Research Article Open Access
Integrins Contributes to Innate Immune Response in Pelteobagrus Fulvidraco
Ming-Ming Han1,2*, Jian-Guo Lu1, Shang-Bin1, Li-Na-Peng1, Shahid Mahboob3,4, Khalid A Al-Ghanim3 and Xiao-Wen Sun1
1Heilongjiang River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Harbin, China
2Shanghai Ocean University, College of Fisheries and Life Science, Shanghai, China
3Department of Zoology, College of Science, King Saud University, Riyadh 11451, Saudi Arabia
4Department of Zoology, GC University, Faisalabad, Pakistan
- *Corresponding Author:
- Ming-Ming Han
Heilongjiang River Fisheries Research Institute
Chinese Academy of Fishery Sciences, Harbin, China
Tel: +86-451-84869349
E-mail: sunxw2002@163.com
Received date: March 15, 2016; Accepted date: April 27, 2016; Published date: May 05, 2016
Citation: Han MM, Lu JG, Bin S, Peng LN, Mahboob S, et al. (2016) Integrins Contributes to Innate Immune Response in Pelteobagrus Fulvidraco. Biochem Physiol 5:204. doi:10.4172/2168-9652.1000204
Copyright: © 2016 Han MM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Biochemistry & Physiology: Open Access
Abstract
Integrins - β are highly conserved pleiotropic cytokine. In this study, we cloned the full - length cDNA sequence of yellow catfish Pelteobagrus fulvidraco integrin - β1 and integrin - β3 gene. The Integrin - β1 cDNA is 2816 bp in length and encodes a protein of 461 amino acids. Integrin - β3 cDNA is 1713 bp in length and encodes 479 amino acids. Integrins - β were classified strictly with the same type of proteins by phylogenetic analysis. Stegastes partitus integrins - β was classified on the same phylogenetic branch compared with other fish. Tissue transcription profile analysis indicated that, in the healthy P. fulvidraco, integrins - β mRNA was predominantly expressed in the liver and brain, follow by gill and spleen. The P. fulvidraco integrins - β shares high homology with the Cypriniformes Danio rerio (83% ~87%). Expression of the two Integrins - β transcripts showed significant expression changes in 4 h after infection with Flavobacterium columnare, which has modulation of expression continuing up to 3 d following pathogen exposure. Both of the integrin - β genes were significantly up - regulated at 24 h in spleen and liver after infection tissue. Integrin - β3 showed the most sensitive to F. columnare, suppressing expression - 6.34 - fold at 24h. This informations elucidates the functions of individual integrins - β genes in regards to pathogen recognition, binding, and their larger immune context.
Keywords
Integrins Β; Pelteobagrus fulvidraco; Gene clone
Introduction
Integrins are mediating cell - cell, cell - matrix, cytoplasmic extracellular matrix interactions and bidirectional signal transduction between the cytosol, which contain type α and β subunits bidirectional signal non - covalent binding of formed between the transduced cell surface heterodimer [1]. Integrins contain 24 different α and β heterodimers receptors [2]. The expression profile of integrins on cancer cells and the composition and organization of extracellular matrix (ECM) of the tumor stroma are major factors in therapy resistance, metastasis and cancer development [3]. Collagen is the most abundant ECM protein in vertebrate organisms, forming the framework of connective tissues and soft interstitial matrices (such as the bone marrow), and is also a critical matrix of the tumor microenvironment [4].
Integrins - β1, associate with 12 different α subunits, is the largest sub - family of integrins [2]. Integrin - β1, as a major collagen receptor, is widely expressed and known to promote cell migration and control tissue homeostasis. Integrin - β1 can either inhibit or promote the cancer cells dissemination process and cancer metastasis regulator [5]. Integrin - β3 plays an important role in a number of physiological and pathological processes such as angiogenesis, tumor invasion and metastasis, and its expression is correlated with disease progression in various tumor types [6,7].
For aquatic animals, the study on integrins - β is few, for wxaple, integrin - β subunits (β - integrin) in shrimps involved in WSSV infection [8]. In order to adapt the continuous improvement of people’s living standard, the scale of aquaculture presents the increasing trend which lead to the farmed animal disease outbreaks [9]. Yellow catfish Pelteobagrus fulvidraco, an omnivorous freshwater fish, is considered as a good candidate for freshwater aquaculture in China for its tasty meat and high market value [10]. Yellow catfish P. fulvidraco integrins - β genes and the distribution of mRNA expression have not been reported.
