Interpretable Machine-Learning Model for Prediction of Convalescent COVID-19 Patients with Pulmonary Diffusing Capacity Impairment
Received: 29-Oct-2022 / Manuscript No. JIDT-22-78654 / Editor assigned: 02-Nov-2022 / PreQC No. JIDT-22-78654 (PQ) / Reviewed: 16-Nov-2022 / QC No. JIDT-22-78654 / Revised: 23-Nov-2022 / Manuscript No. JIDT-22-78654 (R) / Published Date: 30-Nov-2022
Abstract
Introduction: The COVID-19 patients in the convalescent stage noticeably have pulmonary diffusing capacity impairment (PDCI). The pulmonary diffusing capacity is an important indicator of the COVID-19 survivors’ prognosis of pulmonary function, but the current studies focusing on prediction of the pulmonary diffusing capacity of these people are limited. The aim of this study was to develop and validate a machine learning (ML) model for predicting PDCI in the COVID-19 patients using routinely available clinical data, thus assisting the clinical diagnosis.
Methods: The data used in this study were collected from a follow-up study from August to September 2021 of 221 hospitalized COVID-19 survivors 18 months after discharge from Wuhan, including the demographic characteristics and clinical examination. The data were randomly split into a training (80%) data set and a validation (20%) data set. Six popular machine learning models were developed to predict the pulmonary diffusing capacity of COVID-19 patients in the recovery stage. The performance indicators of the model included area under the curve (AUC), Accuracy, Recall, Precision and F1. The model with the optimum performance was defined as the optimal model, which was further used in the interpretability analysis. The MAHAKIL method was utilized to balance the data and optimize the balance of sample distribution, while the RFECV method for feature selection was utilized to select combined features more favorable to machine learning.
Results: A total of 221 COVID-19 survivors discharged from hospitals in Wuhan were enrolled in this study. Of these participants, 117 (52.94%) were female, with a median age of 58.2 years (Standard Deviation (SD)=12). After feature selection, 31 of the 37 clinical factors were ultimately chosen for use in the model construction. Among the six ML models tested, the best performance was accomplished in the XGBoost model, with an AUC of 0.755 and an accuracy of 78.01% after experimental verification. The SHAPELY Additive explanations (SHAP) summary analysis exhibited that Hemoglobin (Hb), Maximal Voluntary Ventilation (MVV), severity of illness, Platelet (PLT), Uric Acid (UA) and Blood Urea Nitrogen (BUN) were the top six most important factors affecting the XGBoost model decision-making.
Conclusion: The XGBoost model reported here showed good prognostic prediction ability for Carbon Monoxide Diffusing Capacity of the lungs (DLCO) of COVID-19 survivors during the recovery period. Of the features selected, Hb and MVV contributed most to the outcome prediction of DLCO of the convalescent COVID-19 survivors.
Keywords: Artificial intelligence; Machine learning; COVID-19; Maximal voluntary ventilation
Introduction
As of October 20, 2022, the global pandemic caused by Corona Virus Disease 2019 (COVID-19) has infected more than 626 million people and claimed 6.57 million lives. Among the COVID-19 survivors, many have shown disastrous effects on multiple organs and systems [1], but the lung is the organ most susceptible to severe damage from COVID-19 [2]. The convalescent COVID-19 patients have demonstrated particularly pronounced PDCI. Our previous study has found that the incidence of DLCO impairment of the COVID-19 patients reached 57.92% in 18 months after discharge [3]. Studies show that pulmonary diffusing capacity of the COVID-19 patients is also significantly impaired in the 1-24 month recovery phase. Studies also suggest impaired gas-blood exchange in patients discharged after admission for COVID-19 [1,4-7], and low DLCO may be the result of interstitial abnormalities or pulmonary vascular abnormalities caused by COVID-19 [8-11]. Therefore, there is an urgent need for a prognostic assessment and early warning system for COVID-19, especially for a model to predict PDCI of the convalescent patients. To solve this problem, establishment of an early warning model to estimate the DLCO of patients is probably an alternative. The current prediction models for COVID-19 are mainly utilized to identify the high-risk groups of the general population [12], diagnose COVID-19 patients [13], and predict the progression of disease severity and mortality [14,15]. However, the prediction models for PDCI of COVID-19 patients are still in deficiency.
Machine learning analysis is based on various data mining algorithms of different data types and formats to characterize the features of data in a more scientific way and gain better insight into data trends and recognized values [16]. The interpretability of ML is essential to help enhance the trust of healthcare professionals because it shows sufficient reasons to make predictions and the way how parameters contribute to the model [17]. However, most ML studies worked hard to improve performance by increasing the model complexity, leading to uncertainties in the way how ML operates and makes decisions [18- 20]. To improve interpretability of the ML models, this study adopted the most popular feature importance estimation in the explainabilty researches [21,22]. We tried to rank the features according to their importance and used the TreeSHAP method proposed by Lundberg et al. to analyze the clinical features [23].
