Inhibition of CTNND1-Mediated Wnt/β-Catenin Signalling by MicroRNA-449b-5p Represses Colorectal Cancer Progression
Received: 14-Jun-2019 / Accepted Date: 26-Jun-2019 / Published Date: 03-Jul-2019 DOI: 10.4172/2161-0681.1000370
Abstract
Several studies have demonstrated the dysregulation of miR-449b-5p in numerous types of human cancers. However, the understanding of the function of miR-449b-5p the mechanisms linking its expression to the development of colorectal cancer (CRC) remains limited. The current work documented the decrease in the level of miR-449b-5p in CRC tissues as well as in CRC-derived cell lines. Functional assays indicated that the overexpression of miR-449b-5p inhibits proliferation, migration, and invasion of CRC cells. Mechanistically, the direct target of miR-449b-5p in CRC cells was identified as the catenin delta 1 (CTNND1) protein. Increased level of miR-449b-5p in CRC cells downregulated β-catenin, WNT11, cyclin D1, and MMP7. Conversely, the restoration of CTNND1 expression abrogated the inactivation of Wnt/β-catenin signaling induced by miR-449b-5p. In conclusion, the present investigation demonstrates for the first time that miR-449b-5p is capable of inhibiting the progression of CRC. This effect is achieved, at least in part, by targeting CTNND1-mediated Wnt/β-catenin signaling. The accumulated data point to miR-449b-5p as a novel target in the treatment of CRC.
Keywords: miR-449b-5p; Colorectal cancer; CTNND1; Wnt/β- catenin signaling; Tumor growth and metastasis
Introduction
Colorectal cancer (CRC) belongs to the most commonly occurring malignancies worldwide. Despite dramatic progress seen in the treatment of CRC, such as novel chemo- and immunotherapies and improved surgical resection techniques, the 5-year survival rate of CRC patients continues to be unsatisfactory. The poor prognosis after the treatment results from tumor invasion and metastasis [1-4]. Therefore, a better understanding of CRC pathogenesis is urgently needed to identify specific therapeutic targets and more effective strategies.
MicroRNAs (miRNAs) constitute a class of small RNA transcripts that are not involved in the coding of proteins. miRNAs regulate numerous developmental and pathological processes in animals and humans by binding to protein-coding transcripts [5]. An increasing body of evidence points to the presence of a link between miRNAs to the initiation, development, and metastasis of various types of tumors, including CRC [6-9]. Although miR-449b-5p is implicated in multiple cancers [10-12], little is known regarding its relationship to the development and progression of CRC.
The current investigation aimed at the identification of the function of miR-449b-5p in CRC and elucidation of underlying mechanisms. miR-449b-5p was found to be downregulated in CRC, and increasing the level of this miRNA species inhibited the growth and metastasis of CRC-derived cells. This anti-tumor effect was mediated, at least in part, by targeting catenin delta 1 (CTNND1) and inactivation of Wnt/ β-catenin signaling pathway.
Materials and Methods
Clinical samples and cell lines
Fifty-one CRC samples and tumor-adjacent tissues from the same patients were collected at the Changhai Hospital (Shanghai, China). The patients did not receive immunotherapy, radiotherapy, or chemotherapy prior to the surgery. All samples were verified to represent CRC by a pathologist and stored in liquid nitrogen before use. All patients signed an informed consent form. The protocol of the study was reviewed and approved by the Institutional Human Experiment and Ethic Committee of the Changhai Hospital and the entire investigation was compliant with the Helsinki Declaration.
The CCD-18Co line of normal intestinal epithelial cells and five human CRC cell lines (HCT116, Lovo, HT29, SW480, and SW620) were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Dulbecco ’ s modified Eagle medium (DMEM; Gibco, Carlsbad, CA, USA) was used in all in vitro experiments. The medium was supplemented with 10% fetal bovine serum (FBS) purchased from Gibco and 1% (v/v) penicillin/streptomycin, purchased from Sigma-Aldrich (St-Louis, MO, USA). All cultures were carried out at 37℃ in a humidified atmosphere containing 5% CO2.