.In the study, we obtained the full - length cDNA sequence of integrinsβ from Yellow catfish, and further make the necessary sequence analysis, tissue expression analyzed the putative proteins encoded by the gene using bioinformatic approaches. The results of our study provided the basic information to identify the function of integrins - β and to study the molecular mechanism of Yellow catfish P. Fulvidraco.
Materials and Methods
RNA isolation
The P. fulvidraco were obtained from the Chinese Academy of Fishery Sciences Pelteobagrus sp. Breeding Engineering Center. All yellow catfish were 1 year old, 31.6+2.8 g weight, 14.5+3.4 cm length. Total RNA of the yellow catfish was extracted with Trizol reagent (Invitrogen, China) following the manufacturer’s instructions. The total RNA concentration was determined by measuring the absorbance at OD260. RNA integrity was checked by electrophoresis. The total RNA was reverse - transcribed into cDNA using the aM - MLV RTase cDNA Synthesis Kit (TaKaRa, Japan). For cloning and expression analysis, various tissue samples, including the gill, blood, muscle, gonad, liver, brain, spleen, kidney, heart, intestine from male and Female,were immediately collected and frozen in liquid nitrogen until RNA isolation.
Integrins - β cloning
Total RNA was extracted from the P. Fulvidraco tissues including the gill, blood, muscle, gonad, liver, brain, spleen, kidney, heart and intestine using a number for RR047A RT Reagent Kit (TaKaRa, Japan) according to the manufacturer’s instructions. The cDNA was used as the template to amplify the gill, blood, muscle, gonad, liver, brain, spleen, kidney, heart and intestine was stored at -20°C. According to Ietalurus Punetaus in Genbank madtoms integrins β (serial number FD303964 and FD290690), we designed the PCR primers β 1 - F1, β 1 - R1, β 2 - F1 and β 2 - R1 for integrins cloning. (Table 1). After the sequence by Heilongjiang River Fisheries Research Institute of products research, development of genomics and molecular breeding research of yellow catfish are found in 99% of similarity in the method of the transcriptome gene sequence and gene sequence designed primers. For integrins cloning, PCR primers β 1 - F1, β 1 - R1, β 2 - F1 and β 2 - R1 were designed (Table 1).
| primer | sequence(5'-3′) | °C Tm | bp size |
|---|---|---|---|
| β 1-F1 | CTGGAAACCTCGACTCACCT | 56 | |
| β 1-R1 | CATCTACATGCTCTCCACTC | 56 | 755 |
| β 1-F2 | TGTGTGGGTGTGTATGTGGA | 58.5 | |
| β 1-R2 | AAACTGCTGGCCATTCTCAG | 58.5 | 789 |
| β 1-F3 | AGTACAACTCCTGCCAAGCT | 56 | 913 |
| β 1-R3 | ACAAACCCTTCCAATTCGCC | 56 | |
| β 1-F4 | GAGGAGGTTGTGATCGCTCT | 59 | |
| β 1-R4 | ACCACAAAACACAATGCATTCA | 59 | 1236 |
| β 1-F5 | CAACAAGCTATGGAGGTC | 60 | 185 |
| β 1-R5 | GAAACTTGGTCGGTGCAT | 60 | |
| Actin-F | CCACCTTCAACTCCATCA | 60 | |
| Actin-R | TACCACCAGACAGAACAG | 60 | 223 |
| β 3-F1 | CCAGACCTGTACTGCCACTT | 58 | 765 |
| β 3-R1 | AGGTATCTGTGCGTGTGGAG | 58 | |
| β 3-F2 | GTGTGTGGAAGCTGTGAGTG | 59.5 | |
| β 3-R2 | ACACAGCACAGAACTAAACCC | 59.5 | 887 |
| β 3-F3 | TGCCCAACAGAGAAGACCAA | 56 | |
| β 3-R3 | GTAGGTAGAGTTCACGGCGT | 56 | 753 |
| β 1-F4 | CCCGAATTTGCAGAGACGAG | 60 | |
| β 1-R4 | GACTTTCCGCTGTTGTCCTC | 60 | 136 |
Table 1: The primer used for cloning and tissue expression of integrins gene in Pelteobagrus fulvidraco.