Accordingly, the aim of this investigation was to develop and validate an interpretable ML model based on clinical variables to assess the risk of PDCI of the COVID-19 patients in recovery.
Methodology
Study design and data set
We conducted from August to September 2021 a follow-up study of COVID-19 survivors 18 months after discharge from Hubei Provincial Hospital of Integrated Traditional Chinese and Western Medicine. A total of 221 survivors were contacted according to their time of discharge. Clinical data related to survivors, including demographic characteristics (age, sex and Body Mass Index (BMI)) and clinical examination indicators (lung function, chest HRCT, antibody titers and various biochemical indicators), were collected by trained physicians. This study was reviewed and approved by the Ethics Committee of Hubei Provincial Hospital of Integrated Traditional Chinese and Western Medicine (2,020,009). All participants had provided their written or verbal consents prior to the study.
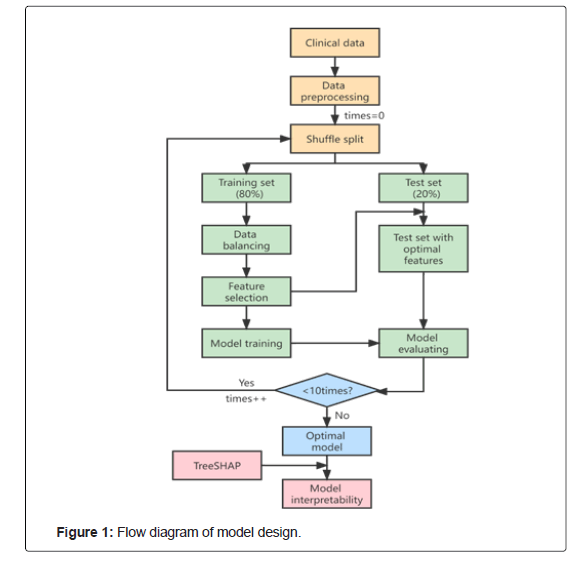
The principal procedures of this study were performed in three main steps. Firstly, we used six popular machine learning models to predict pulmonary diffusing capacity of patients recovering from COVID-19. Secondly, we tested the performance of the six ML models, selected indicators including AUC, Accuracy, Recall, Precision and F1, and defined the model with the optimum performance as the optimal model. Finally, we used the MAHAKIL method for data balance processing to optimize the balance of sample distribution, while the RFECV method for feature selection, which could choose combined features conducive to machine learning. The overall workflow of this study is shown in Figure 1.
Patients and outcomes
The criteria for inclusion of survivors in the study were determined in accordance with the protocols for COVID-19 management of the World Health Organization (WHO) and National Health Commission of the People’s Republic of China [24,25].
The severity of illness of COVID-19 are measured as follows:
• Mild cases: without symptoms and signs of severe and critical infection;
Severe case:
• breathing difficulties, respiratory rate ≥ 30 bpm;
• SpO2 ≤ 93% at rest;
• PaO2/FiO2ratio ≤ 300 mmHg.
Critical cases:
• Respiratory failure requiring mechanical ventilation;
• Shock;
• Multi-organs failure requiring intensive care.
The primary endpoint of our study was the Area Under the Receiver Operating Characteristic curve (AUROC) of the model’s prediction. The secondary endpoints of our study were Accuracy, Recall, Precision and F1 score of the model’s prediction.
Data collection
Clinical data on COVID-19 survivors include demographics, medical history, laboratory tests, severity of illness scoring system and outcomes. Demographic characteristics extracted covered gender, age, height and body weight. Then, we collected data on comorbidities, including heart failure, anemia and Chronic Obstructive Pulmonary Disease (COPD). The laboratory tests abstracted include White Blood Cells (WBC), Hb, PLT, N%, L%, LY#, IgM, IgG, proBNP, Alanine Transaminase (ALT), Aspartate Aminotransferase (AST), Alb, BUN, Cr, UA, HbA1c, Normal unilateral and bilateral score, Forced Vital Capacity (FVC), forced expiratory volume in one second (FEV1), FEV1/FVC, MVV, DLCO, tubercle, Ground-Glass Opacity (GGO), fibrosis, etc. The severity of illness is scaled from 1 to 4.