Cell transfection
miR-449b-5p mimics (miR-449b-5p) and control mimics (control) were purchased from Sigma-Aldrich. CTNND1 expression plasmid (pcDNA3.1-CTNND1), small interfering RNA (siRNA) targeting CTNND1, and non-targeting (NT) siRNA were purchased from Gene Pharma (Shanghai, China). CRC cells were transfected with Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from CRC tissues and cultured cells by the TRIzol reagent (Invitrogen). The cDNA was generated by reverse transcription using a commercially available kit (miScript; QIAGEN, Düsseldorf, Germany). miR-449b-5p and CTNND1 mRNA were amplified utilizing the Sequence Detection System (Model 7500; Applied Biosystems, Foster City, CA, USA). The reagents used in this protocol were miScript SYBR Green PCR kit (QIAGEN) and Sso Advanced Universal SYBR Green Supermix (Bio-Rad, Hercules, CA, USA). Sequences of the primers are shown in Table 1. The relative values of gene expression were computed by the 2-ΔΔCt method. The obtained results were normalized to the expression levels of GAPDH and U6 genes.
| miR-449b-5p | forward, 5'-CTAGCCCGTCGGTTCCGTCGCGCAG-3' |
| reverse, 5'-ATGTTGGCCGGGTAGGATAATGGGT-3' | |
| CTNND1 | forward, 5'-ATGTTTGCGAGGAAGCCGC-3’ |
| reverse, 5'-CGAGTGGTCCCATCATCTG-3’ | |
| GAPDH | forward, 5'-CCATGTTCGTCATGGGTGTG-3' |
| reverse, 5'-GGTGCTAAGCAGTTGGTGGTG-3' | |
| U6 | forward, 5'-GCTTCGGCAGCACATATACTAAAAT-3' |
| reverse, 5'-CGCTTCACGAATTTGCGTGTCAT-3’ |
Table 1: Sequences of the primers used.
Western blot
Following the lysis of CRC cells with the RIPA buffer (Beyotime Biotech, Shanghai, China) the concentration of protein was measured with BCA Protein Assay Kit II (Bio-Rad, Hercules, CA, USA). The samples were then subjected to 10% SDS-PAGE, separated proteins were transferred onto a nitrocellulose membrane (Invitrogen, Waltham, MA, USA), and incubated overnight at 4°C with primary antibodies against CTNND1 (ab92514), WNT11 (ab31962), β-catenin (ab32572), Cyclin D1 (ab134175), and MMP7 (ab5706) (all from Abcam, Cambridge, MA, USA). Antibody against GAPDH (sc-47724, Santa Cruz Biotechnology, Dallas, TX, USA) was used to control loading conditions. Subsequently, the membranes were washed and incubated for 1 h at room temperature with horseradish peroxidase (HRP)-conjugated secondary antibody (Cell Signaling Technology, Beverly, MA, USA). The bound antibodies were visualized with ECL reagents (Millipore, Plano, TX, USA).
Cell migration and invasion assay
Migration and invasion assays were performed in Boyden-type chambers, in which upper part was uncoated (migration assay) or coated (invasion assay) with Matrigel (BD Bioscience, San Jose, CA, USA). Each chamber was seeded with 1×104 cells in DMEM without FBS. Subsequently, the chambers were placed in a multi-well plate containing DMEM supplemented with 10% FBS. After incubating for 24 h at 37°C, the cells contained in the upper compartment were carefully discarded using cotton swabs. The cells that translocated to the lower compartment were fixed with 1% paraformaldehyde for 10 min, hematoxylin-stained for 5 min, and counted under a microscope.
Cell proliferation assay
CRC cells were seeded in a 96-well plate at a density of 1×104/mL. A 10 μL aliquot of the cell counting kit-8 (CCK-8; Dojindo, Tokyo, Honshu, Japan) solution was added to each well every 24 hours for 3 days. After 2 h of incubation, the absorbance at 450 nm was measured by a microplate reader (Thermo Fisher Scientific, Waltham, MA, USA).
Clone formation assay
A total of 500 CRC cells were seeded into six-well plates in triplicate. After 14 days, the formed colonies were visualized with 0.5% crystal violet and counted.