The PCR conditions were as follows: 94°C for 4 min, followed by 35 cycles of amplification (94°C for 1 min, 56°C for 1 min, and 72°C for 1 min 30 s), and 72°C for 10 min. The 25 μL reaction volume contained 50 ng genomic DNA using an Agarose Gel DNA Purification Kit (TaKaRa, Japan), 12.5 μL 2X reaction buffer, 0.2 mM dNTP, 10 pM of each primer and 0.5 U of Taq DNA polymerase(TaKaRa,Japan). Then subcloned into the pMD - 18T cloning vector (TaKaRa) connection at 16°C for the night. Tiangen Top 10 into competent e. coli in LB Amp tablet on 37°C, or 230 r/min, Pick the bacteria , expand training and gas - bacilli PCR detection, expand training and gas - bacilli PCR detection. Subsequently, end to end PCR was used to amplify the length sequences of integrinβ1 and integrinβ3. Following 1.0% agarose electrophoresis, all PCR products from the two genes were purified and isolated using an Agarose Gel DNA Purification Kit ver. 1.0 (TaKaRa, Japan). Positive clones containing inserts of an expected size were sequenced using M13 primers and sequenced at BGI (Shanghai, China). The sequencing results on NCBI homology retrieval. Subsequently, end to end PCR was used to amplify the length sequences of integrin - β1 and integrins - β3. Following 1.0% agarose electrophoresis, all PCR products from the two genes were isolated and purified using an Agarose Gel DNA Purification Kit ver. 1.0 (TaKaRa, Japan).
Sequence analysis of integrins-β
Deduced amino acid sequences of integrins - β cDNA were done using the software Vector NTI Advance 11. Integrins - β sequences from different organisms were obtained using the NCBI BLAST search program. The length cDNA sequences of integrin - β1 and integrin - β3 were assembled using DNAMAN 6.0 software (version 5.0, Lynnon Biosoft, Quebec, Canada). The cDNA sequences of integrin - β1 and integrin - β3 from four species compare to their genomic sequences used by exonic sequences structure analysis. BLAST software (http://www.ncbi.nlm.nih.gov) was used to obtain the integrin - β1 and integrin - β3 exonic structures. Open reading frames (ORFs) were predicted using GENSCAN (http://genes.mit.edu/GENSCAN.html).
Phylogenetic analysis
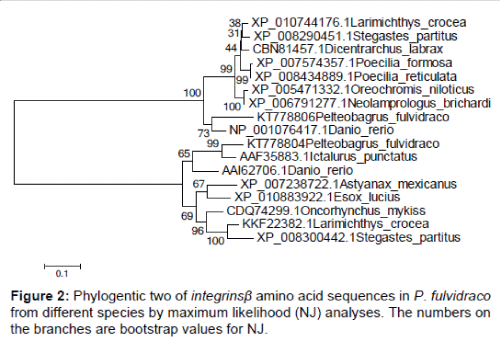
Integrins - β gene cloning CDS coding regions is derived using the Primer 5 software is deduced amino acid sequence, excerpts from GenBank (number for KT778804 and KT778806 GenBank landed). Sequences of integrins - β: angiopoietin of Larimichthys crocea, Poecilia formosa, Dicentrarchus labrax, Stegastes partitus, Poecilia reticulata, Oreochromis niloticus, Neolamprologus brichardi, Ictalurus punctatus, Danio rerio, Oncorhynchus mykiss, Astyanax mexicanus, Esox lucius, Larimichthys crocea and Stegastes partitus. The obtained integrins - β sequences were analyzed using the ClustalW2 program (http://www. ebi.ac.uk/Tools/clustalw2/). Phylogenetic trees were constructed using the neighbour - joining (NJ) method based on the deduced full - length amino acid sequences with 1,000 bootstrapping replications within the Molecular Evolutionary Genetics Analysis (MEGA4.0) [11].
