Research Article Open Access
In-Silico Prediction and Functional Analysis of Salt Stress Responsive Genes in Rice (Oryza sativa)
| Jyotika Bhati1, Pavan Chaduvula K1, Anil Rai1, Kishore Gaikwad2, Soma Marla S3 and Sanjeev Kumar1* | |
| 1Centre for Agricultural Bioinformatics, ICAR-Indian Agricultural Statistics Research Institute, Pusa, New Delhi 110012, India | |
| 2ICAR-National Research Centre Plant Biotechnology, Pusa, New Delhi 110012, India | |
| 3ICAR-National Bureau for Plant Genetic Resources, IARI Campus, New Delhi 110012, India | |
| Corresponding Author : | Sanjeev Kumar Centre for Agricultural Bioinformatics ICAR-Indian Agricultural Statistics Research Institute, Pusa, New Delhi 110012, India Tel: 91-11-25841721 E-mail: sanjeevk@iasri.res.in |
| Received: January 22, 2016; Accepted: February 18, 2016; Published: February 25, 2016 | |
| Citation: Bhati J, Chaduvula KP, Rai A, Gaikwad K, Soma Marla S, et al. (2016) In- Silico Prediction and Functional Analysis of Salt Stress Responsive Genes in Rice (Oryza sativa). J Rice Res 4:164. doi:10.4172/2375-4338.1000164 | |
| Copyright: © 2016 Bhati J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Visit for more related articles at Rice Research: Open Access
Abstract
The area for cultivation of rice on salt affected soil is increasing at very fast rate due to insensitivity towards following proper package and practices of crops production in order to increasing productivity of rice due to high population pressure in Asian countries. Therefore, it is important to identify genes and their regulatory elements for development of salt stress tolerance variety of rice. In order to identify genes responsible for salt stress in rice 7746 expressed sequence tags (ESTs), expressed in salinity stress condition, were mined from the different web recourses. The downloaded ESTs were clustered and assembled into 672 contigs. Biological functions were obtained only for 424 out of 672 contigs through Gene Ontology (GO). The remaining contigs were used for reconstruction, validation and annotation of salt stress genes. These contigs were mapped on to rice genome and full length gene sequences were designed. These designed candidate genes were further validated by means of a) Integrating with known salt stress related quantitative trait loci (QTL) on rice genome and b) Promoter analysis. In this study novel candidate genes were identified which may have a possible involvement in regulation of salt stress mechanism and may be useful in molecular breeding programme in rice for development of salt tolerance varieties through Molecular Assisted Breeding.
| Keywords |
| Rice; Salt stress; Functional annotation; Gene prediction; QTL; Promoter analysis |
| Abbreviations |
| ESTs: Expressed Sequence Tags; QTLs: Quantitative Trait Loci; NCBI: National Centre for Biotechnology Information; UTR: Un Translated Region; CAP: Contig Assembly Program; MRNA: Messenger Ribo Nucleic Acid; GO: Gene Ontology; BLAST: Basic Local Alignment Search Tool; E-value: Expected Value; NR: Non- Redundant; HSP: High-scoring Segment Pairs; DAG: Directed Acyclic Graph; kb: kilobase; TSS: Transcription Start Site; CDS: Coding Sequences; dNTPs: Deoxy Nucleotides; MgCl2: Magnesium Chloride; PCR: Polymerase Chain Reaction |
| Introduction |
