Research Article Open Access
Interaction Effects of Plants and Indigenous Micro-organisms on Degradation of N-Alkanes in Crude Oil Contaminated Agricultural Soil
Toochukwu Ekwutosi Ogbulie1*, Christopher Duru2 and Ferdinand Chidi Nwanebu3
1Department of Biotechnology, School of Science, Federal University of Technology, Owerri (FUTO), Nigeria
2Department of Biological Science, School of Science, FUTO, Nigeria
3Department of Microbiology, School of Science, FUTO, Nigeria
- *Corresponding Author:
- Toochukwu Ekwutosi Ogbulie
Department of Biotechnology
School of Science
Federal University of Technology
Owerri (FUTO), Nigeria
Tel: +23408035472379
E-mail: ogbulie_toochi@yahoo.com
Received Date: August 11, 2015 Accepted Date: August 18, 2015 Published Date: August 20, 2015
Citation: Ogbulie TE, Duru C, Nwanebu FC (2015) Interaction Effects of Plants and Indigenous Micro-organisms on Degradation of N-Alkanes in Crude Oil Contaminated Agricultural Soil. J Ecosys Ecograph 5:166. doi:10.4172/2157- 7625.1000166
Copyright: © 2015 Ogbulie TE, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Ecosystem & Ecography
Abstract
Agricultural soil samples from mapped out areas for the study were aseptically collected with sterile plastic sample containers and microbiologically analyzed to isolate autochthonous microbial flora. Seeds of four annual crops including Vigna unguiculata var unguiculata, Mucuna pruriens, Zea mays and T occidentalis used were planted on the test soil and polluted with Bonny light crude oil twenty eight (28) days after plant growth. Thirty days after pollution, soil samples were collected within the rhizosphere of the test plants and examined microbiologically to isolate persisting microorganisms in the polluted soil. The variation in degradation of n-alkanes was ascertained seven days after pollution using Gas chromatographic analysis on soil samples and compared with the control. The pre microbial lab analysis of the soil under the study revealed culturally, the presence of PenicillIum sp Aspergillus fumigatus, Aspergillus niger, Candida sp, Pseudomonas fluorescence, Acinetobacter baumanni, Bacillus mycoides, Klebsiella sp., Staphylococcus aureus and Escherichia coli whereas the absence of the last two isolates was observed during post microbial analyses. Results of the GC analysis on comparison to the control sample depict that plants kept in the greenhouse were able to degrade alkanes within the range of C7 to C12 and C32 to C40 while samples in the field degraded alkanes within the range C7 to C15 and C36 to C40. M pruriens degraded C13. This study could be a promising tool in conversion of crude oil in contaminated agricultural soil to less toxic substances for enhanced remediation.
Keywords
Plants; Indigenous microorganisms; n-Alkanes; Degradation; Crude oil; Agricultural soil; Nigeria
Introduction
The simplest form of bioremediation is natural attenuation (NA) or bioattenuation, during which the indigenous microbial populations degrade recalcitrants or xenobiotics based on their natural, nonengineered metabolic processes [1-3]. According to the Environmental Protection Agency in the United States [4] NA or intrinsic bioremediation processes include a variety of physical, chemical, and biological processes that act to reduce the mass, toxicity, mobility, volume, or concentration of contaminants. These processes include aerobic and anaerobic biodegradation, dispersion, dilution, sorption, volatilization, radioactive decay, and chemical or biological stabilization, transformation, or destruction of contaminants [5].
Natural attenuation as a tool at this moment is now renewed to develop criteria and methods to follow the process of intrinsic bioremediation and to make this process more predictable and therefore more suitable as a bioremediation tool. In the situation of this study, plants in interaction with microorganisms are now employed. Plant roots release a wide variety of materials to their surrounding soil; these include various alcohols, ethylene, sugars, anion and organic acids, vitamins, nucleotides, polysaccharides and enzymes [6]. Microorganisms in the rhizosphere react to the many metabolites released by plant roots. They (rhizosphere microbes) and their products also interact with plant roots in a variety of positive, negative and neutral ways. Such interactions can influence plant growth and development, change nutrient dynamics, and alter the plant susceptibility to disease and abiotic stress. On the other hand, they serve as liable sources of nutrients for other organisms, thus creating a soil microbial loop in addition to playing critical roles in organic matter synthesis and degradation [6]. A wide range of bacteria in the rhizosphere can however, promote plant growth. The organisms communicate with the plant using complex chemical signals. These chemical signal compounds include auxins, gibberellins, glycolipids and cytokinins.
Furthermore, plants do accumulate non-essential and/or toxic mineral elements such as lead, sodium, in their ionic form when they are present in the soil solution. Their growth, on the other hand, may therefore be limited by the availability of essential elements, as well as by the presence of these toxic elements [7]. However, the interactions between plant roots and organisms within their rhizosphere help them to acquire essential mineral nutrients and prevent the accumulation of toxic elements. Since all the minerals that a plant requires must come from the ground/soil, and as the activity of microbes in the soil are central to the efficient solubilization of these mineral elements, it is not surprising that a series of generalized and specific plant-microbe associations exist to perform this function [8,9]. This study therefore was designed to elucidate the effect of interaction between plant and indigenous micro- organisms on the degradation of n-alkanes in crude oil contaminated agricultural soil.
Methodology
Sample collection
The plant seed samples used for this study are seeds of four annual indigeneous crops including two annual forage leguminous crop, vegetable cowpea (Vigna unguiculata var unguiculata) and velvet bean Mucuna pruriens; a cereal, maize (Zea mays) and a vegetable crop, fluted pumpkin (Telfaira occidentalis). These plant seeds were collected from Umuguma in Owerri West L.G.A of Nigeria. The crude oil used was Bonny light Crude and was collected in sterile containers from Akiri in Oguta, Imo State, whereas the soil sample for microbial analysis was collected from an agricultural soil using sterile containers, at the depth of 15-30 cm, and taken to the laboratory for analysis within 1 hour of collection.
Isolation and characterization of indigenous micro-organisms from the agricultural soil mapped out for the analysis
Soil samples from mapped out areas for the study were aseptically collected with sterile plastic sample containers and microbiologically analyzed to isolate naturally existing microbial flora. After pollution though, soil samples were also collected to know the persisting isolate the polluted soil. These were carried out using spread plate method of Cheesbrough [10] on nutrient agar (Oxoid), MacConkey agar (Oxoid), Mineral salt agar (Lab-M) and Saboraud dextrose agar (Oxoid. The microorganisms isolated were characterized morphologically and biochemically using standard microbiological methods [10]; whereas identification was as described in Berger’s Manual of Determinative Bacteriology [11].
Preparation of seeds for planting
The plant seeds used for the research were surface sterilized to eliminate contaminants as described by Yee [12]. This was done by washing the seeds initially with Tap water, then with 75% ethanol for 30 seconds with continual swirling, rinsed 3 times for 10 minutes each. Thereafter, ethanol was decanted and the seeds were further washed with 5.25% Sodium hypochlorite solution for 15 minutes accompanied with swirling and rinsed 3 times again for 10 minutes each rinse. Following sterilization, the seeds were placed on Petri dishes containing sterile cotton wools pre soaked in a 2.5 % sodium hypochlorite solution for 30 minutes, rinsed with sterile water and autoclaved in a foilcovered beaker containing water. The moistened cotton wool provides moisture needed for the seeds to germinate. The plates were kept in a growth chamber for a period of 72-168 hours with a light cycle of 11h darkness and 13h light and 65% humidity at 25°C for germination to take place before planting.