Tissue expression of integrins β genes
Integrins - β gene in accordance with full design relative fluorescence quantitative PCR primers for β1 - F5, β1 - R5, β3 - F4 and β3 - R4 (Table 1). It can enter the yellow catfish beta actin genes for internal reference. Primers respectively have actins - F and actins - R. The length of amplified fragment is 223 pb. RT - PCR reverse transcription using the reverse transcription kit ABI.Each quantitative real - time PCR was performed on a Stratagene SYBR Premix Ex TaqTMKit (TaKaRa, Japan) Green chemistry . SYBR Premix Ex TaqTMKit was used in 25 μl reactions with 0.5 μl ROX reference dye.The thermal cycling conditions were as follows: (94°C for 3 min, followed by 40 cycles of 94°C for 7 s, 55°C for 10 s, and 72°C for 15 s). Reactions were performed in triplicate, and data were analyzed using MxProTM QPCR software (Stratagene, CA, USA). Experiment according to the instructions, the reaction fluorescence quantitative instrument (ABI 7500).
F. columnare challenges
In the first challenge, the P. fulvidraco, with an average body weight of 28.6 g and average body length of 11.92 cm, kept at 26°C in a flow - through system utilizing heated. Sterile water were used for the challenge.The fish were divided into two groups: (1) F. columnare challenge group (immersed with the dose of 3 ml of 100 CFU ml-1 of F. Columnare for 1 h); (2) control group (without bacteria). To inoculate bacteria for the challenge, a single colony of F. columnare was isolated and cultured in BHI broth at 28°C overnight. The bacterial culture was diluted with PBS (pH 7.4), then 1×105 CFU of bacteria in 200 μL PBS were injected intraperitoneally into the channel P. fulvidraco. The spleen and gill tissues from 60 fish (three pools of 15 fish each) at 4h, 24h, 48h and 3d after F. Columnare treatment were isolated for RNA extraction, respectively. Corresponding, uninfected control samples were taken at each time interval.
Statistical analysis
All data obtained from the qRT - PCR were calculated to the value of 2 - â�?³â�?³CT and differences were evaluated statistically using a t - test in SPSS 20.0 software (IBM, Chicago, USA). The relative expression levels of each gene in different tissues were calculated to the 2 - â�?³â�?³CT of the mean and the standard error (SE) from the three replications. Statistical significance was set with a P value less than 0.01.
Results
Phylogenetic analysis of gene integrins - β
