Special Issue Article Open Access
Inhibition of Cell Growth by Nuclear Receptor COUP-TFI: Possible Involvement of Decorin in Growth Inhibition
| Shinji Morii1,2, Masaki Kato3, Naohiko Seki3, Natsuko Shinmen1, Katsuro Iwase1, Masaki Takiguchi1 and Takaki Hiwasa1* | |
| 1Department of Biochemistry and Genetics, Chiba University Graduate School of Medicine, Japan | |
| 2Department of Medicine and Clinical Oncology, Chiba University Graduate School of Medicine, Japan | |
| 3Department of Functional Genomics, Chiba University Graduate School of Medicine, Japan | |
| *Corresponding Author : | Takaki Hiwasa Department of Biochemistry and Genetics Chiba University, Graduate School of Medicine Inohana 1-8-1, Chuo-ku, Chiba 260-8670, Japan Tel: +81-43-226-2541 Fax: +81-43-226-2037 E-mail: hiwasa_takaki@faculty.chiba-u.jp |
| Received August 05, 2014; Accepted August 07, 2014; Published August 14, 2014 | |
| Citation: Morii S, Kato M, Seki N, Shinmen N, Iwase K, et al. (2014) Inhibition of Cell Growth by Nuclear Receptor COUP-TFI: Possible Involvement of Decorin in Growth Inhibition. Biochem Physiol S4:001. doi:10.4172/2168-9652.S4-001 | |
| Copyright: © 2014 Hiwasa T, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Visit for more related articles at Biochemistry & Physiology: Open Access
Abstract
The transcription factor, Chicken Ovalbumin Upstream Promoter-Transcription Factor I (COUP-TFI) is a member of the steroid/thyroid hormone receptor superfamily and plays important roles especially in development and differentiation. To investigate the effects of COUP-TFI on cell growth, we transfected this factor stably into Ha-rastransformed NIH3T3 (ras-NIH3T3) cells. Ectopic expression of COUP-TFI resulted in reduction of both proliferation rates and anchorage-independent growth. Flow cytometric analysis showed that progression through mid-S to G2/M phase was delayed in COUP-TFI transfectants. cDNA microarray analysis was performed to elucidate candidate genes whose expression levels were changed in response to COUP-TFI introduction. mRNA levels of decorin and cyclin G were elevated in COUP-TFI transfectants. Luciferase reporter assay revealed that the upstream promoter regions between –252 and -205 bp and between -204 and +42 bp of the decorin gene were responsible for COUPTFI. These suggest that decorin transactivated by COUP-TFI is one of the molecules which account for the growthinhibitory effects of COUP-TFI.
|
Abstract
The transcription factor, Chicken Ovalbumin Upstream Promoter-Transcription Factor I (COUP-TFI) is a member of the steroid/thyroid hormone receptor superfamily and plays important roles especially in development and differentiation. To investigate the effects of COUP-TFI on cell growth, we transfected this factor stably into Ha-ras-transformed NIH3T3 (ras-NIH3T3) cells. Ectopic expression of COUP-TFI resulted in reduction of both proliferation rates and anchorage-independent growth. Flow cytometric analysis showed that progression through mid-S to G2/M phase was delayed in COUP-TFI transfectants. cDNA microarray analysis was performed to elucidate candidate genes whose expression levels were changed in response to COUP-TFI introduction. mRNA levels of decorin and cyclin G were elevated in COUP-TFI transfectants. Luciferase reporter assay revealed that the upstream promoter regions between –252 and -205 bp and between -204 and +42 bp of the decorin gene were responsible for COUP-TFI. These suggest that decorin transactivated by COUP-TFI is one of the molecules which account for the growth-inhibitory effects of COUP-TFI.
Keywords
Steroid/thyroid receptor superfamily; Stable transfection; Anchorage-independent growth; Flow cytometry; Cell cycle; cDNA microarray; Decorin
Introduction
Chicken Ovalbumin Upstream Promoter-Transcription Factors (COUP-TFs) [1] are the member of the steroid/thyroid hormone-activated nuclear receptor superfamily. Since no ligand has yet been identified, COUP-TFs are regarded as orphan receptors. Two distinct genes for COUP-TFs, COUP-TFI/Ear3 [1,2] and COUP-TFII/ARP-1 [3], have been identified, and their sequence homology was 98 and 96% in the DNA-binding domain and in the putative ligand-binding domain, respectively [4]. Although COUP-TFI was originally shown to stimulate transcription of the ovalbumin gene [5], a number of studies have demonstrated that COUP-TFI antagonistically represses transactivation by other nuclear receptors [6-10], mainly through competitive occupancy of a target binding sequence. However, it has been recently reported that COUP-TFs can function as transactivators [11-13] as well as an auxiliary cofactor for other transcription factors [14,15].
