD2AP, EPHA1 and CD33. J Alzheimers Dis Parkinsonism 6:276. doi:10.4172/2161-0460.1000276
Copyright: © 2016 Clement N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited
Visit for more related articles at Journal of Alzheimers Disease & Parkinsonism
Abstract
Late onset Alzheimer’s disease (LOAD), the most common cause of late onset dementia, has a strong genetic component. To date, 21 disease-risk loci have been identified through genome wide association studies (GWAS). However, the causative functional variant(s) within these loci are yet to be discovered. This study aimed to identify potential functional splicing mutations in the nine original GWAS-risk genes: CLU, PICALM, CR1, ABCA7, BIN1, the MS4A locus, CD2AP, EPHA1 and CD33. Target enriched next generation sequencing (NGS) was used to resequence the entire genetic region for each of these GWAS-risk loci in 96 LOAD patients and in silico databases were used to annotate the variants for functionality. Predicted splicing variants were further functionally characterised using splicing prediction software and minigene splicing assays. Following in silico annotation, 21 variants were predicted to influence splicing and, upon further annotation, four of these were examined utilising the in vitro minigene assay. Two variants, rs881768 A>G in ABCA7 and a novel variant 11: 60179827 T>G in MS4A6A were shown, in these cell assays, to affect the splicing of these genes. The method employed in the paper successfully identified potential splicing variants in GWAS-risk genes. Further investigation will be needed to understand the full effect of these variants on LOAD risk. However, these results suggest a possible pipeline in order to identify putative functional variants as a result of NGS in disease-associated loci although improvements are needed within the current prediction programme in order to reduce the number of false positives.
Keywords
Late onset Alzheimer’s disease; Next generation sequencing; Splicing; Annotation; Functional variations; Minigene assays
Introduction
Late onset Alzheimer’s disease (LOAD) is an insidious neurodegenerative disease responsible for most cases of dementia in the elderly. Despite being widely studied, the disease still lacks a clear pathogenesis. However, there is a strong genetic component to LOAD with 21 disease-risk genetic regions having been identified through genome wide association studies (GWAS) [1]. Due to the nature of GWAS experimental design, the variants associated through GWAS are frequently not the causal functional variants generating the disease association. Instead, one or several variants in linkage disequilibrium (LD) with the GWAS variant are likely to be causal [2,3]. Therefore, due to the large number of the possible causative functional variants within these loci, the mechanistic role that each of the identified 21 disease-risk genes may play in the progression of LOAD remains unknown.
To identify the disease-causing functional variant(s), a number of next-generation sequencing (NGS) studies have been undertaken for the LOAD GWAS loci. Examples include CLU [4], CLU, PICALM and CR1 [5], ABCA7 [6] and ABCA7, BIN1, CD2AP, CLU, CR1, EPHA1, MS4A4/MS4A6A [7]. All studies, apart from [5], have used targeted exome sequencing, ignoring variants which may be found in the noncoding and intronic regions of the loci. Exome sequencing may also miss exonic variants which are found close to exon/intron borders [8]. Therefore these studies may have overlooked mutations which could affect splicing through disrupting donor and acceptor splice sites.
Splicing plays an important role in human genetic disease. Almost a third of currently known disease-causing variants disrupt splicing, although this is likely to be an underestimate [9,10]. Aberrant splicing is implicated in many neurodegenerative disorders [11] and in LOAD, variants causing dysfunctional splicing have been found in some of the risk genes including PICALM and CD33 [12-14].
The aim of this study was to discover potentially functional causative splicing variants in the original nine genes highlighted through GWAS in 2009: CLU, PICALM [15], CR1 [16], ABCA7, BIN1, the MS4A locus, CD2AP, EPHA1 and CD33 [17]. Target enrichment and next generation sequencing (NGS) were used to sequence the entire GWAS locus for each gene as identified through linkage disequilibrium. Coding and non-coding variants were prioritised for causality using functional annotation. Many of the variants were located close to intron/exon boundaries, suggesting a possible role in splicing. These variants were taken forward for further in silico investigation and experimental assessment of functionality.
Methods
NGS
Informed consent was obtained from all participants and the local Ethics Committee approved the study. 96 CERAD post-mortem confirmed LOAD brain tissue samples were obtained from the University of Nottingham Brain Bank (n=50) and the Manchester Brain Bank (n=46). This sample size gave 80% power to detect variants with a minor allele frequency (MAF) as low as 0.85% at a particular location. All samples were Caucasian, 52.8% female, with average age at onset of 70.8 years and APOE alleles: ε2-9.1%; ε3-57.2%; ε4-33.7%, data taken when consent obtained or upon receipt of the biological samples. DNA was extracted using phenol chloroform, quantified using Quant-iTTM ds DNA Broad Range Assay kit (Invitrogen) and combined into eight equimolar pools of 12 for a total concentration of 6 ug per pool (500 ng per sample).