| Rice (Oryza sativa) is most important cereal crop of Asian countries. More than 90% of the rice is produced in the world is consumed in Asia where 60% of the total population live [1]. In spite of technological development, growth rate of global rice production is not keeping pace with the rate of the growing consumers. Also it faces even greater challenges due to various abiotic stresses such as salinity, drought, high radiation and extreme temperatures in changing climatic conditions. Among these stresses, soil salinity has greater impact for rice productivity as Rice is a highly salt-sensitive crop [2]. During last decades the problem of salt salinity in India is gradually increasing over time due to many factors including climate change, rise in sea levels, excessive irrigation without proper drainage etc. Therefore, it is important to develop salt tolerant varieties of rice in order to increase rice productivity of the saline land and extent its area of cultivation. |
| Development of salt tolerant varieties is one of the important challenges of traditional rice breeding-programs in the recent past. The main problem in the traditional breeding approach is lack of understanding of genetic mechanisms for salt tolerance. The effects of salinity on the growth of rice were found to be related to different stages of plant development, salt concentration, types of salt, and duration of exposure to salt, soil pH, water regime, temperature, humidity and solar radiation. Therefore, it is important to study the salt stress mechanism at molecular level and identify novel genes based on its expression under soil salinity condition for the development salt tolerance in rice varieties. Similar studies on EST sequences were performed in wheat for biotic stress and also in Brassica rapa [3-7]. The identification of highly expressed genes under salinity conditions has manifold significance. First, it provides a more comprehensive understanding of the transcriptional responses to salinity stress. Second, it helps in identification of role of individual genes in stress responses. Third, it aids in the identification of stress responsive promoters and responsible cis elements within them. These identified genes will provide the means of improving/incorporating salt resistance mechanism in rice through genetic engineering and similar work has been performed in Sorghum and Maize also [8,9]. Rice being a model plant for genomic and molecular biology research; its most of the genomic information such as ESTs, expression profiling, updated annotation and QTLs is available. In the present study an attempt has been made to identify putative candidate genes expressed under soil salinity conditions based on EST. These candidate genes were further in-silico validated based on integration of salt stress tolerant QTLs with candidate genes on same map locations and examination of cis regulatory elements in the promoter region. In order to identify role in salt stress mechanism functional characterization has been performed. The finding of the study will be useful in development of salt resistant varieties of rice and other similar crops. |
| Materials and Methods |
| In this study 7746 ESTs pertaining to salt stress in rice were downloaded from NCBI. In order to enhance the effectiveness of the similarity based clustering, ESTs were masked to eliminate sequence parts that would cause incorrect clustering [10]. The processed ESTs sequences were grouped into clusters based on sequences similarity to have stable clusters using CAP3 software, with threshold set at default. Each cluster of ESTs were assembled separately with overlap 80 percent overlap identity cut with minimum number of nucleotide 65 [11] of EG Assembler software [12]. The sequences which cannot be grouped due to their low similarity to other ESTs results in singletons. These singletons may represent genes where only single mRNA has been collected for the expressed gene or may be a result of contamination, were not considered for further analysis in this study. |