Feature selection and data preprocessing
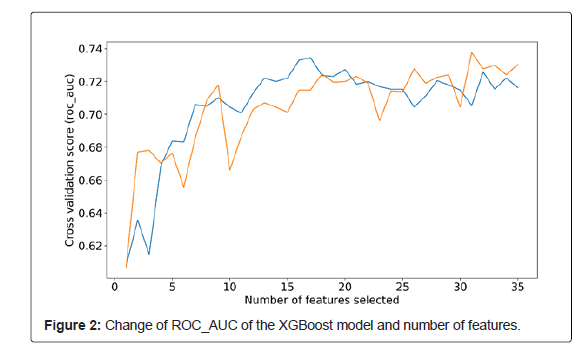
High dimensional data analysis modeling is a challenge for data mining researchers. Feature selection technology provides an effective method to solve this problem by removing irrelevant and redundant data, which can reduce the computation time, improve the learning accuracy and help to better understand the learning model [17]. Cai et al. analyzed and compared some state-of-art feature selection methods on two high-dimensional gene expression data sets through experiments [21], which found that Recursive Feature Elimination (RFE) could achieve higher accuracy than other feature selection methods. In this regard, we chose RFECV, a Cross Validation version of RFE. The purpose of adding Cross Validation is to select the best number of features, which often requires manual trial and error to obtain the best number of features in studies using RFE. In our study, the RFECV method was used to cyclically remove medical features that were detrimental to the ability of the model to learn to predict the pulmonary diffusing capacity until the assembled features enabled the model to perform optimally. After feature selection, 31 of the 37 clinical factors were ultimately chosen for model construction.
Model development and validation
The data were randomly split into a training data set (80%) and a validation data set (20%). Firstly, up-sampling by the MAHAKIL method was utilized to balance the number of samples of different classes in the training set. Secondly, the RFECV method was utilized to select the optimal combination of features. Then, the selected features from the balanced training set were input into the machine learning model for training and modeling, and the grid-search method was utilized to ensure the validity of the combination of parameters during training. Finally, the trained ML model was utilized to predict and evaluate the data results of the test set, and the features in the test set were also processed as the optimal combination of features. In addition, we integrated the overall data, ranked the importance of features by taking XGBoost as the base model and using the TreeExplainer method, and combined with the calculating principle of the SHAP interaction values to further explain the reasons why these features were considered significant.
The XGBoost model ROC_AUC changes corresponding to the number of features are shown in Figure 2. After feature selection via the RFECV method, 31 were selected as the optimal combined features.
Model explainability
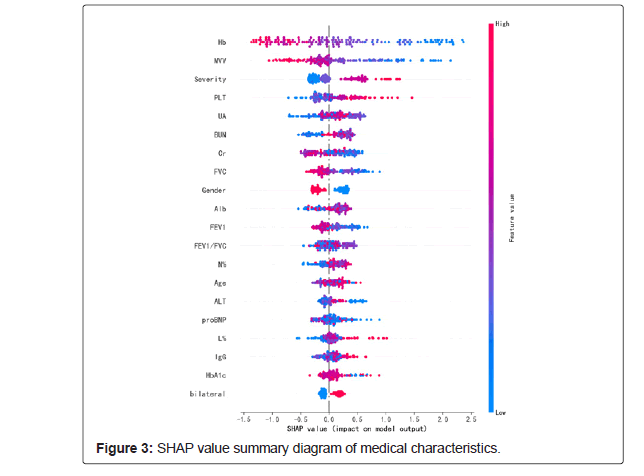
Opening the black box of ML is of great importance to improve the compliance and transparency of the ML decision-making process of healthcare workers [26]. Therefore, we took XGBoost with the best performance in AUC evaluation index as the base model, the optimal combined features after feature selection and labels as the input, and use the Tree Explainer method to sort the SHAP values of features. The SHAP value summary diagram of medical characteristics is shown in Figure 3.
Statistical analysis
The count data were described with the number of cases (%), and Pearson chi-square test was utilized for comparison between groups. Measurement data conforming to the normal distribution were expressed as the median and interquartile range (M (P25, P75)) by t-test or ANOVA, while the Mann-Whitney U test was utilized between groups. After feature selection and data preprocessing, we developed six popular machine learning models to predict PDCI of patients recovering from COVID-19. The overall performance of each model was evaluated by AC, Accuracy, Precision, Recall and F1 measurements. Ultimately, the model was explained by the TreeExplainer method.
SPSS 25.0 (IBM, Armonk, New York, USA) was used for statistical analysis. All statistical tests were two-sided, and P<0.05 was considered statistically significant.