Agilent SureSelect Custom MP3 kit designed on eArray with 5X tiling was used for target enrichment of LOAD associated genes including introns, exons and flanking conserved sequence. For ABCA7, BIN1, CD33, CR1, CD2AP, EPHA1 and the MS4A locus, repetitive elements were masked in SureSelect bait design and the enriched library was sequenced by Source BioScience using 100 bp paired-end sequencing on the Illumina HiSeq 2000 and base called with Illumina Casava 1.9. SureSelect baits for PICALM and CLU were not repeat masked and were sequenced separately on the Illumina GAIIx with 38 bp single-end reads separately. Genomic regions potentially captured by these baiting strategies, as well as the transcript IDs for each of the genes used, are shown in (Table 1).
Raw data was quality control (QC) checked by FastQC prior to alignment to hg19 with BFAST v0.7.0a [18], following the program’s protocol for the read type. Aligned files were manipulated using SAMtools v0.1.18 [19] and the success of the alignment was assessed with SAMtools flagstat function and SAMstat [20]. Variants were called using the pooled data specific program, CRISP [21]. Called variants were filtered following the best practice variant detection suggested by Genome Analysis ToolKit pipeline (GATK v4.0, Broad Institute [22]. Variants were annotated using Ensembl’s Variant Effect Predictor (VEP) [23] supplemented with annotations from Encyclopaedia of DNA Elements (ENCODE) project and PhastCons downloaded from the UCSC genome browser website (http://genome.ucsc.edu/ ENCODE/downloads.html, accessed Nov 2012 [24]) using a custom in-house script. 21 variants identified as potentially affecting splicing (falling within 1-3 bp of exon and 1-12 bp of intron) were carried forward for further investigation. Given the difficulties identifying intronic mutations which affect branch point sites and the large number of intronic variants called, these were not taken forward in this project. Linkage disequilibrium (LD) between the variants of interest and the GWAS variants were calculated with VCFtools [25] using 1000Genomes Phase 1 data for samples of northern European Ancestry [26].
In silico analysis of splicing
Additional in silico programs were used to assess the 21 identified splicing SNPs. Namely, ESEfinder v3 (http://rulai.cshl.edu/cgi-bin/ tools/ESE3/esefinder.cgi?process=home) [27], Berkley Drosophila Genome Project Splice Site Prediction by Neural Network (BDGP) (http://www.fruitfly.org/seq_tools/splice.html) [28] and Human Splicing Finder (HSF) (http://www.umd.be/HSF3/) [29]. All programs were accessed March 2015.
Four potential variants were taken forward that showed differences between the two alleles in all three programs, including a score change of more than 10% between the major and the alternative allele where numerical scores were provided.
In vitro analysis of splicing
As additional brain tissue was not available for all samples in order to perform RNA extractions, as well as the previous DNA extractions already performed, the minigene assay was selected to validate potential splicing variants. This method has been shown to have complete concordance with RT-PCR analysis of patient RNA and is a validated and reliable method for investigating splicing [30].
All methods were performed following manufacturer’s protocol unless otherwise specified. The specific sample in each pool containing the heterozygous splice variant was identified by Sanger sequencing the appropriate exon(s) in each locus.
Minigene primers with the addition of SalI and XbaI restriction enzyme binding sites were designed using the relevant ABCA7, MS4A6A and EPHA1 reference sequences using Primer 3 (v.0.4.0) (http://frodo.wi.mit.edu/primer3/). Amplicons were designed to contain the potential splicing mutations, adjacent exons and intronic sequences (Supplementary Table 1).
| Gene | Transcript ID | Chromosome | Coordinates | BP |
|---|---|---|---|---|
| CLU | NM_001831.3 | 8 | 27450849-27475277 | 24,430 |
| PICALM | NM_007166.3 | 11 | 85665237-85783519 | 118,280 |
| CR1 | NM_000651 | 1 | 207667495-207816719 | 149,224 |
| ABCA7 | NM_019112 | 19 | 1038952âÂ?Â?1066720 | 27,768 |
| BIN1 | NM_139343.2 | 2 | 127778085âÂ?Â?127895723 | 117,638 |
| MS4A LD Locus | NG_016014 NM_152852 XM_005274415 |
11 | 59856028âÂ?Â?60041296 | 185,268 |
| CD2AP | NM_012120 |
6 | 47427281âÂ?Â?47601015 | 173,734 |
| EPHA1 | NM_005232 | 7 | 143082382-143110385 | 28,003 |
| CD33 | NM_001772 | 19 | 51718317-51748546 | 30,229 |
Table 1: Regions of the genome (chromosome number and coordinates) to be targeted by Agilent SureSelect baits for the sequencing project. Coordinates are given relative to hg19. BP refers to how many bases can potentially be covered by the design. The Ensembl transcript IDs presented were utilised in all analysis of the sequence data as well as annotation of variants.
Initial PCR was carried out using 10-100 ng template DNA in 30μl reaction volumes with Expand High Fidelity Taq (Roche). PCR conditions for all reactions were as follows: an initial denaturing step of 2 min at 94°C, followed by 30 cycles of 15 s at 94°C, 30 s at optimized annealing temperature (Ta, Supplementary Table 1) and 40 s, plus 5 s every cycle, at 72°C with a final extension step at 72°C for 7 min.