| Functional analysis of EST-contigs |
| Functional analysis of the EST-contigs was performed using Blast2GO v 2.5 [13]. Blast2GO is Gene Ontology based annotation tool and found to be effective in the functional characterization of plant sequence data [14]. The EST-contigs homologous with annotated proteins in nr database were selected for functional characterisation based on maximum E-value (1E-3) and the minimum alignment size (HSP length 33) using BLASTX. The EST-contigs sequences were then categorized according to the GO vocabularies into three categories i.e. molecular function, biological process and cellular component. The distribution of GO terms was analysed at level 2 of the Directed Acyclic Graphs. Remaining ESTs-contigs, having no BLAST hit and GO prediction were analysed for identification of novel candidate genes related to salt stress in rice. |
| Gene identification from assembled EST-contigs |
| The EST-contigs for which GO terms were not assigned aligned on genome of Oryza sativa using BLAT [15]. The length of the aligned EST-contigs on rice genome was further extended 1 kb up and down stream. These aligned extended sequences were used to predict the structure of gene with TSS, PolyA tails at the extremes and CDS in between by a gene prediction program FGENESH [16]. Further insilico analyses were carried to validate the above candidate genes by two approaches; |
| i) Integrating the annotated salt stress tolerant QTLs and predicted genes on same map locations and |
| ii) Examining the cis regulatory elements in the promoter region of the predicted. |
| PCR and confirmation of candidate genes |
| Partial amplification for all 9 genes mentioned in table was standardized and carried out in Oryza sativa var. indica IR64 in TECHNO gradient thermal cycler in 15 μl reaction containing 10 ng template DNA, 0.2 mM dNTPs, 2 mM MgCl2, 10 pmol of each of nine forward and reverse primer as mentioned in table, 0.5 U of Taq DNA polymerase (vivantis) and 1X PCR buffer. The amplification profile included: initial denaturation for 5 min at 94°C followed by 40 cycles of denaturation for 40s at 94°C, annealing at temperature specific for each target gene for 40s and extension at 72°C for 1 min 20s. Final extension was allowed for 10 min at 72°C and storage at 4°C. Amplified PCR products were resolved on 1.2% agarose gels (Table 1). |
| Results |
| Assembling of ESTs into contigs |
| A total of 7746 EST sequences related to salt stress tolerant of rice were pertaining to different tissues and stress levels downloaded from GenBank. The average length of theses ESTs is 563 base pairs. Out of these 7746 EST sequences, 2136 reads were trimmed and 875 sequences discarded through the sequence cleaning process. The remaining ESTs sequences for Oryza sativa were assembled into 672 EST-contigs. Most of these contigs consists of two or three ESTs. These assembled EST account for only 8.68% of the size of total ESTs. Less abundant or lowly expressed transcripts could not be assembled into larger contigs and remained as 1910 singletons. |
| Functional annotation of EST-contigs |
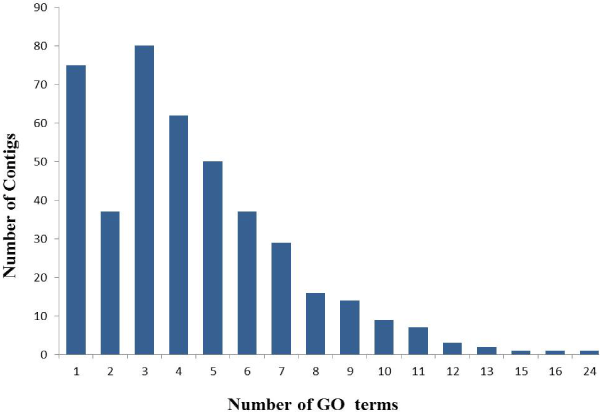
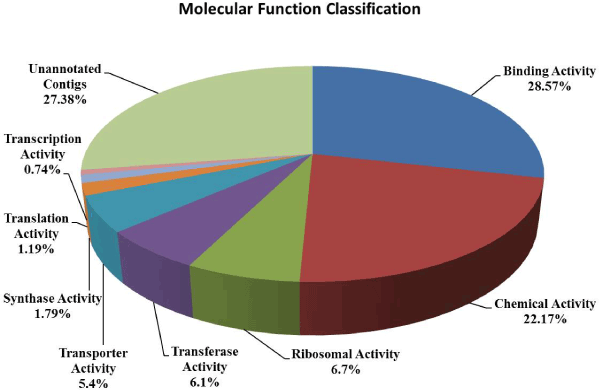
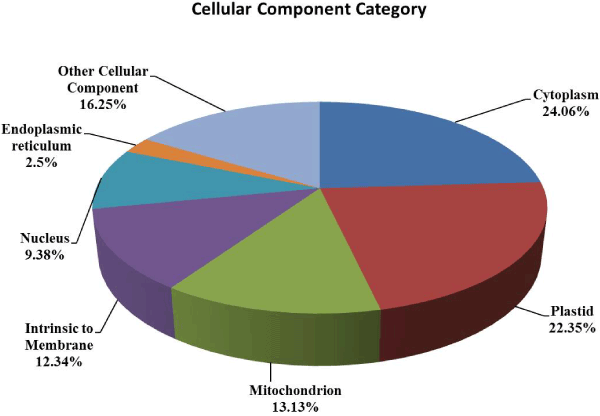
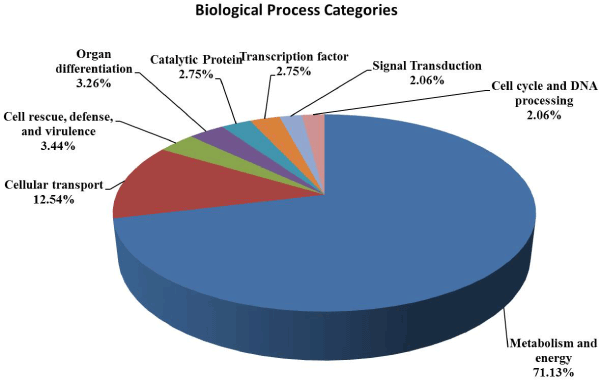
| In order to functional characterized of 672 assembled and translated EST-contigs were compared against NCBI nr database. Out of 672 EST contigs, 488 contigs were selected based on homology search, which were further subjected to GO functional classification. Out of 488 EST contigs GO terms were available only for 424 EST contigs. It has been noticed that overall 1850 GO terms were retrieved which means on an average 4 GO terms per contigs were obtained. The distribution of GO terms per contigs is given in the (Figure 1). Again EST-contigs sequences were categorized according to the GO vocabularies i.e. Molecular Function (Figure 2), Cellular Component (Figure 3) and Biological Process (Figure 4) with obtained number of GO terms as 628, 582 and 640 respectively (Figures 1-4). |