Results
Clinical characteristics
A total of 221 COVID-19 survivors participated in the study (Mild cases, n=93; Moderate cases, n=58; Severe cases, n=54; Critical cases, n=16). The median age of the patients in this study was 58.2 [Standard Deviation (SD)=12]. Among them, 104 survivors (47.06%) were male and 117 (52.94%) were female, with an average BMI of 24.62 [Standard Deviation (SD)=3.5]. The incidence of PDCI in COVID-19 survivors was 57.92% as shown in Table 1.
Table 1: Clinical characteristics of survivors with impaired and normal DLCO.
Model development and validation
After feature selection, we utilized 31 alternative factors for model construction, and among the six ML models tested by the team. Compared with GBDT (A.C. 0.67), KNN (A.C. 0.63), RandomForest (A.C. 0.70) and SVC (A.C. 0.70), MLP (A.C. 0.69), XGBoost (A.C. 0.75) has better DLCO predicting ability for COVID-19 survivors. Table 2 shows that XGBoost has the best performance in AUC, Accuracy, Recall, Precision and F1. After experimental verification, the model has an AUC of 0.755 and an Accuracy of 78.01%. The SHAP summary analysis showed that Hb, MVV, severity of illness, PLT, UA and BUN were the top six most important factors affecting the XGBoost model decision-making.
| Classifier | AUC | Accuracy | Recall | Precision | F1 |
|---|---|---|---|---|---|
| GBDT | 0.6787 | 0.7044 | 0.6787 | 0.6938 | 0.6764 |
| KNN | 0.6392 | 0.6487 | 0.6392 | 0.6378 | 0.6309 |
| RandomForest | 0.7011 | 0.7376 | 0.7011 | 0.7434 | 0.7009 |
| SVC | 0.7085 | 0.7358 | 0.7085 | 0.7224 | 0.7104 |
| MLP | 0.6912 | 0.7066 | 0.6912 | 0.6937 | 0.6872 |
| XGBoost | 0.755 | 0.7801 | 0.755 | 0.7755 | 0.7572 |
Table 2: Experimental results of different classifiers.
Model explainability
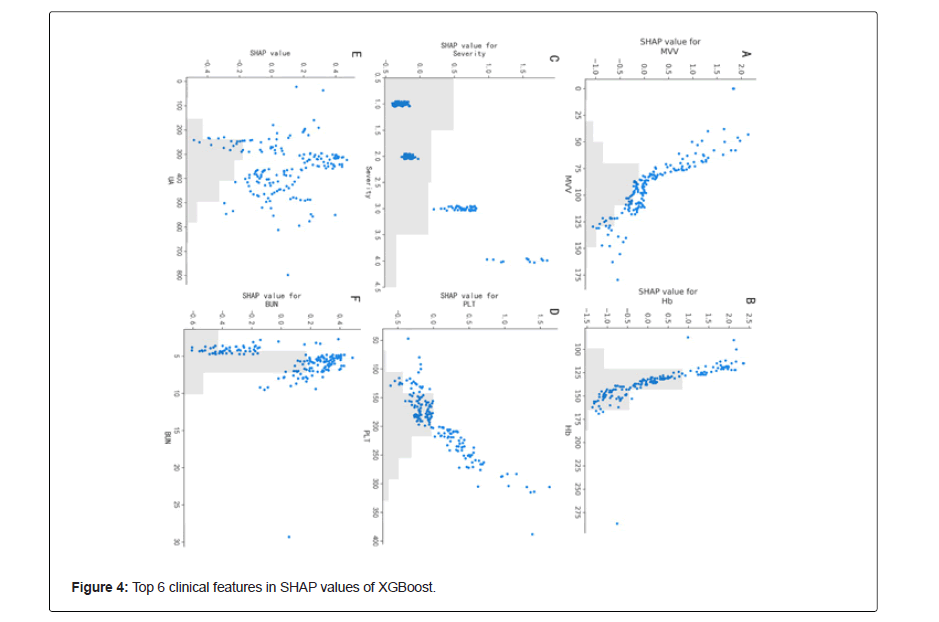
Figure 3 shows the SHAP summary diagram, which ranks the factors according to their importance to the predicted incidence in the validation cohort. The SHAP summary analysis showed that Hb, MVV, level, PLT, UA and BUN were the top six most pivotal factors affecting the XGBoost model decision. Figure 3 also shows the correlation between the six factors and the prediction of PDCI occurred in the COVID-19 survivors. The SHAP values above zero for these six characteristics indicate an increased risk of PDCI. Hb and MVV were negatively correlated with DLCO, while severity of illness, PLT, UA and BUN were positively correlated.