Luciferase reporter assay
Human genomic DNA was utilized to amplify the sequence of CTNND1 3 ’ -UTR containing the putative miR-449b-5p-binding region. The amplified product was cloned into the pGL3 luciferase reporter vector (Promega, Madison, WI, USA). Mutations in the potential miR-449b-5p-binding sites were generated by the Quickchange site-directed mutagenesis kit (New England Biolabs, Beverly, MA, USA). The Lovo and SW620 cells were co-transfected with the wild type (wt) or mutant (mt) CTNND1 3 ’ -UTR vector and miR-449b-5p mimics or miR-449b-5p inhibitors; the transfection was carried out using Lipofectamine 3000 (Invitrogen). The activity of luciferase was determined with a luminometer (Berthold Detection System, Pforzheim, Germany) using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). All luminescence results were normalized to Renilla activity.
Statistical analyses
Continuous variables are shown as the mean ± SD. Statistical analyses were performed using GraphPad Prism software (version 5.0; San Diego, CA, USA). Correlation between miR-449b-5p and CTNND1 mRNA expression was determined by Pearson’s correlation test. Statistical significance of the difference between two groups was calculated by the Student’s t-test, and among multiple groups by the ANOVA method. P-values of less than 0.05 were considered to indicate a statistically significant difference.
Results
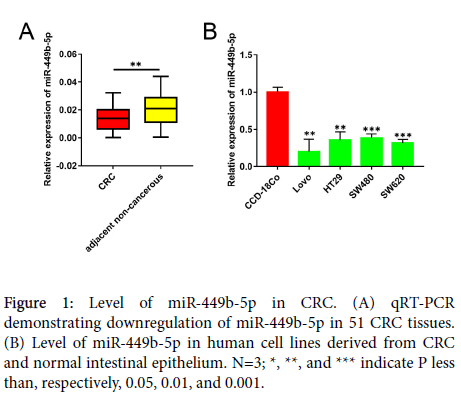
The Level of miR-449b-5p is downregulated in CRC
The levels of expression of miR-449b-5p in human CRC samples and in non-cancerous tissues adjacent to the tumor were compared using qRT-PCR. In comparison with the adjacent non-cancerous tissue, miR-449b-5p expression was significantly downregulated in CRC samples (P<0.05, Figure 1A). The levels of miR-449b-5p in different CRC cell lines were also determined and compared with those of normal intestinal epithelial cell line CCD-18-Co. In a manner similar that observed in tissue samples, all CRC cells lines examined, including Lovo, HT29, SW480, and SW620, exhibited lower level of miR-449b-5p than normal CCD-18Co cells (P<0.05 vs. each CRC cell line, Figure 1B). These data document significant downregulation of miR-449b-5p in CRC.
Decreased miR-449b-5p expression correlates with aggressive clinicopathologic features of CRC patients
To evaluate the association of miR-449b-5p expression levels with clinicopathologic characteristics the 51 patients in the study cohort were divided into high and low miR-449b-5p expression subgroups with its median miR-449b-5p level in CRC tissues as the cut-off. As shown in Table 2, low levels of miR-449b-5p in CRC were significantly correlated with larger tumor size (p=0.045), increased lymph node metastasis (p=0.004), and advanced TNM stage (p=0.001). While, no significant correlations were observed between the level of miR-449b-5p and gender (p=0.714), age (p=0.232), tumor location (p=0.921), tumor differentiation grade (p=0.276), local invasion (p=0.341), or adjuvant chemotherapy (p=0.128). Collectively, these findings indicate that downregulated miR-449b-5p expression may be linked with malignant progression of CRC.