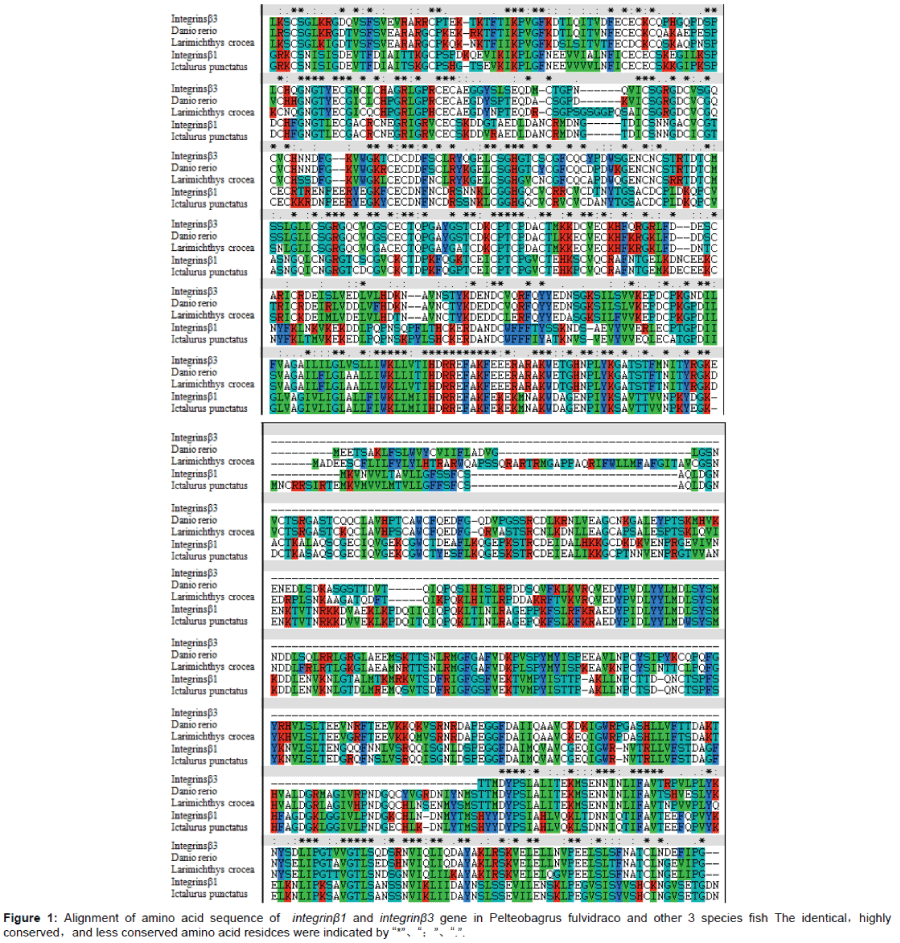
A phylogenetic tree was constructed based on the amino acid sequences of selected integrins - β using the neighbour - joining (NJ) method (Figures 1 and 2). This phylogenetic tree was analyzed by MEGA4 using the Neighbor - Joining method with 1000 bootstrap replicates and the evolutionary distances were calculated using the Poisson correction method The homology similarity of integrins - β (integrins - β1 and integrins - β3) were 83% ~ 87% compared with Cypriniformes Danio rerio (Figure 1). The homology similarity of integrins - β1 was 58% compared with integrins - β3. The homology similarity of integrins - β1 was 90% compared with Ietalurus Punetaus. The homology similarity of integrins - β1 was 79% compared with Esox lucius. The homology similarity of integrins - β3 was 81% compared with Larimichthys crocea. The homology similarity of integrins - β3 was 82% compared with Poecilia formosa.
Tissue expression of integrins - β genes
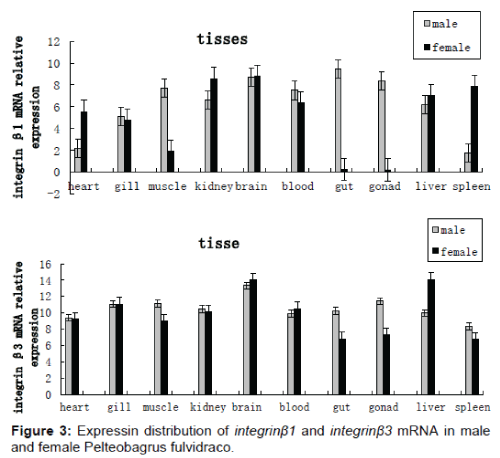
Quantitative real - time PCR was performed to determine the tissue distribution of integrins - β gene expression in both male and female P. fulvidraco. P. fulvidraco integrins - β (integrin - β1 and integrin - β3) were mainly expressed in gut, blood, gonads, brain, liver, kidney, spleen, gills, muscle and heart. Integrin - β genes expression levels in male gonads and gut were higher than those in female. Integrin - β genes expression levels in female liver were higher than those in males P. fulvidraco. Integrin - β1 and integrin - β3 gene expression level in male and females brain and kidney were highest (Figure 3).
Expression of integrin - β after infection of F. Columnare
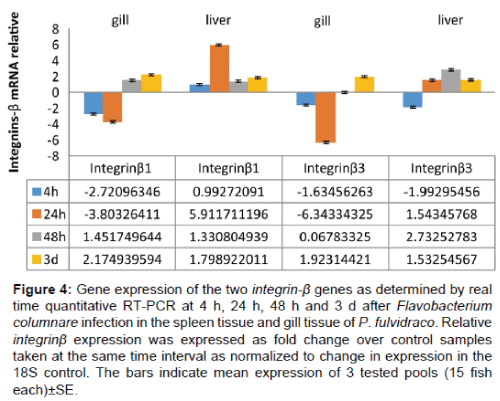
The transcript expression levels of the two integrin - β genes after bacterial infection with F. columnare in liver and gill tissues (Figure 4) was also examined using qRT - PCR. All integrin - β genes were significantly up - regulated at 24 h after columnaris infection in spleen tissue. Integrin - β3 showed the most sensitive response to infection, suppressing expression - 6.34 - fold at 24h (Figure 4). In gill, Integrin - β1 and Integrin - β3 were significantly up - suppressd at 4 h after infection, while integrins - β showed the largest expression changes over the course of the infection. In liver, integrin - β1 and Integrin β3 were significantly up - regulated at 24 h after infection.