The SHAP values in the validation set were uitlized to evaluate the feature importance of the XGBoost classifier. Each dot represents 1 patient and is accumulated vertically to depict density. Colors represent high and low values for each element, with dark colors indicating higher values and light colors lower values. The X-axis of the graph represents the SHAP value. A positive SHAP value indicates that it has a positive contribution to the prediction model and a high probability of PDCI occurrence, and vice versa (Figure 4).
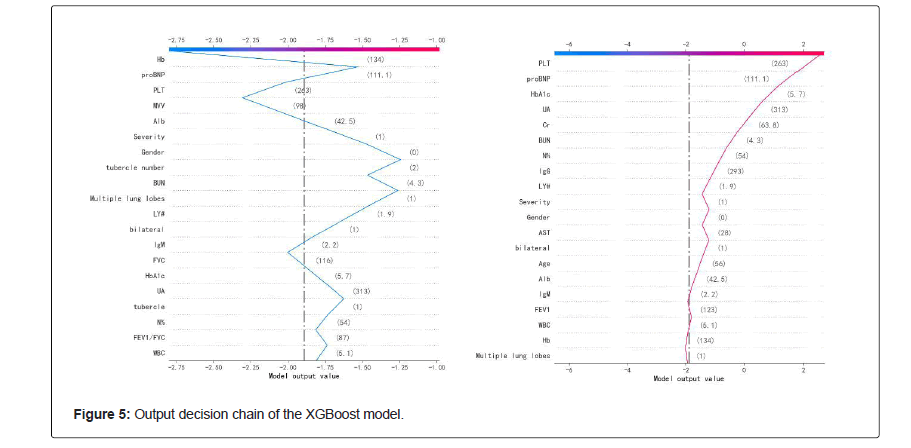
Finally, we plotted the XGBoost decision-making process against the SHAP values, as shown in Figure 5. The gray vertical line in the middle of the decision graph marks the base value of the model, and the colored line is the prediction, indicating whether each feature moves the output value to a value higher or lower than the average prediction. The eigenvalues next to the prediction line can be taken as the reference. Starting at the bottom of the graph, the prediction line shows how the SHAP value accumulates from the base value to the model final score at the top of the graph. The blue broken line is the decision process of predicting a normal object, and the red broken line is the decision process of predicting an exception object.
Discussion
The aim of this ML-based modeling study was to develop a valid, stable and interpretable model for predicting the incidence of PDCI in COVID-19 survivors in the recovery phase. The results manifested that the XGBoost model was the most reliable and accurate among all the tested models, with an AUC of 0.755 and an Accuracy of 78.01%. We also found that Hb, MVV, severity of illness, PLT, UA and BUN were the top six most important factors influencing the XGBoost model decision-making. Overall, our study demonstrated that it is possible to predict the incidence of PDCI in COVID-19 patients using routinely collected clinical data.
As the COVID-19 pandemic continues to be rampant in our world and the number of patients recovering from the disease increases, studies have found that shortness of breath and dyspnea are the most common sequelae among those who have survived hospitalization with COVID-19 due to the presence of PDCI [7]. Therefore, DLCObased pulmonary function testing can be regarded as a useful tool to differentiate those at risk of pulmonary sequelae [27]. However, previous studies on COVID-19 have focused on risk factor analysis and mortality prediction of the mild-to-moderate cases [28,29], without a prediction model for PDCI in COVID-19 survivors. Therefore, it is necessary to develop and validate the risk-level outcome prediction models to evaluate the pulmonary function status of COVID-19 survivors.
Apart from using a machine learning model to predict the pulmonary diffusing capacity in patients recovering from COVID-19, this study further applied the model to interpretability analysis. Because the internal logic and operating mechanisms of ML models are concealed from users, this uncertainty poses challenges for healthcare workers in applying the machine learning systems in reality. In this study, we used the interpretation method based on the importance of the SHAP value features to help medical researchers understand the decision-making criteria of ML models [30-32], enhance the credibility of medical professionals in ML, and coordinate the contradictions or inconsistencies between the knowledge structural elements of machines and human beings with prior knowledge. We adopted the TreeSHAP method [6], which is an effective evaluative method for the importance of tree model features based on the SHAPELY value of classical game theory. The SHAP summary analysis showed the six most important factors of the XGBoost model. Among them, MVV is the most important indicator of lung reserve function, which is closely related to activity endurance. The most serious sequelae of the COVID-19 patients are shortness of breath and dyspnea in the wake of activities, and the significantly decreased pulmonary function reserve [33]. This study confirmed that MVV was positively correlated with the pulmonary diffusing function in COVID-19 patients. The MVV value of COVID-19 patients with normal diffusing function was significantly higher than that of patients with impaired pulmonary diffusing function, which reveals the importance of strengthening pulmonary rehabilitation exercises and increasing pulmonary function reserve in COVID-19 patients during rehabilitation. Studies have found that Hb, a parameter closely related to organ perfusion, alveolar ventilation and blood flow ratio, has greatly contributed to the prediction of pulmonary function outcomes in patients with COVID-19 after recovery. In this study, after the correction of Hb, Hb of COVID-19 patients with decreased pulmonary diffusing function was normal low value or anemia, indicating that there is a long-term imbalance of pulmonary perfusion and ventilation ratio in COVID-19 patients, to which due attention should be paid.