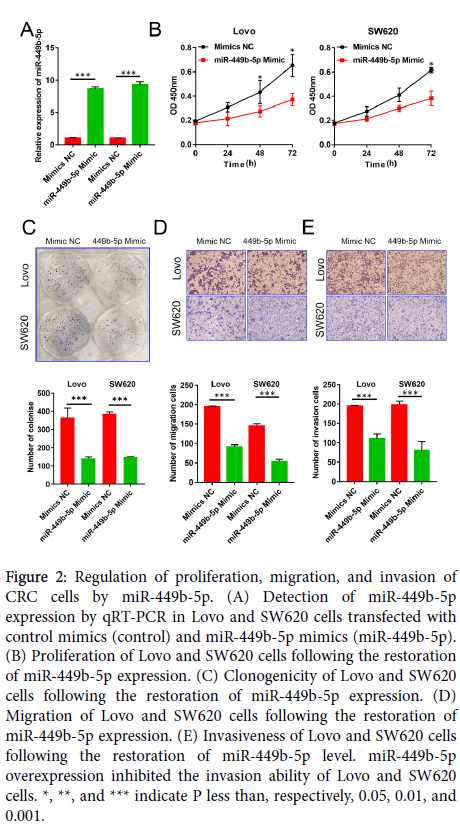
Figure 2: Regulation of proliferation, migration, and invasion of CRC cells by miR-449b-5p. (A) Detection of miR-449b-5p expression by qRT-PCR in Lovo and SW620 cells transfected with control mimics (control) and miR-449b-5p mimics (miR-449b-5p). (B) Proliferation of Lovo and SW620 cells following the restoration of miR-449b-5p expression. (C) Clonogenicity of Lovo and SW620 cells following the restoration of miR-449b-5p expression. (D) Migration of Lovo and SW620 cells following the restoration of miR-449b-5p expression. (E) Invasiveness of Lovo and SW620 cells following the restoration of miR-449b-5p level. miR-449b-5p overexpression inhibited the invasion ability of Lovo and SW620 cells. *, **, and *** indicate P less than, respectively, 0.05, 0.01, and 0.001.
| Characteristics | No of patients (n=51) | p value | ||
|---|---|---|---|---|
| low (n=27) | high (n=24) | |||
| Gender | 0.714 | |||
| Female | 22 | 11 | 11 | |
| Male | 29 | 16 | 13 | |
| Age (years) | 0.232 | |||
| <60 | 15 | 6 | 9 | |
| ≥ 60 | 36 | 21 | 15 | |
| Tumor location | 0.921 | |||
| Rectum | 23 | 12 | 11 | |
| Colon | 28 | 15 | 13 | |
| Differentiation grade | 0.276 | |||
| Well+ Moderate | 39 | 19 | 20 | |
| Poor | 12 | 8 | 4 | |
| Tumor size (cm) | 0.045 | |||
| <5 | 20 | 6 | 14 | |
| ≥ 5 | 31 | 15 | 10 | |
| Local invasion | 0.341 | |||
| pT1-T2 | 8 | 3 | 5 | |
| pT3-pT4 | 43 | 24 | 19 | |
| Lymph node metastasis | 0.004 | |||
| Negative | 32 | 12 | 20 | |
| Positive | 19 | 15 | 4 | |
| TNM stage | 0.001 | |||
| I+II | 32 | 11 | 21 | |
| III | 19 | 16 | 3 | |
| Adjuvant chemotherapy | 0.128 | |||
| No | 24 | 10 | 14 | |
| Yes | 27 | 17 | 10 | |
Table 2: Association between miR-449b-5p expression and clinicopathologic characteristics of CRC patients in the study cohort. A Pearson chisquare test was used for comparison between subgroups. Bold type indicates statistical significance.
MiR-449b-5p inhibits CRC cell growth and metastasis
To establish the function of miR-449b-5p in CRC progression, Lovo and SW620 cells were transfected with miR-449b-5p mimics in order to restore the normal level of expression of this miRNA species. After establishing a significant increase in the level of miR-449b-5p (P<0.05 in both cell lines, Figure 2A), cell proliferation, clonogenicity, migration, and invasiveness were determined. The restoration of miR-449b-5p markedly inhibited proliferation of CRC cells as measured by the CCK-8 assay (P<0.05 in both cell lines, Figure 2B). Moreover, this intervention resulted in the inhibition of the rate of clone formation in Lovo and SW620 cells (P<0.05 in both cell lines, Figure 2C) and reduction in their migration and invasion (P<0.05 in both cell lines and for both properties, Figure 2D and E). Together, these results demonstrate that miR-449b-5p is a potent suppressor of malignant features of CRC cells.