Planting of the seeds
After 72-168 hours, germinated seeds were planted in 450 g of soil contained in sterile bottom perforated plastic pots. They were transported from the laboratory to the green house and were watered with 300 mls of sterile water using sterile measuring cylinder [13] at 48 hours interval as described by Yee [12].
Pollution of plants using crude oil
After twenty- eight (28) days of plant growth, 100 mls of Crude oil was poured evenly at the base of each plant (surface pollution), that is on the surface of the soil using sterile measuring cylinder as described by Ogbulie [13], together with 50 ml of sterile water. Subsequently other concentrations of crude oil (200, 400, and 800) ml were used for surface pollution of the other potted plants. However, no additional water was added during the remaining period of the experiment as described by Yee [12].
Assay for n-alkane degradation
After 30 days of pollution, the n-alkane present in the test samples was determined using the Gas Chromatograph (GC) with GC recorder interfaced with a computer to ascertain the various C- chains that were removed successfully through the interaction. This was carried out by Anal Concept Nigeria Limited in PortHarcourt, River State.
Results and Discussion
The results of the microbial analysis of the study soil sample are as shown in Tables 1 and 2. The consortia of microorganisms present in the soil under the study (before pollution), were identified to be Aspergillus fumigatus, Aspergillus niger, Penicillium sp, Candida sp, Pseudomonas fluorescens, Acinetobacter baumanni, Bacillus mycoides, Klebsiella sp., Staphylococcus aureus and Escherichia coli. (Table 3) on the other hand, depicts the prevalence of the isolates from the agricultural soil under study before and after pollution hence, the disappearance of S. aureus and E. coli was observed after pollution.
| Isolates | Colour (physical morphology) | Microscopic description | Probable genera |
|---|---|---|---|
| 1 | White colony with slightly green pigmentation. Colourless mycelium at the periphery mycelium at the periphery | Dense brush-like spore bearing structures with branched conidiophores terminated by clusters of flask-shaped phialides | Penicillium. sp. |
| 2 | Whitish colony with central part greenish | A dense club- like spore bearing structures of the conidiophores was seen | Aspergillus. fumigatus |
| 3 | Cream coloured pasty colony with yeast odour | Budding cells | Candida sp |
| 4 | White to greenish colony with central part dark green in colour with black pigmentation | Large flask-shaped Large flask-shaped | Aspergillusniger |