Amplicons were cloned into pCR 2.1-TOPO vectors using the TOPO TA Cloning kit (Invitrogen, Life Technologies) before being ligated into the exon trap vector, pET01 (MoBiTech) and transformed into NEB High Efficiency 5-alpha chemically competent E. coli cells C2987I. Vector DNA was then extracted following the NucleoBond Xtra Midi Plus EF Kit in an endotoxin free environment as well as being sequenced to confirm the insert sequence was accurately cloned.
Two cell lines were obtained from the European Collection of Cell Cultures (ECCC): CV-1 in Origin, carrying SV40 (COS-7) derived from monkey African green kidney cells and human Caucasian neuroblastoma cells (BE(2)-C). COS-7 is typically used for testing minigene splicing assays as it is easy to transfect and grow, as well as being known to accurately represent the complex eukaryotic splicing environment seen in vivo. BE(2)-C cells were selected in order to investigate possible neurological-specific splicing effects due to the brain’s unique spliceosome. COS-7 were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 10% Foetal Bovine Serum (FBS), 2 mM L-Glutamine, 100 U/ml penicillin-streptomycin and 100 U/ml fungizone and BE(2)-C were cultured in 1:1 Eagle’s Minimal Essential Medium (EMEM):Ham’s F12 with 1% non-essential amino acids, 2 mM Glutamine, 15% FBS, 100 U/ml penicillin-streptomycin and 100 U/ml fungizone. The COS-7 and BE(2) cells were plated out at 3.5 × 105 cells per plate and 6 × 105 cells per plate, respectively. Cells were transfected using TransFact (Promega) and incubated for 24 h. The transfection was repeated in triplicate in each cell line. Total RNA was then extracted utilising the RNeasy Mini Kit (Qiagen) with the additional on column DNase treatment (TURBO DNA-free Kit, Ambion). Total cDNA was synthesised with AffinityScript Multiple Temperature cDNA Synthesis kit (Agilent) using oligodT primers. The cDNA of interest was then PCR amplified in a 30 μl reaction volume using LongAmp Taq (New England BioLabs) and primers specific to the pET01 vector (Forward: GATCGATCCGCTTCCTG, Reverse: CACTGGAGGTGGCCCG). The thermal cycling conditions were: 30 s at 94°C for initial denaturation, followed by 30 cycles of 15 s at 94°C for denaturation, 30 s at 59°C for annealing and 50 s at 65°C for extension and 10 m at 65°C for the final extension. Amplicons were compared by electrophoresis as well as Sanger sequenced in order to undertake a more detailed comparison.
Results
NGS study
An average of 245.5 million reads per pool was obtained. The eight pools of twelve passed all FastQC parameters apart from sequence duplication which is to be expected given the sequencing strategy employed.
The flagstat analysis in SAMtools showed that 99% of reads were mapped correctly and 95-99% of reads were properly paired. CRISP called 3205 variants within the nine loci, with 760 novel variants. The minor allele frequency estimates from CRISP were strongly positively correlated with frequencies found in the 1000 Genomes project indicating successful variant calling (Spearman Correlation Coefficient=0.60, p<0.0001 across all gene regions examined). Following annotation, only 126 exonic variants were called with the majority of variants being non-coding. There were 43 variants in untranslated regions (UTRs) and 44 missense variants (Supplementary Table 2). As defined above, 21 variants were predicted to affect splicing (Supplementary Table 2).
In silico analysis of splicing
All 21 variants were further annotated by three in silico programs, the results of which can be seen in (Supplementary Table 3). From this original list of 21 variants, four were predicted to be splice site variants by at least two programs by altering the donor or acceptor sites (predicted by the BDGP and HSF programs) as well as altering splicing factor binding sites (predicted by ESEfinder) (Table 2). These variants (19:1054696 and rs881768 in ABCA7, 11:60179827 in MS4A6A and rs6967117 in EPHA1) were therefore analysed in vitro in order to assess the accuracy of these predictions.
In vitro analysis of splicing
For two of the variants analysed (19:1054696 in ABCA7 and rs6967117 in EPHA1) there was no difference between the RNA extracted from cells transfected with the wild-type insert and the mutant insert in both BE(2)-C and COS-7 cell lines. This was apparent on both gel electrophoresis and sequencing of the RT-PCR products.