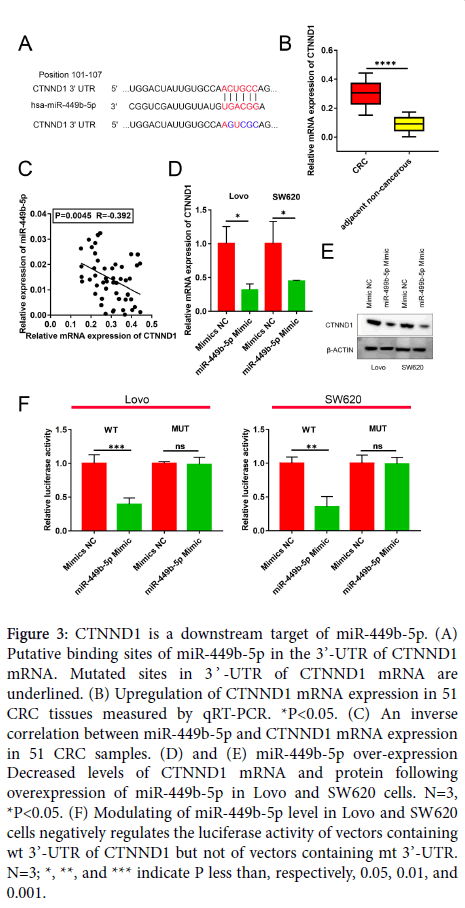
CTNND1 is a downstream target of miR-449b-5p in CRC
Candidate target genes of miR-449b-5p were searched for utilizing a public database TargetScan (http://www.targetscan.org/vert_72/). The performed query identified that the well-characterized and important CRC oncogene CTNND1 contains a complementary sequence for miR-449b-5p (Figure 3A). In view of this finding, the levels of expression of CTNND1 mRNA was measured by qRT-PCR in human CRC samples and adjacent non-tumor tissues. As illustrated in Figure 3B, the expression of CTNND1 mRNA was markedly higher in CRC than in normal tissue (P<0.05). Moreover, the Spearman correlation analysis indicated the presence of a negative correlation between miR-449b-5p and CTNND1 mRNA expression in CRC specimens (r= −0.392, P=0.0045, Figure 3C). This negative correlation was confirmed in experiments in which the restoration of miR-449b-5p expression in Lovo and SW620 cells downregulated CTNND1 at both mRNA and protein levels (P<0.05 for both cell lines; Figure 3D and E). The luciferase reporter assay documented that the luciferase activity in Lovo and SW620 cells transfected with wt CTNND1 3’-UTR was diminished by the restoration of miR-449b-5p levels (P<0.05 for both cell lines, Figure 3F). In a control experiment, modifying the level of miR-449b-5p had no effect on the luciferase activity in Lovo and SW620 cells transfected with mt CTNND1 3’-UTR (Figure 3F). These results demonstrate that the expression of CTNND1 in CRC is under direct control of miR-449b-5p.
Figure 3: CTNND1 is a downstream target of miR-449b-5p. (A) Putative binding sites of miR-449b-5p in the 3’-UTR of CTNND1 mRNA. Mutated sites in 3 ’ -UTR of CTNND1 mRNA are underlined. (B) Upregulation of CTNND1 mRNA expression in 51 CRC tissues measured by qRT-PCR. *P<0.05. (C) An inverse correlation between miR-449b-5p and CTNND1 mRNA expression in 51 CRC samples. (D) and (E) miR-449b-5p over-expression Decreased levels of CTNND1 mRNA and protein following overexpression of miR-449b-5p in Lovo and SW620 cells. N=3, *P<0.05. (F) Modulating of miR-449b-5p level in Lovo and SW620 cells negatively regulates the luciferase activity of vectors containing wt 3’-UTR of CTNND1 but not of vectors containing mt 3’-UTR. N=3; *, **, and *** indicate P less than, respectively, 0.05, 0.01, and 0.001.