Table 1: Mycological characterization of probable fungal isolates.
| Colony Morphology | Grams rxn | Citrate | Oxidase | Catalase | Indole | MR | VP | Glucose | Sucrose | Glucose | Lactose | Fructose | Maltose | Mannitol | Probable Genus | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Round creamy flat colony with a glistering surface | +vecocci | + | + | + | - | + | - | + | NT | NT | NT | NT | NT | A | Staphylo coccussp | |
| Irregular creamy flat colony | -ve rod | - | - | + | + | + | - | + | AG | AG | AG | NR | A | A | Escherichia coli | |
| Roundish wavy flat cream colony | -ve rods | + | - | + | - | + | - | + | AG | AG | AG | AG | A | A | Klebsiellasp | |
| Round to spreading colony with yellow-green pigment. | -ve rods | + | + | - | - | + | - | + | NR | A | NR | NR | AG | NR | Pseudomonas fluorescens | |
| Grey-white Irregularcolonies with wavy edge. Non haemolytic on blood agar. | +ve sporing rods | + | - | + | - | NR | NR | + | NR | AG | NR | NR | AG | NR | Bacillus mycoides | |
| Roundish cream colony with entire edge, grows on 4% NaCl. | -ve rods | + | - | + | + | + | - | + | AG | AG | AG | NR | NR | NR | Acinetobacter baumanni | |
Legend: -ve = negative; +ve = positive; + = positive reaction; - = negative reaction, NR = no reaction; AG = acid and gas production; A = acid production, sp =specie, NT = not tested