Targeted disruption of the COUP-TFI gene resulted in defects in neurogenesis, axon guidance, and arborization [16], whereas targeted deletion of COUP-TFII resulted in embryonic lethality with defects in angiogenesis, vascular remodeling, and fetal heart development [17]. Therefore, COUP-TFs are essential for early development and cell differentiation [18,19], and could be associated with cell proliferation. Recent studies have reported that COUP-TFI was required for growth inhibition and apoptosis in response to retinoic acid in some cancer cell lines [20], and that ectopic expression of COUP-TFII in breast cancer cell lines caused growth arrest associated with modulation in expression of cell-cycle regulatory proteins such as p21Cip1 and cyclin D1 [21]. On the other hand, COUP-TFI stimulates proliferation [22] and migration [23] of breast cancer cells by activating estrogen signaling or EGF signaling. In the present study, we stably introduced COUP-TFI into Ha-ras-transformed NIH3T3 cells and examined its effect on cell growth. Ectopic expression of COUP-TFI led to decrease in cell proliferation rates, reduction of anchorage-independent growth, and delay of progression through mid-S to G2/M phase. Results of cDNA microarray analysis and reporter assay suggested that decorin is one of the transcriptional targets of COUP-TFI and mediates the growth-inhibitory effects of COUP-TFI. Materials and Methods
Cells and culture
A cell strain, Ha-ras-transformed NIH3T3 (ras-NIH3T3, clone F25) was kindly supplied by T. Sekiya [24] and cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 5% calf serum.
Stable transfection
Transfection of plasmids into ras-NIH3T3 cells and selection of stable transfectants were done essentially as described [25]. Cells were plated at a density of 2×105 cells in 35 mm dishes. Twenty-four hours later, the cells were co-transfected with the plasmid expressing human COUP-TFI cDNA [7] under the control of the elongation factor 1α promoter of pEF-BOS [26], in addition to neo gene-bearing plasmid pcDNA3 (Thermo Fisher Scientific, Waltham, MA) at a ratio of 10:1. Control cells were co-transfected with pEF-BOS and pcDNA3. For transfection, FuGENE6 reagent (Roche Diagnostics, Mannheim, Germany) was used according to manufacturer's protocol. The next day, the cells were replated onto 100 mm dishes in growth medium containing 400 μg/ml G418 (Thermo Fisher Scientific). After 3 weeks, the G418-resistant colonies were isolated.
Analysis of cell proliferation rates and cell cycle
Cells (2×104) were seeded in 35 mm plates in triplicate. After the culture for various periods, cells were trypsinized and counted using Coulter Counter ZM (Beckman Coulter, Fullerton, CA). For cell cycle analysis, cells were collected by centrifugation and washed three times with Phosphate-Buffered Saline (PBS). The cells were then resuspended in 0.9 ml of staining solution (0.1% Triton X100, 100 μg/ml of RNase A, 40 μg/ml of propidium iodide and 4 mM sodium citrate), and incubated at room temperature for 20 min in the dark. Following addition of 0.1 ml of 1.5 M NaCl, samples was analyzed with FACScan (Becton Dickinson Immunocytometry Systems, San Joe, CA) as described previously [27]. Twenty thousand events of individual samples were collected for each analysis.
Semi-synchronized cell cycle analysis
To synchronize cell cycle at least partially, cells were incubated in DMEM containing 0.2% calf serum for two days, and then released from serum starvation by addition of calf serum to 5%. After the culture for various periods, cell cycle distribution was measured by the flow cytometry using FACScan.
Anchorage-independent growth
Anchorage-independent growth was tested by colony-forming assay using soft agar medium essentially as described [28]. The top layer in a 60 mm dish contained 600 cells in 2 ml of 0.4% Sea Plaque Agarose (BioWhittaker Molecular Applications, Rockland, ME) in DMEM supplemented with 5% calf serum, while the bottom layer consisted of 3 ml of 0.6% agarose in DMEM supplemented with 5% calf serum. Cells were incubated at 37°C in 5% CO2 for 3 weeks, and colonies with diameters larger than 0.1 mm were scored under a phase-contrast microscope.
Northern blot analysis
Total cellular RNA was prepared by the acid-guanidine thiocyanate-phenol/chloroform extraction method [29]. The RNA was subjected to denaturing agarose gel electrophoresis and transferred to a nylon membrane. Hybridization with digoxigenin-labeled antisense RNA probes followed by chemiluminescent detection on X-ray films was done according to manufacturer's protocol (Roche Diagnostics) as described previously [30].
Western blot analysis
Cells were lysed in the protein lysis buffer (0.5% Nonidet P-40, 20 mM Tris-HCl (pH 7.5), 1 mM EDTA, 1 mM EGTA, 1 mM phenylmethanesulfonyl fluoride, 50 μM leupeptin, 50 μM antipain, 50 μM pepstatin A and 50 μM N-acetyl-leucyl-leucyl-norleucinal), and protein concentrations were determined using the Bradford method [31]. Equal amounts of sample lysates were separated by SDS-polyacrylamide gel electrophoresis and electrically transferred onto a nitrocellulose membrane. The membrane was blocked with 5% nonfat dry milk in a buffer consisting of 20 mM Tris-HCl (pH 7.6), 137 mM NaCl and 0.1% Tween 20, and incubated with specific primary antibodies against COUP-TFI (a gift from MJ Tsai) [4]. After incubation with horseradish peroxidase-conjugated secondary antibody, immunoreactivity was detected with the ECL kit (GE Healthcare, Buckinghamshire, England) as described previously [32].