| Candidate gene prediction from EST contigs |
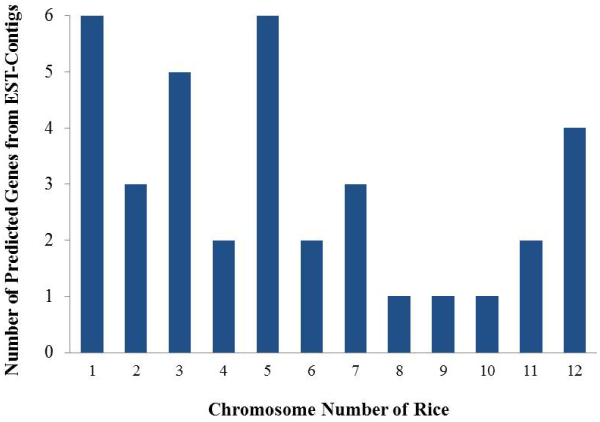
| Out of 672 the remaining 184 EST-contigs for which no BLASTX hit and GO terms were found were considered for candidate gene prediction of the salt stress tolerant genes. These EST-contigs were aligned on genome of Oryza sativa using BLAT. The score range of the alignments of these EST-contigs on rice genome is given in Table 2. Perusal of table reveals that 9 out of 184 contigs have not aligned on rice genome. Rest 175 contigs that aligned on genome with different score ranges were further extended up to 1kb up and down stream on the genome. Altogether, 36 of such genomic region were obtained, with TSS (transcription start site), PolyA tails at the extremes and CDS (coding sequences) in between, as novel candidate genes. Further it was found that these novel candidate genes were distributed among Chromosome number 1 to 12 where chromosome number 1 and 5 contains maximum 6 genes each whereas chromosome number 8, 9,10 contains minimum gene that is one each (Figure 5). |
| Integration of putative gene position with QTLs |
| QTLs having controlled complex physiological traits related to rice salt-tolerance under salt stress were obtained. The information regarding physical position, chromosome number, start / stop position and sequence of the QTL marker were collected (Table 2) based on the position of flanking markers from the Gramene annotated Nipponbare Sequence 2009 map set (available at Gramene website: http://www. gramene.org). These salt stress QTLs were identified by Lin et al. [4] in the shoot and the root of rice, using F2 and F3 generations, derived from a cross between a high salt-tolerance variety and a sensitive variety of salt treatment. Further in order to integrate these QTLs with the candidate genes coding position along with other details such as length, TSS, CDS position and polyA tail are given in Table 3. The physical map position of predicted genes and salt stress QTLs were integrated on rice genome and the consensus of integration between them has been considered here as an in-silico validation of the candidate genes. Perusal of Table 2 reveals that there are two QTLs (start / stop position) i.e. AQEM001 (33956950 / 37713775) and AQEM006 (9820009 / 11232822) lies on chromosome number one of rice genome. On the other hand, it can be observed from Table 3 that there are four candidate genes namely contigs 1, 2, 4 and 54 lies in chromosome number one of the rice genome with start / stop positions as 34161530/34165992, 9873777/9876475, 9874877/9875695 and 10748403/10750424 respectively. A close examination of columns start/stop position of Tables 3 and 4 reveals that Contig 1 lays inside the QTL-AQEM001 whereas Contig 2, 4 and 54 in QTL-AQEM006. Similarly, Contig 138 and 314 lies within the QTL AQEM004 region on chromosome number 7 while Contig 215 and 558 in QTL AQEM007 region on chromosome number 9. Therefore, eight novel candidate genes among 36 predicted candidate genes were identified which lays within the annotated salt stress QTLs regions on rice genome (Tables 3 and 4). |
| Promoter analysis of novel candidate genes |