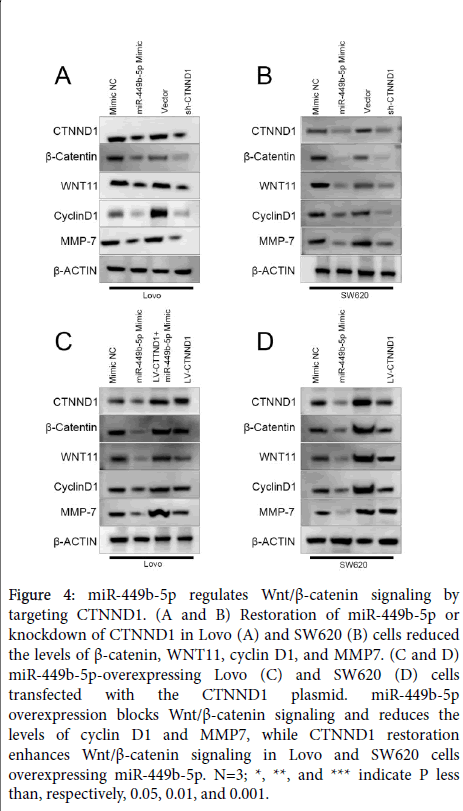
CTNND1-mediated Wnt/β-catenin signaling is necessary for the biological function of miR-449b-5p in CRC
Since CTNND1 is an upstream regulator of Wnt/β-catenin signalling [13], and in view of the above-presented results, the possibility was advanced that miR-449b-5p can regulate this signalling pathway as well. To test this hypothesis, the impact of the restoration of miR-449b-5p and knockdown of CTNND1 on the expression of CTNND1, β-catenin, WNT11, cyclin D1, and MMP7 was determined in Lovo and SW620 cells. Both interventions resulted in a marked decrease in the levels of these components of Wnt/β-catenin signaling (Figure 4A and B). These results were complemented by the finding that the normalization of CTNND1 expression activated Wnt/β- catenin signalling in Lovo and SW620 cells overexpressing miR-449b-5p (Figure 4C and D). Together, the accumulated results demonstrated unequivocally that ability of miR-449b-5p to suppress tumor-related properties of CRC cells depends, at least in part, on its inhibitory effect on CTNND1-mediated Wnt/β-catenin signalling pathway.
Figure 4: miR-449b-5p regulates Wnt/β-catenin signaling by targeting CTNND1. (A and B) Restoration of miR-449b-5p or knockdown of CTNND1 in Lovo (A) and SW620 (B) cells reduced the levels of β-catenin, WNT11, cyclin D1, and MMP7. (C and D) miR-449b-5p-overexpressing Lovo (C) and SW620 (D) cells transfected with the CTNND1 plasmid. miR-449b-5p overexpression blocks Wnt/β-catenin signaling and reduces the levels of cyclin D1 and MMP7, while CTNND1 restoration enhances Wnt/β-catenin signaling in Lovo and SW620 cells overexpressing miR-449b-5p. N=3; *, **, and *** indicate P less than, respectively, 0.05, 0.01, and 0.001.
Discussion
Extensive research on the function of miRNAs in CRC led to the conclusion that these molecules are critical regulators of the initiation and progression of CRC [14]. The multiple effects exerted by miRNAs in CRC cells include the regulation of proliferation, apoptosis, autophagy, migration, invasion, metastasis, epithelial-mesenchymal transition, angiogenesis, and drug resistance [15]. Deranged level of miR-449b-5p was demonstrated in multiple types of human cancer [10-12]. The present study demonstrated significant downregulation of miR-449b-5p in CRC in comparison with the level of its expression in adjacent non-tumor tissue.