Figure 4: Gene expression of the two integrin-ß genes as determined by real time quantitative RT-PCR at 4 h, 24 h, 48 h and 3 d after Flavobacterium columnare infection in the spleen tissue and gill tissue of P. fulvidraco. Relative integrinß expression was expressed as fold change over control samples taken at the same time interval as normalized to change in expression in the 18S control. The bars indicate mean expression of 3 tested pools (15 fish each)±SE.
Discussion
In this study, we firstly get the cDNA of P. fulvidraco integrins - β gene. Through sequence analysis, it can be seen that yellow catfish P. fulvidraco integrin - β gene encodes a protein with high homology to other integrins - β fishes. This implied that the integrins - β genes were highly conserved in some fishes and the yellow catfish integrins - β gene might have similar functions as the integrins - β genes of other fishes. It can be also found that the yellow catfish integrins - β protein does not show complete identity to other fishes.
Integrin - β inhibitors can inhibit the occurrence of bone tumor by inhibiting the expression of osteoclast activity and tumor derived factors [12]. In colorectal cancer, integrin - β1 mediated ERK activation has been shown to regulate lineage commitment in multipotent human colorectal cancer cells [13]. Integrin - β1 can also inhibit differentiation and promote a stem cell - like phenotype in human colorectal carcinoma cells, which was associated with formation and expression of stem cell markers Bmi1 and CD133 [14]. P. fulvidraco integrins - β and ERK contact with colorectal cancer cells of innate immune strategies system has certain help by an as yet undisclosed mechanism(s). Currently in human clinical trials, there are alpha2 and beta 3 integrin inhibitors for the treatment of a variety of cancer [15]. Yelow catfish P. fulvidraco integrins - β3 thus blocking cell sensing this influence bone metastasis ability of tumor cells to transition a new therapeutic opportunity. Importantly, For future research and development, yelow catfish integrins - β3 inhibitors would be used to other diseases in the clinic, and may be a promising way to prevent the progression of bone tumors.
In mouse model, integrin - β1 cells constitute the basal layer andp75 neurotrophin receptor (p75NTR) cells constitute the undercoat structure. Interestingly, in human patients with chronic skin ulcers, p75NTR+ cells were significantly decreased. These results indicate that integrin - β1 cells may play an important role in wound healing process [16]. It may be a chronic skin ulcers catfish of new therapeutic targets. Integrin - β1 involved in the wound healing process: inflammation, collagen synthesis stimulating angiogenesis, fibroblast proliferation and deposition and remodeling of the new extracellular matrix [17]. The subsequent yellow catfish integrin - β1 research, such as constitute the basal layer cells constitute the undercoat structure, would reduce the yellow catfish body skin breakdown and mention rapid healing capabilities. The suitable explanation for this under current conditions is that at the same time those biological activities related to the mRNA expression of yellow catfish integrins - β gene were presented diversely in different tissues. To further understand the function of this novel gene, more research based on these primary results is needed.
Integrins - β Genes have a putative role domain and aconserved. However, in kidney and gill, the increase in expression of integrins - β was following F. columnare stimulation, implying a tissue - specific modulation of expression. It should be noted that the two regulatory regions of the integrins - β exhibited different to previously identified integrin - β1 and integrin - β2 sequences.
In summary, based on the sequence from cDNA of the integrins - β belong to integrin family in P. fulvidraco, we analyzed the sequence, detected tissue transcription profile analysis, and predicted and analyzed the putative protein using bioinformatics tools. These results of our study will pave the way to further elucidate the functions of individual integrins - β genes in regards to pathogen recognition, binding, and their larger immune context.
Acknowledgment
The research was funded and supported by grants from National Science Foundation of China (31302177), and The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for funding this Research No. RG 1435 - 012.
References
- Takada Y, Ye X, Simon S (2007) Theintegrins. Genome Biol 8: 215.