Table 2: Phenotypic and biochemical characterization of probable bacterial isolates.
| Isolates | Before pollution | After pollution |
|---|---|---|
| Aspergillusfumigatus, | + | + |
| Aspergillusniger, | + | + |
| Penicilliumsp, | + | + |
| Candida albicans, | + | + |
| Klebsiellasp., | + | + |
| Bacillusmycoides | + | + |
| Pseudomonas fluorescens | + | + |
| Ainetobacterbaumani | + | + |
| Staphylococcus aureus | + | - |
| Escherichia coli | + | - |
| + = Present, - = Absent | ||
Table 3: Prevalence of isolates from soil under study before and after pollution.
Interactions between plant and indigenous microorganisms in degradation of crude oil
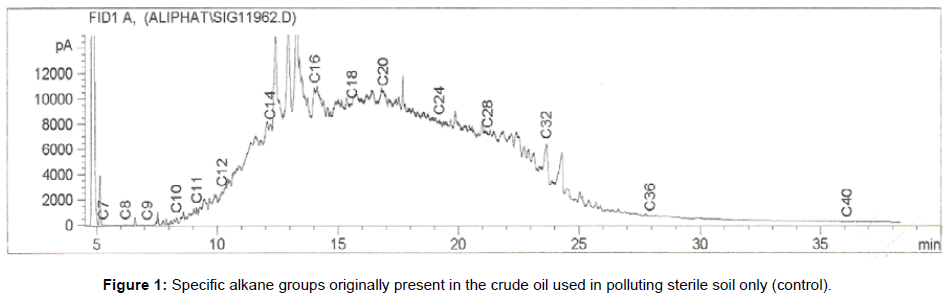
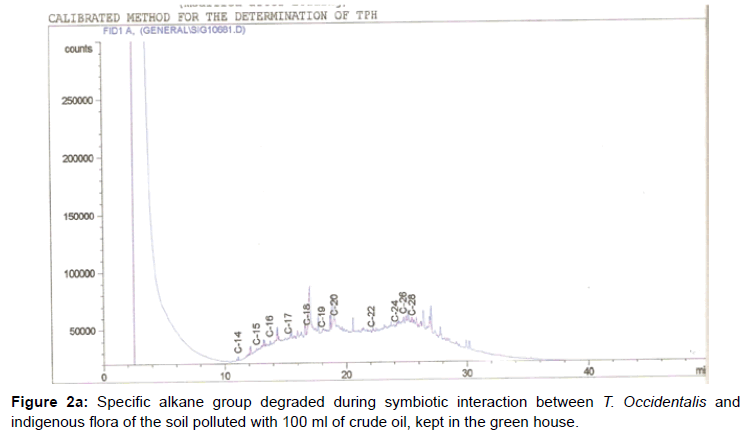
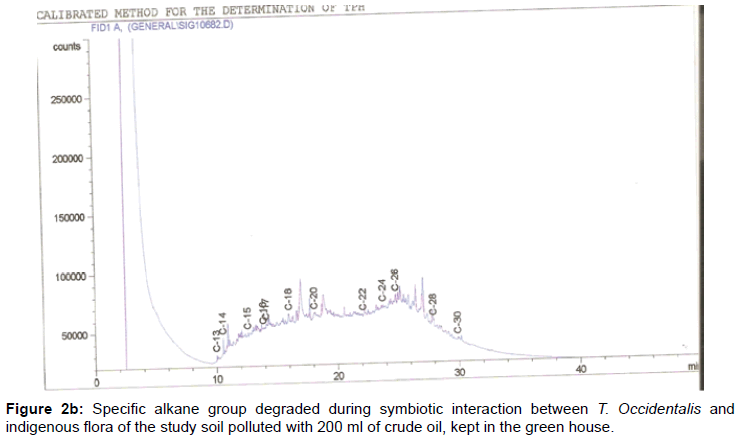
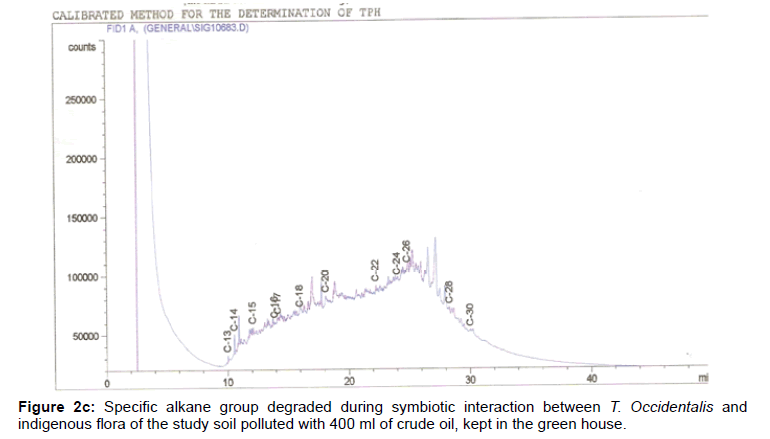
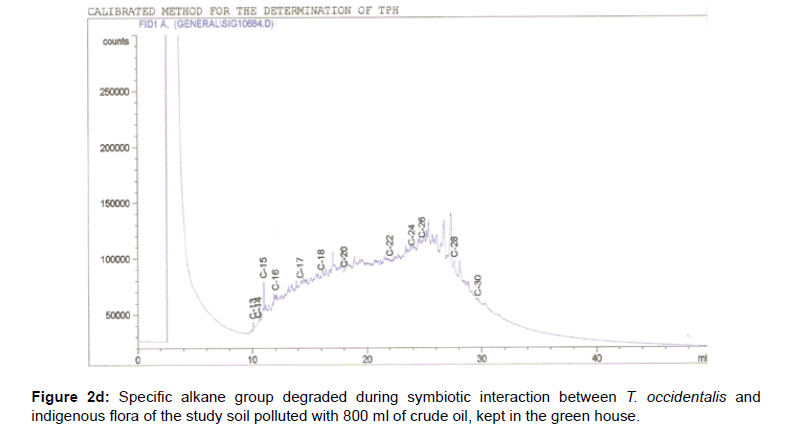
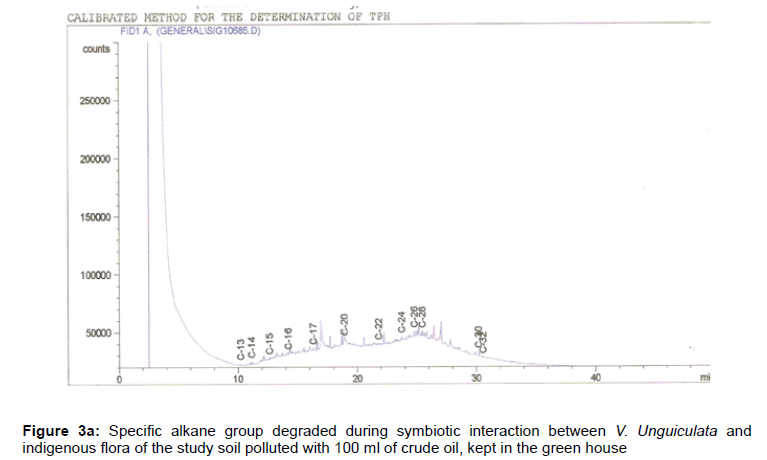
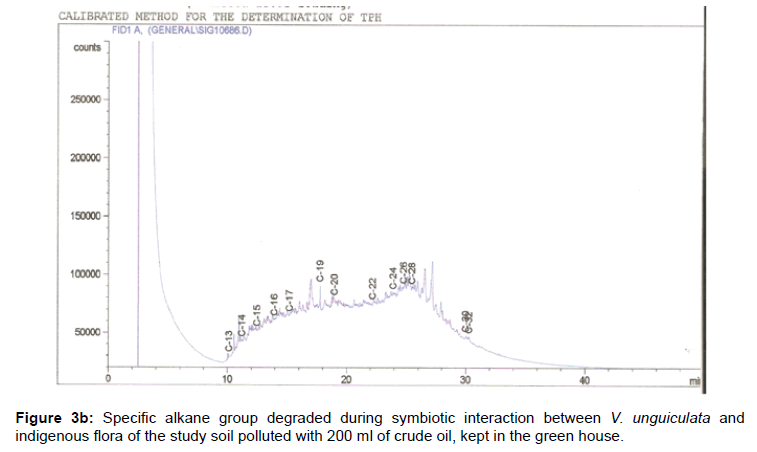
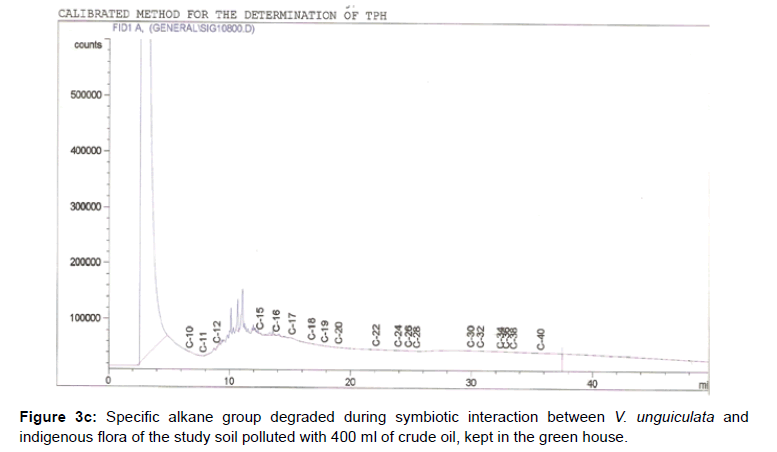
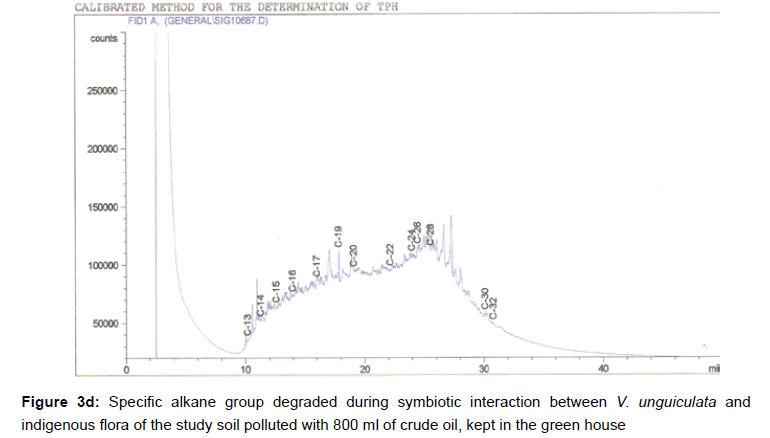
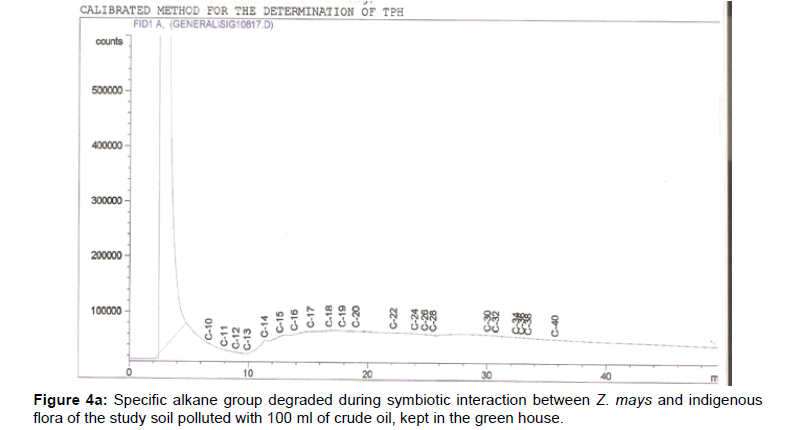
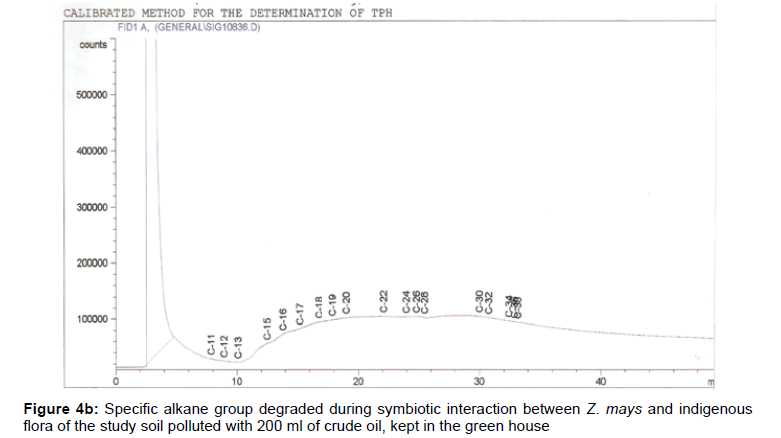
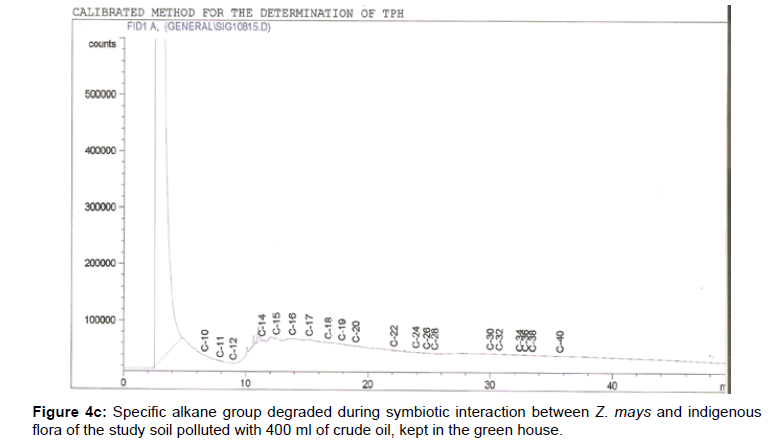
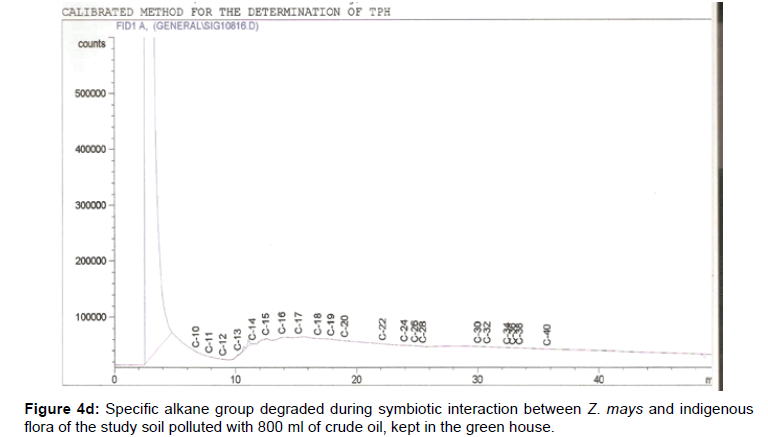
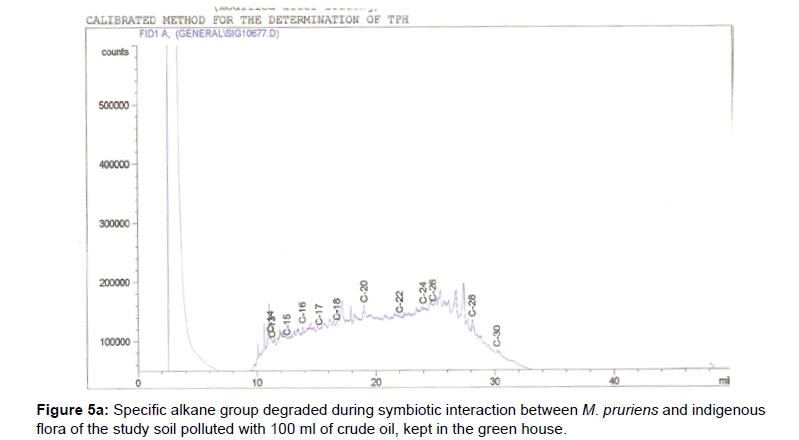
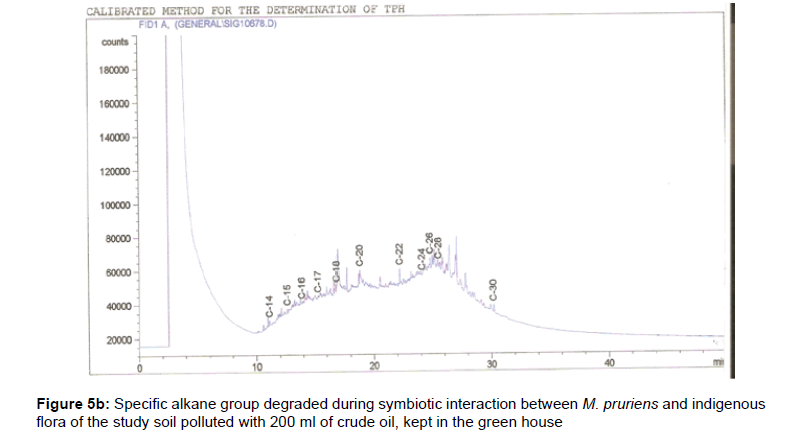