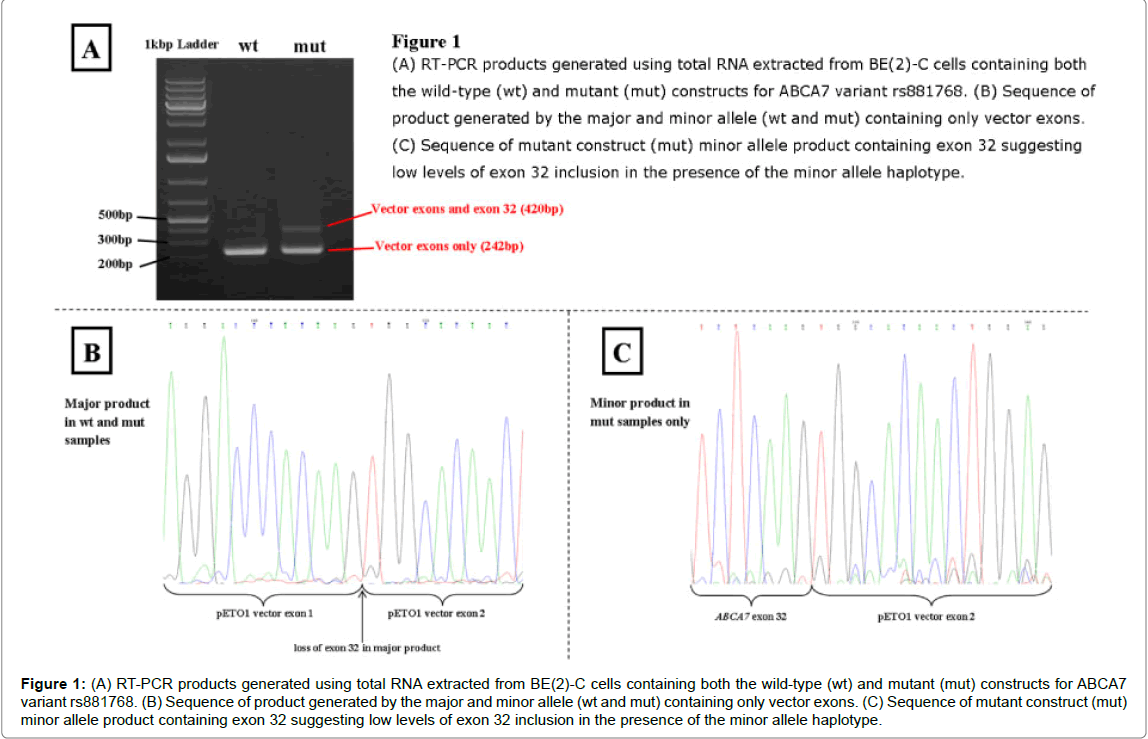
The ABCA7 variant rs881768 A>G, however, produced different results between the wild-type and mutant constructs upon analysis of RT-PCR products, as shown in (Figure 1). The major product with both constructs (when either the A or G allele was present) had exon 32 spliced out, leaving only vector sequence. With the mutant construct there was also a minor product, where exon 32 was included, suggesting low levels of exon 32 inclusion with the minor allele haplotype. This was evident in both COS-7 and BE(2)-C cell lines.
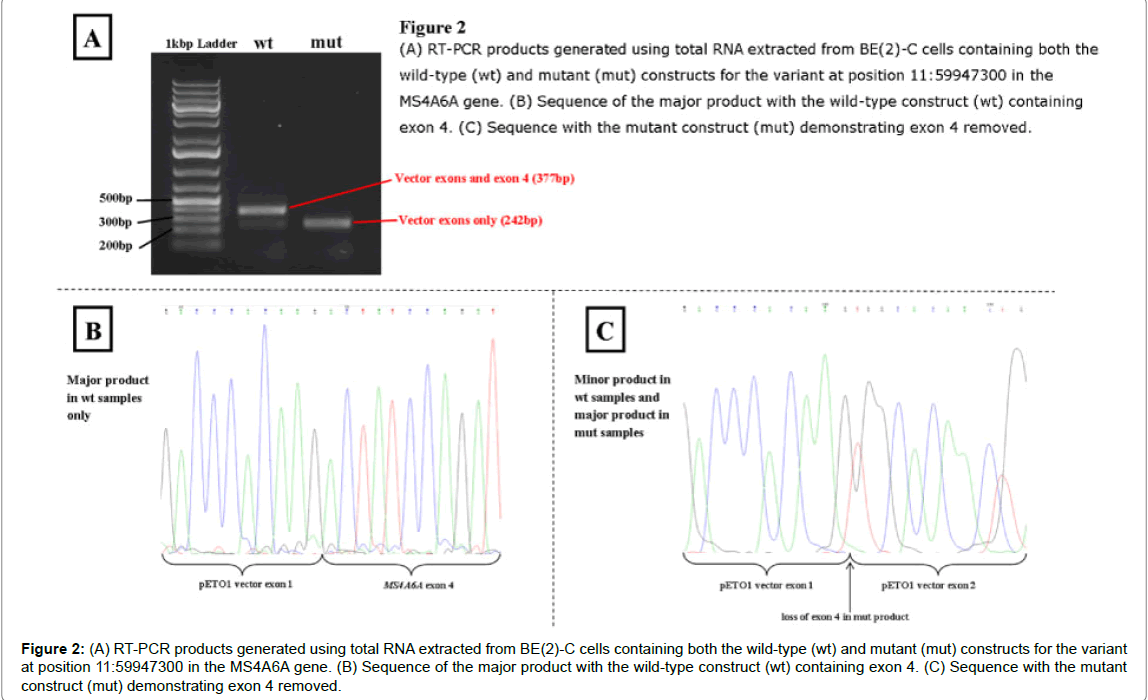
The MS4A6A variant at genomic position 11: 60179827 T>G also showed some differences between constructs, presented in (Figure 2). The major product with the wild-type construct, as predicted, contained both the vector exons and exon 4 of the gene. There was also a minor product present which contained just the vector sequence, suggesting low levels of a gene product with this exon spliced out. The only product with the mutant construct showed exon 4 spliced out. These results were obtained with both COS-7 and BE(2)-C cells lines.