In addition, PLT and severity of illness were negatively correlated with pulmonary diffusing function. The more severe the disease was, the higher the normal value of PLT was, and vice versa. Studies have confirmed that PLT activation is involved in the formation of inflammatory micro-vascular thrombosis in COVID-19 patients and is closely related to respiratory failure in COVID-19 patients [10,34-38]. However, one and a half years later, our study found that PLT was still closely related to PDCI of COVID-19 survivors. Consistent with our previous study [3], these observations suggest that clinically obtained MVV, PLT, Hb and severity of illness are the key factors for using the XGBoost model to predict pulmonary function status in COVID-19 survivors. Besides, compared with the indicators directly affecting pulmonary function, the SHAP pooled analysis exhibited that the increased UA and BUN may be associated with an increased risk of the retrogressive pulmonary diffusing capacity of the COVID-19 patients.
Currently, the combination of high-frequency biological data streams and artificial intelligence offers a promising application for predicting the diffusing capacity of lungs, which could allow early identification of pulmonary function recovery in patients with COVID-19 [39-41]. However, there are still some limitations in this study. First, the modeling and retrospective design of this study do not allow causal inferences to be drawn about the association between variables and the ability of the pulmonary diffusing capacity. Second, the predictive efficiency of the current models may be affected by racial and ethnic differences. Moreover, it is difficult to obtain more relevant data due to the privacy of COVID-19 patients, leading to a lack of proper external validation of our prediction model, which will affect the credibility of the XGBoost model. Finally, although the findings showed that the model had learned the medical rules in the data, the expansion of data is an urgent ness in the future to improve the performance of the model.
Conclusion
The XGBoost model showed desired predictive ability for PDCI of COVID-19 survivors during recovery. Among the selected features, Hb and MVV contributed the most to the prediction of PDCI outcomes in survivors recovering from COVID-19. The significance of the SHAP values could help to improve the interpretation of ML model results.
Funding
This study was funded by Clinical Study on Prevention and Treatment of COVID-19 by Integrated Traditional Chinese and Western Medicine (2020YFC0841600) under the National Science and Technology Emergency Project.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published. All authors revised the manuscripts and approved the final manuscript.
Author Contributions
BX and JXZ designed the study and had full access to all of the data in the study; CY F and CH took responsibility for the data collection. FQMand HRY were responsible for the data analysis. ZWH HRM and YQ conducted the manuscript writing.
Declaration of Competing Interest
None
References
- Huang L, Li X, Gu X, Zhang H, Ren L et al. (2022) Health outcomes in people 2 years after surviving hospitalisation with COVID-19: A longitudinal cohort study. Lancet Respir Med 10:863-876.
[Crossref] [Google Scholar] [PubMed]
- Korompoki E, Gavriatopoulou M, Hicklen RS, Ntanasis-Stathopoulos I, Kastritis E, et al. (2021) Epidemiology and organ specific sequelae of post-acute COVID19: A narrative review. J Infect 83:1-16.
[Crossref] [Google Scholar] [PubMed]
- Xu B, Ma FQ, He C, Wu ZQ, Fan CY, et al. (2022) Incidence and affecting factors of pulmonary diffusing capacity impairment with COVID-19 survivors 18 months after discharge in Wuhan, China. J Infect 84:16.
[Crossref] [Google Scholar] [PubMed]
- Huang L, Yao Q, Gu X, Wang Q, Ren L, et al. (2021) 1-year outcomes in hospital survivors with COVID-19: a longitudinal cohort study. Lancet 398:747-758.
[Crossref] [Google Scholar] [PubMed]
- Shah AS, Wong AW, Hague CJ, Murphy DT, Johnston JC, et al. (2021) A prospective study of 12-week respiratory outcomes in COVID-19-related hospitalisations. Thorax 76:402-404.
[Crossref] [Google Scholar] [PubMed]
- Huang Y, Tan C, Wu J, Chen M, Wang Z, et al. (2020) Impact of coronavirus disease 2019 on pulmonary function in early convalescence phase. Respir Res 21:163.