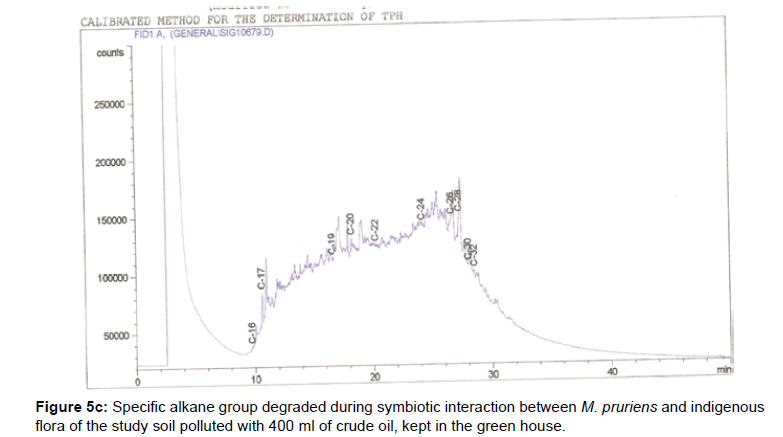
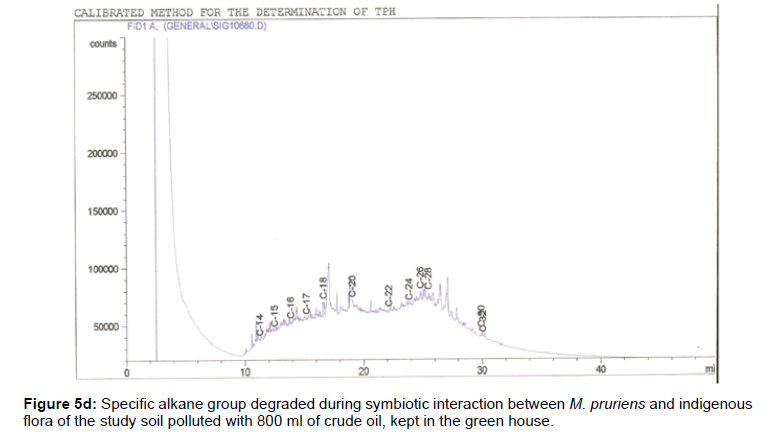
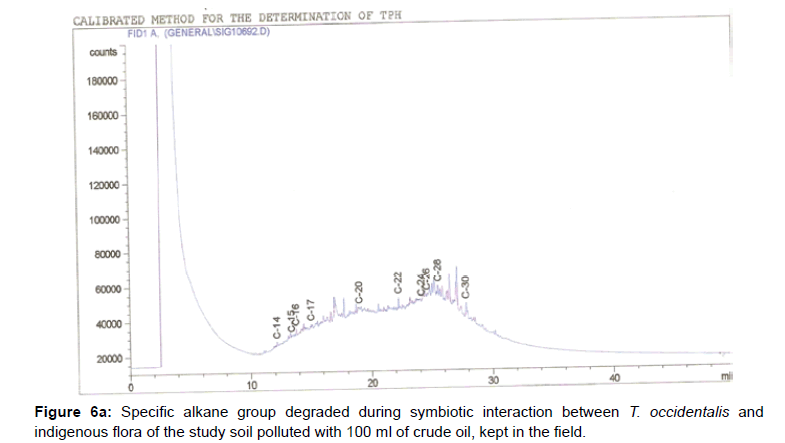
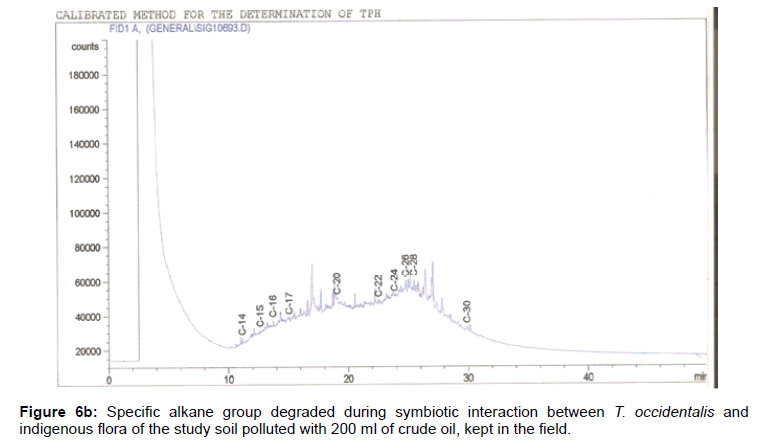
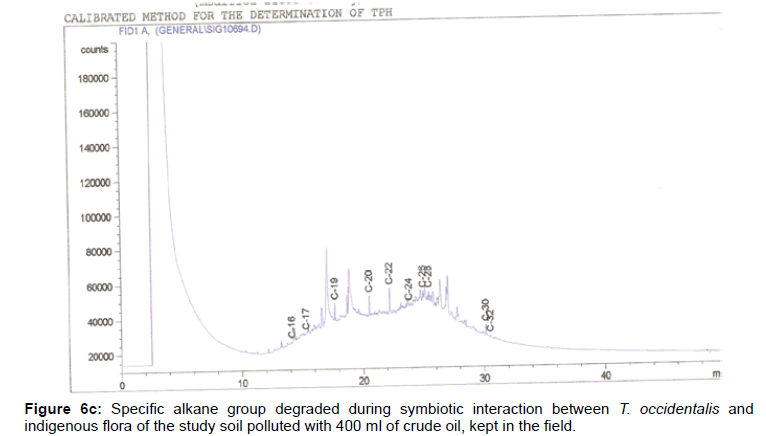
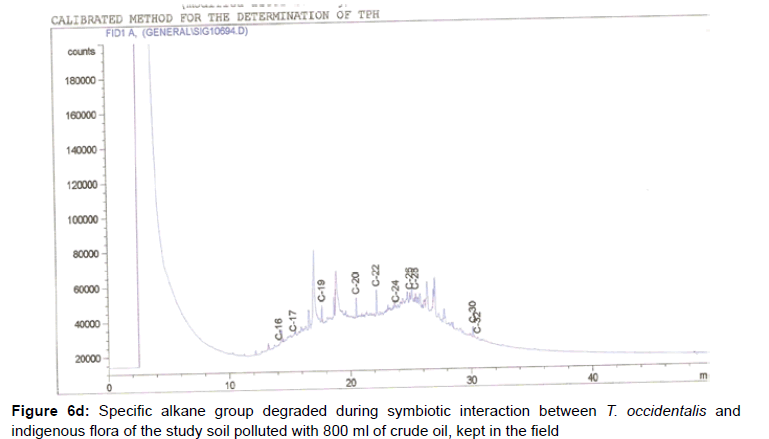
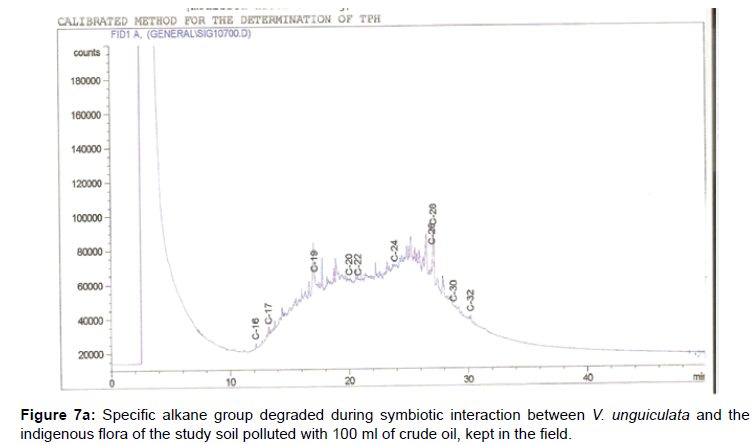
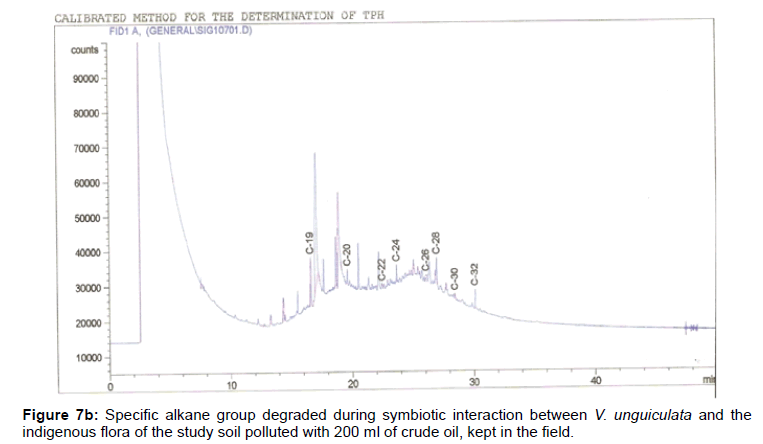
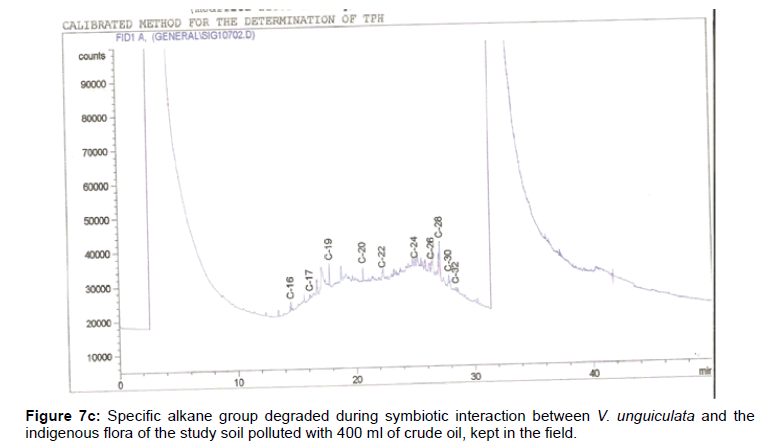
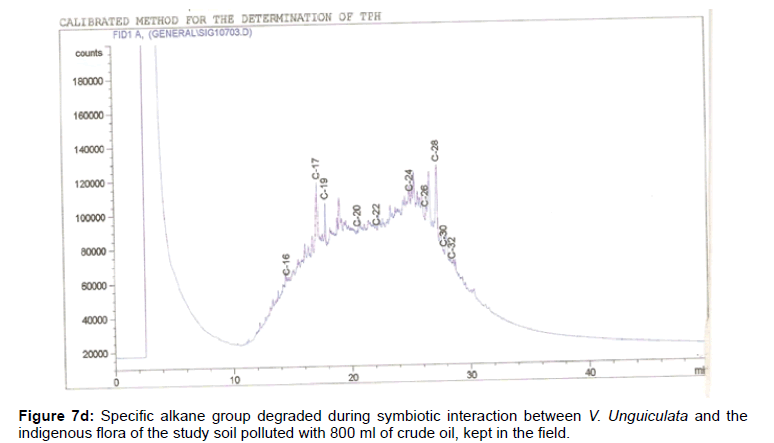
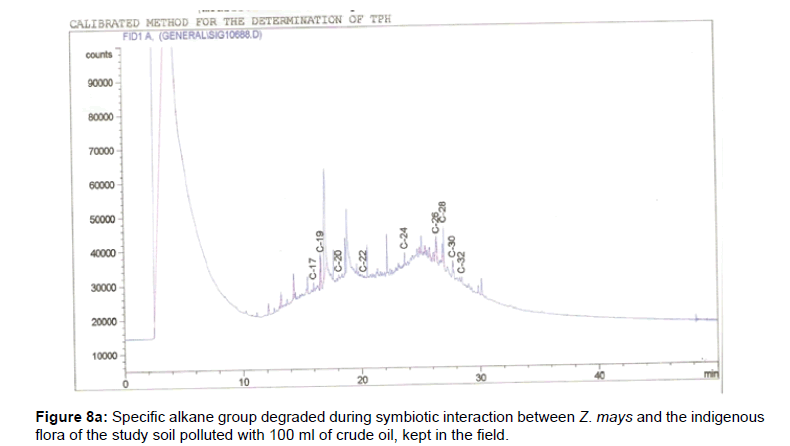
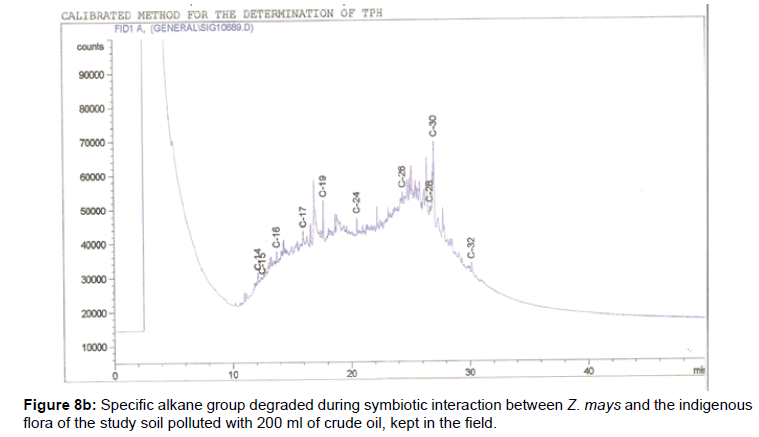
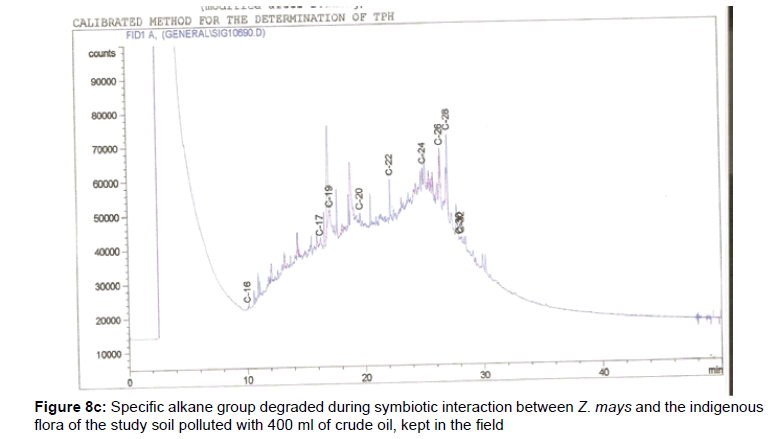
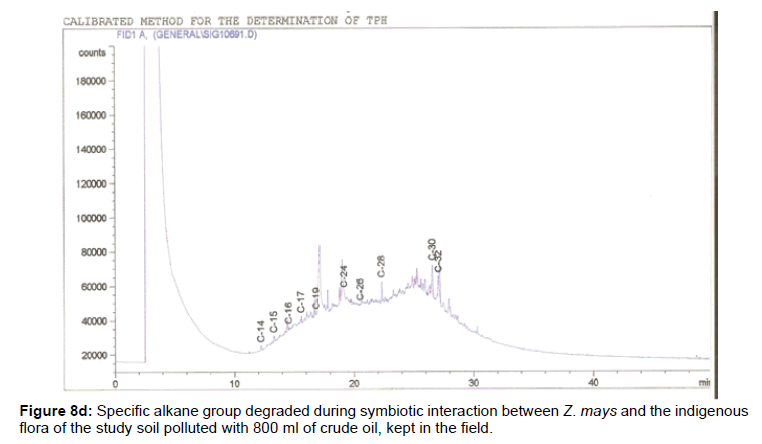
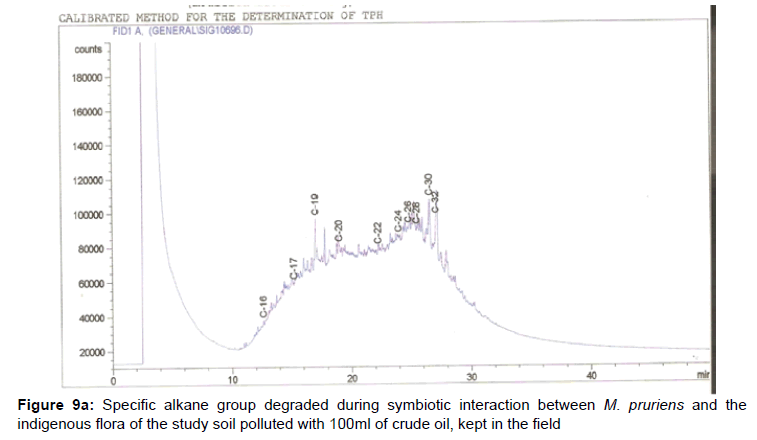
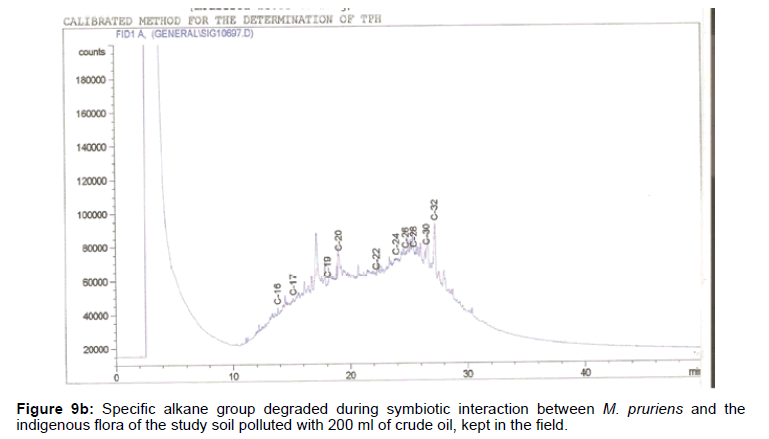
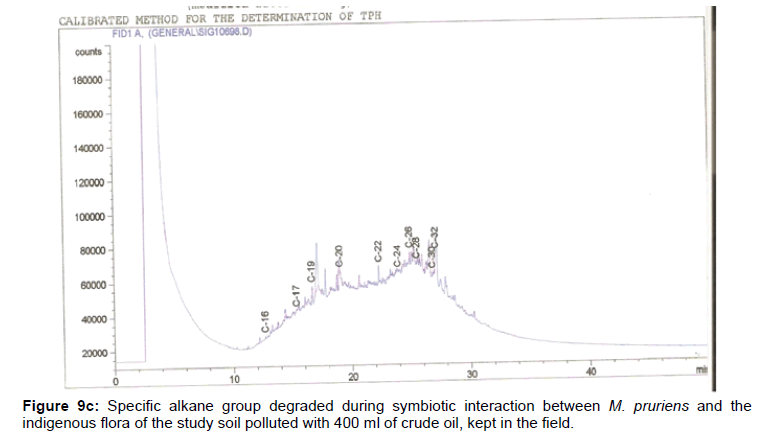
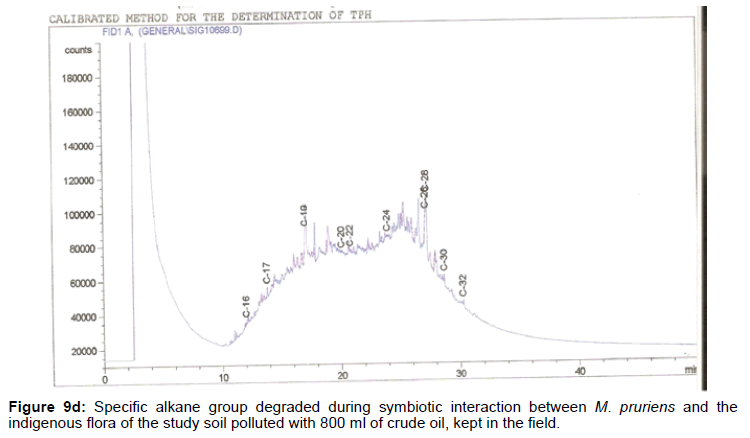
The removal of straight chain hydrocarbon from the test samples in comparison to that of the control sample (Figure 1) showed that there was degradation of n-alkanes from the crude oil sample and the various C chains removed are as shown in Figures 2a-9d. With comparison to the control sample, plants kept in the greenhouse were able to degrade alkanes within the range of C7 to C12 and C32 to C40 except Z mays samples that were able to degrade C7-C9 alkane goup. M pruriens plants on the other hand were able to degrade C13 alkanes (Figures 2a-5d). Comparatively, the field samples were able to degrade alkanes within the range C7 to C15 and C36 to C40 (Figures 6a-9d).
Microbiological analysis carried out in this study revealed the isolation of fungal and bacterial genera identified as Aspergillus fumigatus, Aspergillus niger, Penicillium sp, Candida albicans, Pseudomonas fluorescens, Acinetobacter baumanni, Bacillus mycoides, Klebsiella sp., from the treated natural soil under the study. The isolation of Aspergillus fumigatus, Aspergillus niger, Penicillium sp. and Candida albicans from the sample which persisted even after treatment supports the findings of Sutherland and da Silva [14] who reported the degradation of polycyclic aromatic hydrocarbons (PAHs) by Aspergillus niger and Penicillium janthinellum among others. It also lend more weight to the studies made by Nkwelang [15] on the diversity, abundance and succession of hydrocarbon utilizing microorganisms in the tropical soil polluted with oil sludge. They also isolated bacterial as Pseudomonas sp., Bacillus sp., Acinetobacter sp. and fungal genera as Aspergillus sp, Penicillium sp, Candida sp., Mucor, Rhizopus sp., Sporobolomyces. The report also showed that Pseudomonas sp., Bacillus sp., Aspergillus sp., and Penicillium sp were present in the polluted soil throughout the experimental period [15].