Discussion
A 2013 GWAS meta-analysis for LOAD used over 74000 samples to identify 11 new disease-risk genes and confirmed 10 previous susceptibility loci (CLU, PICALM, CR1, BIN1, ABCA7, the MS4A locus, CD2AP, EPHA1, the HLA-DRB5-HLA-DRB1 locus, PTK2B, SORL1, SLC24A4-RIN3, INPP5D, MEF2C, NME8, ZCWPW1, CELF1, FERMT2 and CASS4 [1]). The population attributable risk for each of these loci range from 1 to 8% indicating further genetic risk factors remain to be discovered for LOAD [1].
This study used targeted sequencing to identify 21 potential splicing variants located in nine of the original genetic loci associated with LOAD in 96 patients. Following the in silico assessment of the variants’ impact on splicing, four variants were put forward for in vitro confirmation using hybrid minigenes in the pET01 vector. Only two of the variants were confirmed to be functional; rs881768 A>G in ABCA7 and novel variant 11: 60179827 T>G in MS4A6A, part of the MS4A locus. This suggests that the methods currently used to prioritize potential functional variants need further refinement.
| Gene (Transcript ID) | Genomic Location (rsID) | Transcript location | Ref/Alt | MAF | D’ | ESEfinder | BDGP | Human Splicing Finder | Combined predicted consequence |
|---|---|---|---|---|---|---|---|---|---|
| ABCA7 (NM_019112) |
19:1054696 (-) |
Intron 28-29 +3bp |
G/C | 0.010 | NA | SRSF2 site gained | Known functional donor site with a score of 0.91 reduced to a score of 0.38 | Alteration of an intronic ESS site. | Change in ESE and intronic ESS site could change transcript isoform ratio. |
| 19:1056066 (rs881768) |
Exon 32 1bp |
A/G | 0.250 | -0.824 | SRSF1 site gained, SRSF6 site lost | Novel donor site created with a score of 0.98 Strengthens known functional acceptor site score from 0.56 to 0.76 |
A cryptic donor site is activated. | Activation of donor site at start of exon might result in exon skipping. | |
| MS4A6A* (NM_152852) |
11: 60179827 (-) |
Intron 4-5 +4bp |
T/G | 0.480 | NA | SRSF5 sites lost | Known functional donor site with a score of 0.66 lost | Activation of intronic cryptic acceptor site, and intronic cryptic donor site and creation of intronic ESE site. | Loss of existing donor site could result in exon skipping. However, activating intronic acceptor or donor site could result in a change in the sequence included in the exon. |
| EPHA1* (NM_005232) |
7:143391774 (rs6967117) |
Exon 17 +1bp |
T/C | 0.060 | 0.75 | SRSF1 site gained | Novel donor site created with score of 0.49. | A cryptic donor site is activated. | Activation of donor site at start of exon might result in exon skipping. |
Genomic location is relative to GRCh38 and rsID is provided where available, (-) defines no rsID for this genomic location. Genes located on the reverse strand are indicated (*) and transcripts of these genes used are provided, defined by Ensembl.org. The reference and alternative allele for the forward strand as called in CRISP are shown (Ref/Alt) as is the minor allele frequency (MAF). The Linkage Disequilibrium between the SNP presented and the GWAS tag SNP for that gene is also presented in the D’ format. “NA” defines the LD is unavailable, most likely due to the variant in question not being present in the CEU population in the 1000Genomes database. All variants resulted in results indicating they have the potential to alter splicing by all three in silico programmes used (ESEfinder, BDGP and Human Splicing Finder). SRSF = Serine/arginine-rich splicing factor, ESE=Exonic Splicing Enhancer and ESS=Exonic Splicing Suppressor.
Table 2: Prioritised splicing variants from the NGS study selected for further experimental investigation.
Figure 1: (A) RT-PCR products generated using total RNA extracted from BE(2)-C cells containing both the wild-type (wt) and mutant (mut) constructs for ABCA7 variant rs881768. (B) Sequence of product generated by the major and minor allele (wt and mut) containing only vector exons. (C) Sequence of mutant construct (mut) minor allele product containing exon 32 suggesting low levels of exon 32 inclusion in the presence of the minor allele haplotype.