| The objective of promoter analysis is to identify the possible cisacting DNA sequences that may be responsible for the regulation of candidate gene expression. Cis-acting sequences are the regulatory sequences that are part of the gene, which influence only the expression of the gene that contains them. Thus, the cis regulatory elements in the promoter region of the candidate genes were obtained from PLACE (http://www.dna.affrc.go.jp/PLACE/), i.e. a database of motifs found in plant cis-acting regulatory DNA elements all from previously published reports. In contrast, salt stress responsive cis elements of the promoter regions in cereals were collected from the publish literature. The collected salt stress responsive cis elements viz. ABRE, ARR, DOF, DRE, E2F, MYB, MYC, NOD, RAV and WRKY along with their conserved cis motif sequences are given in the Table 5. Table 6 provide the list of all reported salt responsive cis elements present in candidate genes. Perusal of Table 6 reveals that contig 1, 2, 43, 215 and 314 having all the ten salt stress responsive reported cis elements whereas contig 138, 545 have nine and contig 54 has eight among their cis elements present in promoter region. The presence of these cis elements in the promoter region of candidate genes indicates their possible involvement in salt stress mechanism of rice (Tables 5 and 6). |
| Confirmation of candidate genes by partial amplification |
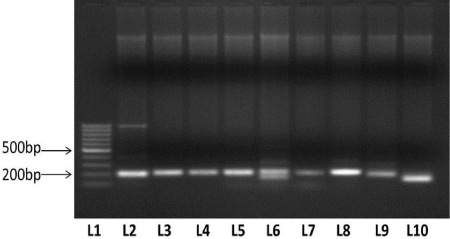
| All nine abiotic stress responsive candidate genes were partially amplified in Rice cultivar IR64. Approximate amplicon size of all the nine genes was ~200 bp for rice cultivar IR64. The genes amplified are characterized as DNA directed RNA polymerase, ferritin superfamily, plant peroxidase superfamily, ATPase expression protein, microtubule associated protein, exonuclease, major latex protein and protein kinase. |
| Discussion |
| Development of salt tolerant varieties is one of the important challenges of rice breeding-programs in the recent past [3]. An important genomic approach to identify salt stress related genes is based on ESTs generated from different cDNA libraries representing stress treated tissues collected at various stages of development. The clustering of EST sequences generated from abiotic stress-treated cDNA libraries provides information on gene number, gene content and possible number of gene families involved in stress responses. Putative functions are assigned to such stress-responsive genes by BLASTX comparison to the protein database. This type of analysis provides a valuable resource of information regarding a gene index associated with stress-responsive genes. Analysing the various EST collections enabled us to find stress-regulated genes. |
| Further these EST contigs were classified with respect to different molecular functional activities. It has been observed that 51% of the total EST contigs belongs to binding activity (28.57%) and chemical activity (22.17%). Distributions of molecular functional activities were presented in Figure 2. It may be noted that the binding activity has been closely associated with regulation of salt-stress-modulated gene expression in various plant species [17-19]. Also it has been reported that proteins involved in various chemical activities like kinases, pyro phosphatases, reductases, and hydrogenases play key roles in detecting and relaying salt stress signals for the regulation of specific genes and thus mediate cellular responses to salt stress [20]. The other EST contigs are classified into different molecular functional activities which are responsible for salt stress mechanism such as ribosomal activity (6.7%), transferase (6.1%), transporter activity (5.4%), synthesis (1.79%), translation (1.19%) and transcription (0.74%). Rice ESTs for ribosomal proteins were found to be up-regulated after exposure to 150mM NaCl for 1 hour and returned to the original expression levels after 3 hours [21] further ribosomal protein S4 homologue to Chrysodidymus synuroideus [22] was detected under salt stress. The plant glutathione transferase (GST) enzyme is responsible for detoxification of cytotoxic endogenous and xenobiotic compounds as well as hormone metabolism of tomato for improving salt stress tolerance [23]. Arabidopsis thaliana glutathione S-transferase U17 (AtGSTU17) plays role in adaptive responses to salt stresses by reverse functioning of stress-mediated signal transduction pathways [24]. |
| Zhao et al. [25] proved that the disruption of nitric oxide synthase (NOS) dependent nitric oxide production is associated with salt tolerance in Arabidopsis thaliana. It was also observed that overexpression of helix-loop-helix (bHLH) encoding gene, OrbHLH2, localized in the nucleus, conferred increased tolerance to salt and osmotic stress, and the stress-responsive genes DREB1A/CBF3, RD29A, COR15A and KIN1 were up regulated in transgenic plants [26]. |
| Out of GO term pertaining to cellular component category 72% belongs to cytoplasm (24.06%), Plastid (22%), mitochondrion (13.13%) and intrinsic to membrane (12.34%) (Figure 3). It has been already reported that these four cellular components are related to salt stress in plant. Gene AtLecRK2 in Arabidopsis which is located on the cytoplasm contains a signal peptide (an extracellular lectin-like domain and a cytoplasmic protein kinase domain) is induced by salt stress [27]. It has been reported that plastid-expressed choline monooxygenase gene improves salt tolerance through accumulation of glycine betaine in tobacco [28]. It is also observed that 50% of the plastid showed cellular composition to chloroplasts. A chloroplast enhancing stress tolerance protein (CEST) overexpressing in Arabidopsis and shown to have salt tolerance which causes photo-oxidative stress [29]. Mitochondria have uncoupling proteins (UCPs) that uncouple electron transport from ATP synthesis. Higher levels of Arabidopsis thaliana uncoupling proteins (AtUCP1) improves tolerance to salt stress, so the cellular localization of the EST-contigs in the mitochondria confers the putative role of these contigs in salt stress. The manipulation of the UCP protein expression in mitochondria may leads to improved crop varieties with greater salt tolerance [30]. The known aquaporins subfamily plasma membrane intrinsic proteins (PIPs) such as OsPIP gene from rice, showed enhanced tolerance to salt [31]. The other less prominent cellular locations were nucleus (9.38%), endoplasmic reticulum (2.5%) and others organelles (16.25%). A stress-responsive cyclophilin gene (CYP1, enriched significantly in the nucleus) expression is highly inducible by salt in Thellungiella halophila [32]. Many rice heat shock proteins (HSPs) have been reported to be regulated by salt stress and localized in the endoplasmic reticulum [33]. |
| Out of GO terms pertaining to biological process category 71.13% belongs to metabolism and energy (Figure 4). It has been reported that many genes, involved in various metabolism and energy processes such as bioenergetics, secondary metabolism, lipid metabolism, amino acid metabolism, DNA metabolism, nucleotide metabolism, and N, S, and P metabolism, under salt stress. A total of 12.54% of the EST-contigs were found to be actively involved in the cellular transport. Vitart et al. [34] reported based on analysis of phenotypes caused by the semi-dominant aha4-1 mutation that the activity of vacuolar type H+-ATPase and vacuolar pyro phosphatase (responsible for cellular transport of ions) is increased by salt treatment and induced gene expression for the upregulation. There were many other biological process found amongst the EST-contigs involved in cell defense (3.44%), catalytic activity (2.75%), DNA processing (2.06%), signal transduction (2.06%) and presence of transcription factors (3.26%) and all these has some or other role in salt stress responses [35-39]. |