Additionally, the conducted experiments have shown that restoration of the normal level of miR-449b-5p results in the inhibition of proliferation, migration, and invasion of CRC cells, indicating that this miRNA species performs a function of tumor suppressor in CRC. The primary mechanism by which miRNAs control the expression of their target mRNAs is binding to their 3'-UTR [16]. Previous studies utilizing various cell types have documented several targets of miR-449b-5p including SOX4 [10], CREPT [17], HMGB1 [18], and FOXP1 [19]. The current study expanded the list of known miR-449b-5p targets by CTNND1. This conclusion was supported by several experiments. In CRC tissues, the level of CTNND1 mRNA was inversely correlated with miR-449b-5p expression, and changes in miR-449b-5p level in CRC cells negatively regulated the abundance of CTNND1. Most importantly, the luciferase reporter assay demonstrated that CTNND1 is a direct target of miR-449b-5p in CRC cells. Earlier reports have shown that CTNND1 is regulated by various species of miRNAs. For example, miR-145 and miR-29c target CTNND1 and inhibit metastasis of gastric cancer [20-22]. Direct regulation of CTNND1 by miR-409c has also been documented in osteosarcoma [20]. In CRC, high expression of CTNND1 is required for tumor growth and metastasis [23]. The function of CTNND1 as an oncoprotein depends on its ability to indirectly activate Wnt/β-catenin signalling, and this pathway is critical for the initiation and progression of CRC [23,24]. In this regard, the current work documented that both CTNND1 knockdown and miR-449b-5p restoration counteract the activation of Wnt/β-catenin cascade and inhibit its downstream targets such as cyclin D1 and MMP7. Of these, Cyclin D1 is required for the promotion of CRC progression by Wnt/β- catenin signalling [25], and MMP7 activity enables migration and invasion of CRC cells [26]. Consistent with this notion, the current study demonstrated that the restoration of CTNND1 reversed miR-449b-5p-induced inactivation of Wnt/β-catenin signalling, promoting the growth and metastasis of miR-449b-5p-overexpressing Lovo and SW620 cells. Together, the accumulated results provide new insights into the function and significance of the miR-449b-5p/ CTNND1 axis in CRC.
Given the limited number of samples available for this investigation, no attempt was made to correlate the level of miR-449b-5p and clinical characteristics of the patients. Moreover, the prognostic value of the level of miR-449b-5p in tumor tissue was not analyzed. These clinically relevant issues will be addressed in future studies on the significance of miR-449b-5p expression in CRC.
Conclusion
In summary, the findings of the present study document that CRC is characterized by the downregulation of miR-449b-5p, and this alteration contributes to tumor progression. miR-449b-5p represses post-transcriptionally the expression of CTNND1, inhibiting proliferation and invasiveness of CRC cells in vitro. Mechanistically, miR-449b-5p acts as a tumor suppressor by targeting CTNND1- mediated Wnt/β-catenin signaling in CRC. The clinical relevance of miR-449b-5p and its potential role in the treatment of CRC warrant further studies in this field, which may lead to the discovery of novel therapeutic strategies for CRC.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
This study was supported by a grant from the Natural Science Foundation of the Shanghai Science and Technology Commission (No. 16ZR1400800).
References
- Popat S, Hubner R, Houlston RS (2005) Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol 23: 609-618.
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet Tieulent J, et al. (2015) Global cancer statistics, 2012. CA Cancer J Clin 65: 87-108.
- Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, et al. (2017) Colorectal cancer statistics, 2017. CA Cancer J Clin 67: 177-193.
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, et al. (2016) Cancer statistics in China, 2015. CA Cancer J Clin 66: 115-132.
- Ebert MS, Sharp PA (2012) Roles for microRNAs in conferring robustness to biological processes. Cell 149: 515-524.
- Hayes J, Peruzzi PP, Lawler S (2014) MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol Med 20: 460-469.
- Adams BD, Kasinski AL, Slack FJ (2014) Aberrant regulation and function of microRNAs in cancer. Curr Biol 24: R762-R776.
- Deng X, Hou C, Liang Z, Wang H, Zhu L, et al. (2017) miR-202 Suppresses cell proliferation by targeting FOXR2 in endometrial adenocarcinoma. Dis Markers 2017: 2827435.
- Rupaimoole R, Slack FJ. (2017) MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 16: 203-222.
- Sandbothe M, Buurman R, Reich N, Greiwe L, Vajen B, et al. (2017) The microRNA-449 family inhibits TGF-beta-mediated liver cancer cell migration by targeting SOX4. J Hepatol 66: 1012-1021.