The synonymous variant rs881768 is located at the first base of exon 32 in ABCA7 (transcript ID NM_019112) and is in linkage disquilibrium with the ABCA7 GWAS variant (D’=0.824, Table 2). In silico predictions suggest that the G allele creates a new donor site with a similar score to the original donor site, located only 126 bp away (a score of 0.98 compared with 0.99) and changes the binding of two exon splicing enhancer (ESE) proteins (Table 2). These predictions suggest the G allele could activate a cryptic splice site or cause a change in protein isoform ratios. However, population data from 1000 Genomes shows that the G allele is actually the ancestral allele with a European population allele frequency of 0.44. Additionally, score predictions for the acceptor site shows that the A allele has a lower score than the G allele (0.56 compared to 0.76) which could lead to exon skipping. This makes in silico functional prediction of this variant very difficult.
Figure 2: (A) RT-PCR products generated using total RNA extracted from BE(2)-C cells containing both the wild-type (wt) and mutant (mut) constructs for the variant at position 11:59947300 in the MS4A6A gene. (B) Sequence of the major product with the wild-type construct (wt) containing exon 4. (C) Sequence with the mutant construct (mut) demonstrating exon 4 removed.
In the minigene assays the G allele causes low levels of exon 32 to be incorporated into the transcript, while the A allele causes exon 32 to be spliced out. Expressed sequence tag (EST) databases such as GTEx portal [31] show exon 32 is incorporated in most transcripts and utilising this transcript data to examine the effect of rs881768 shows that this variant has no effect on splicing in this database. ABCA7 has an interesting pattern of alternative splicing, with many introns consisting of multiples of three, potentially allowing in-frame addition and deletion of introns, although no such transcripts have been identified. Exon 32, however, does results in a frame-shift change creating a termination codon in the next exon and causing early truncation of the protein. This truncated protein is non-functional as it lacks the last five transmembrane domains and the second nucleotide binding domain and may be targeted for nonsense mediated decay. This would prevent any truncated proteins being expressed causing sole expression of the complete isoform, explaining the lack of this truncated protein in databases such as GTEx.
The novel variant identified in MS4A6A, part of the MS4A locus, 11:60179827 T>G is found 4 bp into the intron between exon 4 and 5. The G allele removes the native donor site for exon 4/intron 4 possibly causing exon skipping (Table 2). However, a cryptic donor site is activated and the binding of two ESE proteins are affected, potentially changing the sequence included in the transcript. The minigene assays show that the G allele does appear to cause exon skipping, creating transcripts without exon 4. However, the T allele, while mostly producing transcripts which contain exon 4, also produces a small proportion of transcripts without exon 4. Transcripts without exon 4 appear to be produced by both alleles despite the removal of exon 4 generating a truncated protein which lacks two transmembrane domains and one noncytoplasmic domain. Although the G allele does cause exon 4 to be excluded through obliterating the donor site, there must be additional alternative splicing regulation mechanisms occurring within the in vitro assay, as well as in vivo, which affect the levels of each isoform being produced explaining the presence of both isoforms in the presence of the T allele.
To fully explore the role both of these variants play in splicing, direct analysis of RNA samples from affected tissues from LOAD individuals with the variants should be examined. Unfortunately for rare variants in LOAD this tissue is not always readily available. Epstein- Barr transformed cell lines used for 1000 Genomes project, however, are a possible alternative. Targeted sequencing of RNA extracted from these cell lines selected to be homozygous for different alleles could be compared to the results of the minigene assays. This would determine the role of these splicing variants in an environment where the whole genome is present, rather than just the fragment inserted in the minigene assay. There is also publicly available RNA-seq data from these cell lines (see the GEUVADIS project [32]); unfortunately coverage for many of the LOAD risk loci is poor.
While only two variants show aberrant splicing in this study, dysfunctional splicing has been previously implicated in LOAD. Distinctive patterns of alternative splicing and promoter use were discovered in LOAD brain tissue through comparing the transcriptome profiles of the frontal, temporal and parietal lobes of AD patients with controls [33,34]. Additionally, small nuclear and heterogeneous nuclear ribonucleoproteins have been shown to be disrupted in LOAD [35,36]. Therefore the role of splicing in LOAD disease pathology should not be ignored.
Identifying variants that have a functional role in complex diseases is a challenge [37]. This is particularly true for next generation sequencing studies which identify large numbers of potential functional variants. The problem arises while attempting to prioritise the variants for further functional assays or experiments. Current in silico databases have minimal experimental information available. The annotation programs used need to be chosen carefully in order to correctly assign potential functionality and ensure pathogenic variants are found.
Prediction algorithms for exonic and intronic splicing enhancer and silencer sites are less robust than programs predicting disruptions of 3’ and 5’ consensus splice site regions. This is largely because more is known about the 3’ acceptor and 5’ donor consensus sequences, thus allowing better prediction models to be built [38]. Due to this, the initial selection of splicing variants in this study selected variants within 1-3 bp of an exon or 1-12 bp of an intron, biasing the selection towards variants that may disrupt consensus splice sites. The BDGP and HSF prediction programs used in this study both examine the effects of mutations on 3’ (acceptor) and 5’ (donor) consensus sequences. The two variants that had an effect on splicing in vitro were both predicted to affect acceptor and or donor consensus sites by BDGP and HSF (Table 2).