The individual carbon (C) chain removed from the crude oil contaminated soil samples as a result of hydrocarbon degradation shown in this study depicts that aside C6 which was originally absent in the crude oil sample used, this bioremediation processes employed, irrespective of the location of the study, was able to degrade C7 to C15 and C32 to C40. Z mays possibly was not able to degrade more carbon chain during the study due to the phytotoxic effect of the crude oil on the plant, hence the plant died eight days after pollution as reported by Ogbulie and Nnopuechi, [16] who studied on the phytoremediation of crude oil polluted soil and its effect on the growth rate of test plants. Nonetheless, similar biodegradation of n-alkanes with molecular chain lengths up to n-C44 have been demonstrated which according to Atlas [17] normally proceeds by monoterminal attack resulting in the formation of a primary alcohol, an aldehyde and a monocarboxylic acid. These attacks have also been reported elsewhere [18] to be initiated by monogenase enzymes produced by microorganisms such as Corynebacterium sp. Similar enzymes have also been detected in other bacteria and yeasts such as Acinetobacter calcoaceticus, Pseudomonas putida and Candida tropicalis, Candida, rugosa and Candida lipolytica respectively [17,18]. This however is in line with the findings of this study and supports the findings of Kastueur and Mahro [19], Onwurah [20] and Heidelberg et al. [21] who reported that biodegradation pathways for each petroleum hydrocarbon or its derivatives are catalyzed by enzymes and cannot be degraded by a single strain of bacterium; hence it requires a consortium of microbes [22].
Furthermore, in line with the findings of this study, the degraded carbon chain was within the two major categories of petroleum hydrocarbon including i) gasoline range organics (GRO) which corresponds to small chain alkanes (C6-C10) with low boiling point (60-170°C) such as isopentane, 2,3-dimethyl butane, n-butane and npentane, and volatile aromatic compounds such as the monoaromatic hydrocarbons benzene, toluene, ethylbenzene, and xylenes (BTEX); ii) diesel range organics (DRO) including longer chain alkanes (C10–C40) and hydrophobic chemicals such as polycyclic aromatic hydrocarbons (PAH) [23].
This study therefore could be a promising tool in remediation of crude oil present in contaminated soil to a less toxic substance or to a permissible level.
Acknowledgement
The author acknowledges the effort of Mr Vincent Agu of Technology Partners International (TPI) Port Harcourt for his help during the Gas Chromatographic analysis of this research.
References
- Widada J, Nojiri H, Omori T (2002) Recent developments in molecular techniques for identification andmonitoring of xenobiotic-degrading bacteria and their catabolic genes in bioremediation . Applied Microbiology and Biotechnology 60: 45-59.
- Kuiper I,LagendijkEL,BloembergGV, LugtenbergBJ(2004)Rhizoremediation. A beneficial plant-microbe interaction. Molecular Plant-Microbe Interactions17: 6-15.
- Sivasubramaniam L, Seshadri M, Shah N (2005) Bioremediation: Biotransformations of toxic wastes toharmless products. Special Article Reviews, Career Development.Technical Free full text article.Katangulathur, Chennai, India.Pharmainfro.net.
- United States Environmental Protection Agency (USEPA) (1999) Use of monitored natural attenuation at superfund, RCRA Corrective action and underground storage tank sites.
- Rittmann BE (2004) Definition, objectives and evaluation of natural attenuation. Biodegradation15: 349-357.
- Prescott LM, Harley JP, Klein DA (2005) Microorganisms in terrestrial environment. In:Microbiology, sixthed., McGraw-Hill Limited, New York651-660.
- White Jr PM, Wolf DC, Thoma GJ, Reynolds CM (2006) Phytoremediation of alkylated polycyclic aromatic hydrocarbon in a crude oil-contaminated soil. Water, Air and Soil Pollution169:207-220.
- Morgan JAW, Bending GD, White PJ (2005) Biological cost and benefits to plant- microbe interactions in the rhizosphere. Journal of Experimental Botany56: 1729-1739.
- Lines-Kelly R (2005) Therhizosphere: soil biology basics. Profitable and sustainable primary industries.http://www.agric.nsw.gov.au/reader/soil-biology
- Cheesbrough M (1994) District laboratory practice in Tropical countries. Part 2 CambridgeUniversitypress 479.
- Holt JG (1984) The Shorter Berger’s Mannual of Determinative Bacteriology, WilliwmsWilkiins, Baltimore 356.
- Yee DC, Maynard JA, Wood TK (1998) Rhizoremediation of trichloroethylene by a recombinant root–colonizing Pseudomonas fluorescens strain expressing toluene ortho- monoxygenase constitutively.Applied and Environmental Microbiology 64: 112-118.
- Ogbulie TE, Nwigwe HC, Okpokwasili GC, Iwuala MOE (2011) Comparative Study on the effect ofsymbiotic interaction between plants and non-indigenous isolates on crude oil remediation. AnaleleUniversitãtii din Oradea- FasciculaBiologie Tom XVIII: 15-22.
- Sutherlan KP, da Silva I (2003) Fungal Diversity and use in decomposition of environmental pollutants. Critical Reviews in Microbiology 31: 197-212.
- Nkwelang G, Kamga HF, Nkeng GF, Antai SP (2008) Studies on the Diversity, abundance and succession of hydrocarbon utilizing microorganisms in tropical soil polluted with oily sludge. AfricanJournal of Biotechnology7: 1075-1080.
- OgbulieTE, Nnopuechi R (2012) Studies on the Phytoremediation of Crude Oil Contaminated Soil and its Effect on the Growth rate of Test Plants. International Journal of Environmental Health andHuman Development 13: 32-40.
- Atlas RM (1981). Microbial degradation of petroleum hydrocarbons: an environmental perspectiveMicrobiological Reviews 45:180-209.
- Higgins IJ, Gilbert PD(1989) The Biodegradation of Hydrocarbons, Cambridge University press, Cambridge, UK 80-106.
- Kastner M,Mahro B (1996) Microbial degradation of polycyclic aromatic hydrocarbon in soils affected by the organic matrix of compost. Applied Microbiology andBiotechnology44: 668-675.
- Onwurah INE (2000) Bioremediation technologies for oily wastes. In: APerspective ofIndustrial and Environmental Biotechnology. Snaap Press Ltd., Enugu, Nigeria 53-71.
- Heidelberg JE, Paulsen II, Nelson KE, Gaidos EJ, Nelson WC, et al. (2002) Genome sequence of thedissimilatory metal ion-reducing bacterium. Shewanellaoneidensis.Nature and Biotechnology1:1-6.
- Ogbulie TE, Nwigwe HC, Iwuala MOE, Okpokwasili GC (2010) Study on theuse of Monoculture andMultispecies in Bioaugumentation of crude oil contaminated Agricultural soil. Nigerian Journal ofMicrobiology 24: 2160-2167.
- Ezeonu CS,Onwurah INE,Oje OA (2012) Comprehensive Perspectives in Bioremediation of CrudeOil Contaminated Environments, Introduction to Enhanced Oil Recovery (EOR) Processes and Bioremediation of Oil-Contaminated Sites, Dr. Laura Romero-Zerón (Ed.), ISBN: 978-953-51-0629-6,InTech.
Relevant Topics
- Aquatic Ecosystems
- Biodiversity
- Conservation Biology
- Coral Reef Ecology
- Distribution Aggregation
- Ecology and Migration of Animal
- Ecosystem Service
- Ecosystem-Level Measuring
- Endangered Species
- Environmental Tourism
- Forest Biome
- Lake Circulation
- Leaf Morphology
- Marine Conservation
- Marine Ecosystems
- Phytoplankton Abundance
- Population Dyanamics
- Semiarid Ecosystem Soil Properties
- Spatial Distribution
- Species Composition
- Species Rarity
- Sustainability Dynamics
- Sustainable Forest Management
- Tropical Aquaculture
- Tropical Ecosystems
Recommended Journals
Article Tools
Article Usage
- Total views: 16237
- [From(publication date):
September-2015 - Mar 31, 2025] - Breakdown by view type
- HTML page views : 11552
- PDF downloads : 4685