[Crossref] [Google Scholar] [PubMed]
- Wu X, Liu X, Zhou Y, Yu H, Li R, et al. (2021) 3-month, 6-month, 9-month, and 12-month respiratory outcomes in patients following COVID-19-related hospitalisation: A prospective study. Lancet Respir Med 9:747-754.
[Crossref] [Google Scholar] [PubMed]
- Huang L, Wang Y, Li X, Ren L, Gu X (2021) 6-month consequences of COVID-19 in patients discharged from hospital: A cohort study. Lancet 397:220-232.
[Crossref] [Google Scholar] [PubMed]
- Zhao YM, Shang YM, Song WB, Li QQ, Xie H, et al. (2020) Follow-up study of the pulmonary function and related physiological characteristics of COVID-19 survivors three months after recovery. E Clinical Medicine 25:100463.
[Crossref] [Google Scholar] [PubMed]
- Lang M, Som A, Mendoza DP, Flores EJ, Reid N, et al. (2020) Hypoxaemia related to COVID-19: vascular and perfusion abnormalities on dual-energy CT. Lancet Infect Dis 20:1365-1366.
[Crossref] [Google Scholar] [PubMed]
- Hanidziar D, Robson SC (2021) Hyperoxia and modulation of pulmonary vascular and immune responses in COVID-19. Am J Physiol Lung Cell Mol Physiol 320:12-16.
[Crossref] [Google Scholar] [PubMed]
- Carr E, Bendayan R, Bean D, Stammers M, Wang W, et al. (2021) Evaluation and improvement of the National Early Warning Score (NEWS2) for COVID-19: A multi-hospital study. BMC Med 19:23.
[Crossref] [Google Scholar] [PubMed]
- Jin C, Chen W, Cao Y, Xu Z, Tan Z, et al. (2020) Development and evaluation of an artificial intelligence system for COVID-19 diagnosis. Nat Commun 11:1-4.
[Crossref] [Google Scholar] [PubMed]
- Abdulaal A, Patel A, Charani E, Denny S, Mughal N, et al. (2020) Prognostic modeling of COVID-19 using artificial intelligence in the United Kingdom: Model development and validation. J Med Internet Res 22:20259.
[Crossref] [Google Scholar] [PubMed]
- Pan P, Li Y, Xiao Y, Han B, Su L, et al. (2020) Prognostic assessment of COVID-19 in the intensive care unit by machine learning methods: Model development and validation. J Med Internet Res 22:23128.
[Crossref] [Google Scholar] [PubMed]
- Zampieri FG, Salluh JI, Azevedo LC, Kahn JM, Damiani LP, et al. (2019) ICU staffing feature phenotypes and their relationship with patients’ outcomes: An unsupervised machine learning analysis. Intensive Care Med 45:1599-1607.
[Crossref] [Google Scholar] [PubMed]
- Ploug T, Holm S (2020) The four dimensions of contestable AI diagnostics-A patient-centric approach to explainable AI. Artif Intell Med 107:101901.
[Crossref] [Google Scholar] [PubMed]
- Roscher R, Bohn B, Duarte MF, Garcke J (2020) Explainable machine learning for scientific insights and discoveries. Ieee Access 8:42200-42216.
[Crossref] [Google Scholar] [PubMed]
- Reddy S (2022) Explainability and artificial intelligence in medicine. Lancet Digit Health 4:214-215.
[Crossref] [Google Scholar] [PubMed]
- McCoy LG, Brenna CT, Chen SS, Vold K, Das S (2022) Believing in black boxes: Machine learning for healthcare does not need explainability to be evidence-based. J Clin Epidemiol 142:252-257.
[Crossref] [Google Scholar] [PubMed]
- Cai J, Luo J, Wang S, Yang S (2018) Feature selection in machine learning: A new perspective. Neurocomputing 300:70-79.
- Lundberg S, Lee SI (2017) A unified approach to interpreting model predictions. Neural Inf Process Syst.
- Lundberg SM, Erion G, Chen H, DeGrave A, Prutkin JM, et al. (2020) From local explanations to global understanding with explainable AI for trees. Nat Mach Intell 2:56-67.
[Crossref] [Google Scholar] [PubMed]
- Liu X, Zhou H, Zhou Y, Wu X, Zhao Y, et al. (2020) Temporal radiographic changes in COVID-19 patients: Relationship to disease severity and viral clearance. Sci Rep 10: 10263.
[Crossref] [Google Scholar] [PubMed]
- Liu X, Zhou H, Zhou Y, Wu X, Zhao Y, et al. (2020) Risk factors associated with disease severity and length of hospital stay in COVID-19 patients. J Infect 81:95-97.