The recognition of exon and intron borders in pre-mRNA by the spliceosome does not simply involve identifying the correct consensus 3’ and 5’ splicing sequences. A multitude of splicing regulatory proteins and ribonucleoproteins interact to precisely control splicing and influence the levels of different transcript isoforms created [39]. Many variants that disrupt splicing are found deep within exons and introns and affect splicing through the disruption of exonic or intronic splicing enhancers or silencers [40]. Through limiting the analysis to variants located near consensus splice sites, as in this study, these mutations will be missed. With the advent of additional functional experimental studies, predictions of mutations influencing the binding of RNA proteins can only improve and would be useful for future work. This issue has recently been addressed by ANNOVAR, with the option to include bulk annotations from the SPIDEX dataset. This dataset provides an improved algorithm for detecting potential splicing variants [41]. However, a review of the method suggests that two other prediction methods for exonic splicing regulatory elements may perform better [42].
Conclusion
Pooled NGS with target enrichment successfully identified potential functional splicing variants in nine gene regions associated with LOAD. Through targeting the entire genomic sequence, we were able to investigate variants in these regions which potentially affect consensus splice sites in silico and in vitro. Improvements are needed in current splicing prediction programs to reduce the number of false positives which are taken forward for analysis as well as reducing the number of false negatives discarded prior to further investigation. Further work is needed to fully clarify the role that the variants rs881768 (ABCA7) and 11: 60179827 T>G (MS4A6A) may have in LOAD in vivo.
Acknowledgement
The Nottingham Group are funded by Alzheimer’s Research UK and the Big Lottery Fund. NC was funded by a scholarship from The Jean Shanks Foundation. MA was funded by the MASTER it! Scholarship Scheme; this scholarship is partfinanced by the European Union – European Social Fund.
References
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, et al. (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for AlzheimerâÂ?Â?s disease. Nat Genet 45:1452-1458.
- Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M (2012) Linking disease associations with regulatory information in the human genome. Genome Res 22: 1748-1759.
- Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, et al. (2010) Integrating common and rare genetic variation in diverse human populations. Nature 467: 52-58.
- BettensK, Brouwers N, Engelborghs S, Lambert JC, Rogaeva E, et al. (2012) Both common variations and rare non-synonymous substitutions and small insertion/deletions in CLU are associated with increased Alzheimer risk. MolNeurodegener 7: 3.
- Lord J, Turton J, Medway C, Shi H, Brown K, et al. (2012) Next generation sequencing of CLU, PICALM and CR1: Pitfalls and potential solutions. Int J MolEpidemiol Genet 3: 262-275.
- Cuyvers E, De Roeck A, Van den Bossche T, Van Cauwenberghe C, Bettens K, et al. (2015) Mutations in ABCA7 in a Belgian cohort of AlzheimerâÂ?Â?s disease patients: a targeted resequencing study. Lancet Neurol 14: 814-822.
- Vardarajan BN, Ghani M, Kahn A, Sheikh S, Sato C, et al. (2015) Rare coding mutations identified by sequencing of Alzheimer disease genome-wide association studies loci. Ann Neurol78: 487-498.
- Belkadi A, Bolze A, Itan Y, CobatA, Vincent QB, et al. (2015) Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants.ProcNatlAcadSci USA 112: 5473-5478.
- Lim KH, Ferraris L, Filloux ME, Raphael BJ, Fairbrother WG (2011) Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. ProcNatlAcadSci USA 108: 11093-11098.
- Sterne-Weiler T, Howard J, Mort M, Cooper DN, Sanford JR (2011) Loss of exon identity is a common mechanism of human inherited disease. Genome Res 21: 1563-1571.
- Mills JD, Janitz M (2012) Alternative splicing of mRNA in the molecular pathology of neurodegenerative diseases. Neurobiol Aging 33: 1012.e11âÂ?Â?1012.e24.
- Schnetz-Boutaud NC, Hoffman J, Coe JE, Murdock DG, Pericak-Vance MA, et al. (2012) Identification and confirmation of an exonic splicing enhancer variation in exon 5 of the Alzheimer disease associated PICALM gene. Ann Hum Genet 76:448-453.
- Malik M, Simpson JF, Parikh I, Wilfred BR, Fardo DW, et al. (2013) CD33 AlzheimerâÂ?Â?s risk-altering polymorphism, CD33 expression and exon 2 splicing. J NeurosciOff J SocNeurosci 33: 13320-13325.
- Raj T, Ryan KJ, Replogle JM, Chibnik LB, Rosenkrantz L, et al. (2014) CD33: Increased inclusion of exon 2 implicates the Ig V-set domain in AlzheimerâÂ?Â?s disease susceptibility. Hum Mol Genet 23: 2729-27336.
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, et al. (2009) Genome-wide association study identifies variants at CLU and PICALM associated with AlzheimerâÂ?Â?s disease. Nat Genet 41: 1088-1093.