| In-silico functional annotation was carried out by using BLASTX for the 8 EST-contigs that were confirmed to be candidate salt responsive genes. The size of all the nine isolated genes from Oryza sativa cultivar was about 200 bp using primers designed for showing match with predicted candidate gene. Among this, Contig 1 showed functional similarity with DNA directed RNA polymerase; the same function which has also been reported to be associated with salt stress in Rice [40]. The functional annotation of Contigs 2 and 43 predicted carboxylate protein belonging to Ferritin superfamily which is related to salt stress [41]. Peroxidase proteins, distributed widely in plants, shows diverse expression profiles, indicating their involvement in salinity response [42] and Contig 54 shows similarity with this protein. Contig 138 has been found to be related to ATPase expression protein which responds to salt stress with increased expression and enzyme activity [43]. Contig 215 is found to be similar with microtubules which are best suited for surviving in high salinity [44]. Exonuclease activity is associated with salinity in Arabidopsis [45] and Contig 314 is likely to be associated with it. Contig 558 showed homology with ribosomal protein which are already been acknowledged to play active role in salt response [21]. Each gene showed their presence in the rice cultivar IR64 (Figure 6). |
| Conclusion |
| Present study was undertaken with an objective to identify salt stress responsive genes in Oryza sativa. Nine salt stress responsive candidate genes previously validated for their significance in stress responses in various model crops and other plant species, were amplified in Oryza sativa and sequenced after purification using gene specific primer pairs. Therefore, present study provided basic information about some salt stress responsive genes in rice that can be exploited in overcoming various salt stress related problems limiting its production and subsequently in breeding superior varieties giving better yield under salt stress conditions by pyramiding favourable alleles for stress response in single variety using modern molecular breeding approaches [45-57]. |
| Acknowledgements |
| We acknowledge support from National Agricultural Bioinformatics Grid (NABG), National Agricultural Innovation Project (NAIP) and National Agricultural Science Fund, Indian Council for Agricultural Research (ICAR). |
References |
|
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 |
| Table 4 | Table 5 | Table 6 |
Figures at a glance
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 |
 |
 |
 |
| Figure 4 | Figure 5 | Figure 6 |
Relevant Topics
- Basmati Rice
- Drought Tolerence
- Golden Rice
- Leaf Diseases
- Long Grain Rice
- Par Boiled Rice
- Raw Rice
- Rice
- Rice and Aquaculture
- Rice and Nutrition
- Rice Blast
- Rice Bran
- Rice Diseases
- Rice Economics
- Rice Genome
- Rice husk
- Rice production
- Rice research
- Rice Yield
- Sticky Rice
- Stress Resistant Rice
- Unpolished Rice
- White Rice
Recommended Journals
Article Tools
Article Usage
- Total views: 12339
- [From(publication date):
February-2016 - Apr 04, 2025] - Breakdown by view type
- HTML page views : 11081
- PDF downloads : 1258