- Hynes RO (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110: 673-687.
- Eke I, Cordes N (2015) Focal adhesion signaling and therapy resistance in cancer. Semin Cancer Biol 31: 65-75.
- Narunsky L, Oren R, Bochner F, Neeman M (2014) Imaging aspects of the tumorstroma with therapeutic implications. PharmacolTher 141: 192-208.
- Naci D, Vuori K, Aoudjit F (2015) Alpha2beta1 integrin in cancer development and chemoresistance. Semin Cancer Biol 35: 145-153.
- Gruber G, Hess J, Stiefel C (2005) Correlation between the tumoral expression of ß3-integrin and outcome in cervical cancer patients who had undergone radiotherapy. British journal of cancer 92: 41-46.
- Shao N, Lu Z, Zhang Y, Wang M, Li W, et al. (2015) Interleukin-8 upregulates integrin β3 expression and promotes estrogen receptor-negative breast cancer cell invasion by activating the PI3K/Akt/NF-κB pathway. Cancer Lett 364: 165-172.
- Zhong R, Tang X, Zhan W. Expression kinetics of ß-integrin in Chinese shrimp (Fenneropenaeuschinensis) hemocytes following infection with white spot syndrome virus. Fish & Shellfish Immunology 35: 539-545.
- Sun X, Zhong Y, Huang Z, Yang Y (2014) Selenium accumulation in unicellular green alga Chlorella vulgaris and its effects on antioxidant enzymes and content of photosynthetic pigments. PLoS One 9: e112270.
- Tan X, Luo Z, Xie P (2009) Effect of dietary linolenic acid/linoleic acid ratio on growth performance, hepatic fatty acid profiles and intermediary metabolism of juvenile yellow catfish Pelteobagrusfulvidraco. Aquaculture 296: 96-101.
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. MolBiolEvol 24: 1596-1599.
- Goodman SL, Picard M (2012) Integrins as therapeutic targets. Trends PharmacolSci 33: 405-412.
- Kirkland SC, Ying H (2008) Alpha2beta1 integrin regulates lineage commitment in multipotent human colorectal cancer cells. J BiolChem 283: 27612-27619.
- Kirkland SC (2009) Type I collagen inhibits differentiation and promotes a stem cell-like phenotype in human colorectal carcinoma cells. Br J Cancer 101: 320-326.
- Page J M, Merkel A R, Ruppender N S (2015) Matrix rigidity regulates the transition of tumor cells to a bone-destructive phenotype through integrin ß3 and TGF-ß receptor type II. Biomaterials 64: 33-44.
- Iwata Y, Akamatsu H, Hasegawa S, Takahashi M, Yagami A, et al. (2013) The epidermal Integrin beta-1 and p75NTR positive cells proliferating and migrating during wound healing produce various growth factors, while the expression of p75NTR is decreased in patients with chronic skin ulcers. J DermatolSci 71: 122-129.
- Penn JW, Grobbelaar AO, Rolfe KJ (2012) The role of the TGF-β family in wound healing, burns and scarring: a review. Int J Burns Trauma 2: 18- 28.
Relevant Topics
- Analytical Biochemistry
- Applied Biochemistry
- Carbohydrate Biochemistry
- Cellular Biochemistry
- Clinical_Biochemistry
- Comparative Biochemistry
- Environmental Biochemistry
- Forensic Biochemistry
- Lipid Biochemistry
- Medical_Biochemistry
- Metabolomics
- Nutritional Biochemistry
- Pesticide Biochemistry
- Process Biochemistry
- Protein_Biochemistry
- Single-Cell Biochemistry
- Soil_Biochemistry
Recommended Journals
- Biosensor Journals
- Cellular Biology Journal
- Journal of Biochemistry and Microbial Toxicology
- Journal of Biochemistry and Cell Biology
- Journal of Biological and Medical Sciences
- Journal of Cell Biology & Immunology
- Journal of Cellular and Molecular Pharmacology
- Journal of Chemical Biology & Therapeutics
- Journal of Phytochemicistry And Biochemistry
Article Tools
Article Usage
- Total views: 12089
- [From(publication date):
June-2016 - Jul 04, 2025] - Breakdown by view type
- HTML page views : 11118
- PDF downloads : 971