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, et al. (2009) Genome-wide association study identifies variants at CLU and CR1 associated with AlzheimerâÂ?Â?s disease. Nat Genet 41: 1094-1099.
- Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, et al. (2011) Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset AlzheimerâÂ?Â?s disease. Nat Genet 43: 436-441.
- Homer N, Merriman B, Nelson SF (2009) BFAST: an alignment tool for large scale genome resequencing. PloS One 4:e7767.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. (2009) The sequence alignment/map format and SAMtools. BioinformaOxfEngl 25: 2078-2079.
- Lassmann T, Hayashizaki Y, Daub CO (2011)SAMStat: Monitoring biases in next generation sequencing data. BioinformaOxfEngl27: 130-131.
- Bansal V, Libiger O, Torkamani A, Schork NJ (2010) Statistical analysis strategies for association studies involving rare variants. Nat Rev Genet 11: 773-785.
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, et al. (2010) The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297-1303.
- McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, et al. (2010) Deriving the consequences of genomic variants with the Ensembl API and SNP effect predictor. BioinformaOxfEngl26: 2069-2070.
- Consortium TEP (2011) A userâÂ?Â?s guide to the encyclopedia of DNA elements (ENCODE). PLoSBiol9:e1001046.
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, et al. (2011)The variant call format and VCFtools. BioinformaOxfEngl 27: 2156-2158.
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. (2012) An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56-65.
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR (2003)ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res 31: 3568-3571.
- Reese MG,Eeckman FH, Kulp D, Haussler D (1997) Improved splice site detection in genie. J ComputBiol4: 311-323.
- Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, et al. (2009) Human splicing finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37: e67.
- Steffensen AY, Dandanell M, Jønson L, Ejlertsen B, Gerdes AM, et al. (2014)Functional characterization of BRCA1 gene variants by mini-gene splicing assay. Eur J Hum Genet EJHG 22: 1362-1368.
- Ardlie KG, Deluca DS, Segre A V, Sullivan TJ, Young TR, et al. (2015) The genotype-tissue expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 348: 648-660.
- Lappalainen T, Sammeth M, Friedländer MR, t Hoen PAC, Monlong J, et al. (2013) Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501: 506-511.
- Twine NA, Janitz K, Wilkins MR, Janitz M (2011) Whole transcriptome sequencing reveals gene expression and splicing differences in brain regions affected by AlzheimerâÂ?Â?s disease. PloS One6:e16266.
- Mills JD, Nalpathamkalam T, Jacobs HIL, Janitz C, Merico D, et al. (2013) RNA-Seq analysis of the parietal cortex in AlzheimerâÂ?Â?s disease reveals alternatively spliced isoforms related to lipid metabolism. NeurosciLett536: 90-95.
- Bai B, Hales CM, Chen PC, Gozal Y, Dammer EB, et al. (2013) U1 small nuclear ribonucleoprotein complex and RNA splicing alterations in AlzheimerâÂ?Â?s disease.ProcNatlAcadSci USA 110: 16562-16567.
- Berson A, Barbash S, Shaltiel G, Goll Y, Hanin G, et al. (2012) Cholinergic-associated loss of hnRNP-A/B in AlzheimerâÂ?Â?s disease impairs cortical splicing and cognitive function in mice. EMBO Mol Med 4: 730-742.
- MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, et al. (2014) Guidelines for investigating causality of sequence variants in human disease. Nature 508: 469-476.
- Jian X, Boerwinkle E, Liu X (2014)Insilico tools for splicing defect prediction: A survey from the viewpoint of end users. Genet Med Off J Am Coll Med Genet 16: 497-503.
- DeConti L, Baralle M, Buratti E (2013) Exon and intron definition in pre-mRNA splicing. Wiley Interdiscip Rev RNA 4: 49-60.
- Sterne-Weiler T, Sanford JR (2014) Exon identity crisis: Disease-causing mutations that disrupt the splicing code. Genome Biol15:201.
- Xiong HY, Alipanahi B, Lee LJ, Bretschneider H, Merico D, et al. (2015)The human splicing code reveals new insights into the genetic determinants of disease. Science 347:1254806.
- Soukarieh O, Gaildrat P, Hamieh M, Drouet A, Baert-Desurmont S, et al. (2016)Exonicsplicing mutations are more prevalent than currently estimated and can be predicted by using in silico tools. PLoS Genet 12:e1005756.
Relevant Topics
- Advanced Parkinson Treatment
- Advances in Alzheimers Therapy
- Alzheimers Medicine
- Alzheimers Products & Market Analysis
- Alzheimers Symptoms
- Degenerative Disorders
- Diagnostic Alzheimer
- Parkinson
- Parkinsonism Diagnosis
- Parkinsonism Gene Therapy
- Parkinsonism Stages and Treatment
- Stem cell Treatment Parkinson
Recommended Journals
Article Tools
Article Usage
- Total views: 3805
- [From(publication date):
November-2016 - Jul 09, 2025] - Breakdown by view type
- HTML page views : 2904
- PDF downloads : 901