[Crossref] [Google Scholar] [PubMed]
- Hu C, Li L, Li Y, Wang F, Hu B, et al. (2022) Explainable machine-learning model for prediction of in-hospital mortality in septic patients requiring intensive care unit readmission. Infect Dis Ther 11:1695-1713.
[Crossref] [Google Scholar] [PubMed]
- Blanco JR, Cobos-Ceballos MJ, Navarro F, Sanjoaquin I, de Las Revillas FA, et al. (2021) Pulmonary long-term consequences of COVID-19 infections after hospital discharge. Clin Microbiol Infect 27:892-896.
[Crossref] [Google Scholar] [PubMed]
- Cen Y, Chen X, Shen Y, Zhang XH, Lei Y, et al. (2020) Risk factors for disease progression in patients with mild to moderate coronavirus disease 2019: A multi-centre observational study. Clin Microbiol Infect 26:1242-1247.
[Crossref] [Google Scholar] [PubMed]
- Zhou F, Yu T, Du R, Fan G, Liu Y, et al. (2020) Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395:1054-1062.
[Crossref] [Google Scholar] [PubMed]
- Price WN (2018) Big data and black-box medical algorithms. Sci Transl Med 10: 5333.
[Crossref] [Google Scholar] [PubMed]
- Lancet Respiratory M (2018) Opening the black box of machine learning. Lancet Respir Med 6:801.
- Musolf AM, Holzinger ER, Malley JD, Bailey-Wilson JE (2022) What makes a good prediction? Feature importance and beginning to open the black box of machine learning in genetics. Hum Genet 141:1515-1528.
[Crossref] [Google Scholar] [PubMed]
- Arnold DT, Hamilton FW, Milne A, Morley AJ, Viner J, et al. (2021) Patient outcomes after hospitalisation with COVID-19 and implications for follow-up: results from a prospective UK cohort. Thorax 76:399-401.
[Crossref] [Google Scholar] [PubMed]
- Patel BV, Arachchillage DJ, Ridge CA, Bianchi P, Doyle JF, et al. (2020) Pulmonary angiopathy in severe COVID-19: Physiologic, imaging, and hematologic observations. Am J Respir Crit Care Med 202:690-699.
[Crossref] [Google Scholar] [PubMed]
- Taus F, Salvagno G, Canè S, Fava C, Mazzaferri F, et al. (2020) Platelets promote thromboinflammation in SARS-CoV-2 pneumonia. Arterioscler Thromb Vasc Biol 40:2975-2989.
[Crossref] [Google Scholar] [PubMed]
- Chao Y, Rebetz J, Bläckberg A, Hovold G, Sunnerhagen T, et al. (2021) Distinct phenotypes of platelet, monocyte, and neutrophil activation occur during the acute and convalescent phase of COVID-19. Platelets 32(8):1092-102.
[Crossref] [Google Scholar] [PubMed]
- Nicolai L, Leunig A, Brambs S, Kaiser R, Weinberger T, et al. (2020) Immunothrombotic dysregulation in COVID-19 pneumonia is associated with respiratory failure and coagulopathy. Circulation 142:1176-1189.
[Crossref] [Google Scholar] [PubMed]
- Manne BK, Denorme F, Middleton EA, Portier I, Rowley JW, et al. (2020) Platelet gene expression and function in patients with COVID-19. Blood 136:1317-1329.
[Crossref] [Google Scholar] [PubMed]
- Topalovic M, Das N, Janssens W (2019) Artificial intelligence for pulmonary function test interpretation. Eur Respir J 53: 1900782.
[Crossref] [Google Scholar] [PubMed]
- Topalovic M, Das N, Burgel PR, Daenen M, Derom E, et al. (2019) Artificial intelligence outperforms pulmonologists in the interpretation of pulmonary function tests. Eur Respir J 53.
[Crossref] [Google Scholar] [PubMed]
- Mekov E, Miravitlles M, Petkov R (2020) Artificial intelligence and machine learning in respiratory medicine. Expert Rev Respir Med 14:559-564.
[Crossref] [Google Scholar] [PubMed]
Citation: Fu-qiang MA, Cong HE, Yang H, Zuo-wei HU, He-rong Mao, et al. (2022) Interpretable Machine-Learning Model for Prediction of Convalescent COVID-19 Patients with Pulmonary Diffusing Capacity Impairment. J Infect Dis Ther.S6:005.
Copyright: © 2022 Fu-qiang MA, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Usage
- Total views: 2674
- [From(publication date): 0-2022 - Nov 27, 2025]
- Breakdown by view type
- HTML page views: 2198
- PDF downloads: 476