- Fang Y, Gu X, Li Z, Xiang J, Chen Z (2013) miR-449b inhibits the proliferation of SW1116 colon cancer stem cells through downregulation of CCND1 and E2F3 expression. Oncol Rep 30: 399-406.
- Yang X, Feng M, Jiang X, Wu Z, Li Z, et al. (2009) miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev 23: 2388-2393.
- Tang B, Tang F, Wang Z, Qi Z, Liang X, et al. (2016) Overexpression of CTNND1 in hepatocellular carcinoma promotes carcinous characters through activation of Wnt/beta-catenin signaling. J Exp Clin Cancer Res 35: 82.
- Liu G, Li B (2019) Role of miRNA in transformation from normal tissue to colorectal adenoma and cancer. J Cancer Res Ther 15: 278-285.
- Chen H, Oho A, Xu Z, Liu D (2019) Small non-coding RNA and colorectal cancer. J Cell Mol Med 23: 3050-3057.
- Wong CM, Tsang FH, Ng IO (2018) Non-coding RNAs in hepatocellular carcinoma: Molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol 15: 137-151.
- Jiang J, Yang X, He X, Ma W, Wang J, et al. (2019) MicroRNA-449b-5p suppresses the growth and invasion of breast cancer cells via inhibiting CREPT-mediated Wnt/beta-catenin signaling. Chem Biol Interact 302: 74-82.
- Zhang Y, Lv J, Wu G, Li W, Zhang Z, et al. (2019) MicroRNA-449b-5p targets HMGB1 to attenuate hepatocyte injury in liver ischemia and reperfusion. J Cell Physiol 234: 16367-16375.
- Cheng L, Shi X, Huo D, Zhao Y, Zhang H (2019) MiR-449b-5p regulates cell proliferation, migration and radioresistance in cervical cancer by interacting with the transcription suppressor FOXP1. Eur J Pharmacol 856: 172399.
- Wu S, Du X, Wu M, Du H, Shi X, et al. (2016) MicroRNA-409-3p inhibits osteosarcoma cell migration and invasion by targeting catenin-delta1. Gene 584: 83-89.
- Wang Y, Liu C, Luo M, Zhang Z, Gong J, et al. (2015) Chemotherapy-Induced miRNA-29c/Catenin-delta signalling suppresses metastasis in gastric cancer. Cancer Res 75: 1332-1344.
- Xing AY, Wang YW, Su ZX, Shi DB, Wang B, et al. (2015) Catenin-delta1, negatively regulated by miR-145, promotes tumour aggressiveness in gastric cancer. J Pathol 236: 53-64.
- Tang B, Tang F, Wang Z, Qi G, Liang X, et al. Overexpression of CTNND1 in hepatocellular carcinoma promotes carcinous characters through activation of Wnt/beta-catenin signaling. J Exp Clin Cancer Res 35: 82.
- Rahmani F, Avan A, Hashemy SI, Hassanian SM (2018) Role of Wnt/beta-catenin signalling regulatory microRNAs in the pathogenesis of colorectal cancer. J Cell Physiol 233: 811-817.
- Zhang X, Sukamporn P, Zhang S, Min KW, Baek SJ. (2017) 3,3'-diindolylmethane downregulates cyclin D1 through triggering endoplasmic reticulum stress in colorectal cancer cells. Oncol Rep 38: 569-574.
- Duan L, Wu R, Ye L, Yang X, Wang H, et al. (2013) S100A8 and S100A9 are associated with colorectal carcinoma progression and contribute to colorectal carcinoma cell survival and migration via Wnt/beta-catenin pathway. PLoS One 8: e62092.
Citation: Jin X, Hong Y, Li P, Hao L, Chen M (2019) Inhibition of CTNND1-Mediated Wnt/β-Catenin Signalling by MicroRNA-449b-5p Represses Colorectal Cancer Progression. J Clin Exp Pathol 9: 370. DOI: 10.4172/2161-0681.1000370
Copyright: © 2019 Jin X, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 3034
- [From(publication date): 0-2019 - Apr 16, 2025]
- Breakdown by view type
- HTML page views: 2290
- PDF downloads: 744