cDNA microarray analysis
A microarray chip harboring 2,304 cDNA clones derived from the mouse embryo was prepared essentially as described [33]. Total RNA was prepared from COUP-TFI-transfected cells, and from parental cells. Poly(A)+ RNA was isolated, using oligo(dT)-paramagnetic beads Dynabeads Oligo(dT)25 (Dynal, Oslo, Norway). Two μg of poly(A)+ RNA and 4.5 μg of oligo(dT) in 15 μl of solution were heat-denatured at 70°C for 10 min, and chilled on ice immediately. The solution was made up to 30 μl of the mixture containing 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 10 mM DTT, 0.5 mM each of dATP, dCTP, and dGTP, 0.2 mM dTTP, 0.1 mM Cy3- or Cy5-dUTP, and 400 units of reverse transcriptase SuperScript II (Thermo Fisher Scientific). After the incubation at 42°C for 1 h, template mRNAs were degraded at 65°C for 10 min in the presence of 1.5 μl of 1 N NaOH/20 mM EDTA, and then neutralized with 270 μl of 10 mM Tris-HCl (pH 8.0)/1 mM EDTA and 1.5 μl of 1 N HCl. The Cy3- or Cy5-labeled cDNA was purified with Centricon-30 microconcentrators (Millipore, Bedford, MA). Hybridization was performed in a solution containing 2 μg/μl of yeast RNA, 2 μg/μl of poly(A), 3.4×SSC (1×SSC consists of 0.15 M NaCl/15 mM sodium citrate) and 0.3% SDS at 65°C overnight under humidified condition. After the hybridization, the arrays were washed twice for 5 min with 2×SSC/0.1% SDS at room temperature, twice for 5 min with 0.2 x SSC/0.1% SDS at 40°C, and finally rinsed with 0.2×SSC. The fluorescent images were scanned with a laser-scanning device (ScanArray 4000, GSI Lumonics, Bedford, MA).
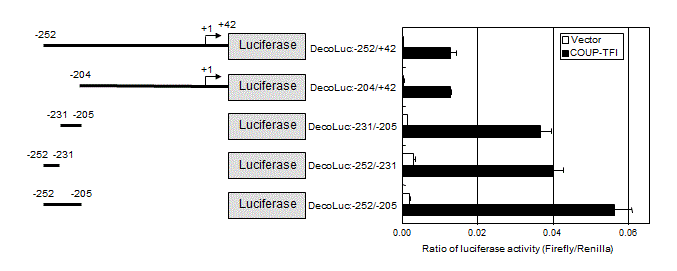
Luciferase reporter assay
Genomic DNA was isolated from T.Tn human esophageal carcinoma cell line [34] and used as a template. The promoter regions of –1301 to –205, –252 to +42, and –204 to +42 of the decorin gene were amplified by PCR and inserted into the XhoI-HindIII site of the firefly luciferase reporter plasmid, pGL3-Basic (Promega, Madison, WI), to construct DecoLuc:-1338/-205, DecoLuc:-252/+42 and DecoLuc:-204/+42. By using them as templates, DecoLuc:-252/-205, DecoLuc:-252/-231, DecoLuc:-231/-205 were constructed by reverse PCR method using KOD-Plus-Mutagenesis kit (Toyobo, Osaka, Japan) according to the manufacturer’s instruction. Base sequences of these constructs were confirmed by sequencing analysis.
The cells (1×105 cells/well) seeded on 24-well plates were transfected with COUP-TFI expression plasmid [7] or control empty vector pEF-BOS (0.1 μg) together with firefly luciferase reporter plasmid (0.4 μg) and Renilla luciferase reporter plasmid CMV-Rluc [25] by using LipofectAMINE-Plus (Thermo Fisher Scientific). Two days after the transfection, firefly and Renilla luciferase activities were measured with a Dual Luciferase Assay System (Promega) and a luminescencer (Atto, Tokyo, Japan). Firefly luciferase activities were normalized with respect to parallel Renilla luciferase activities, to correct for differences in transfection efficiency. Results
Isolation of stable transfectants of COUP-TFI
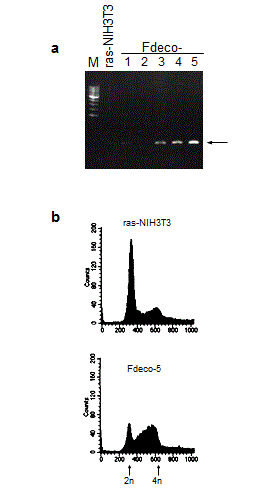
To examine the biological activity of COUP-TFI, Ha-ras-transformed NIH3T3 (ras-NIH3T3) cells were stably transfected with COUP-TFI-expressing plasmids. Among G418-selected clones, COUP-TFI-transfected clones (FCO-3, 7 and 10) showed higher expression of COUP-TFI at both mRNA and protein levels than the parental cells and a vector-transfected clone (Figure 1a and b).
Reduced proliferation rates and delay of cell cycle progression through mid-S to G2 phase in COUP-TFI transfected cells
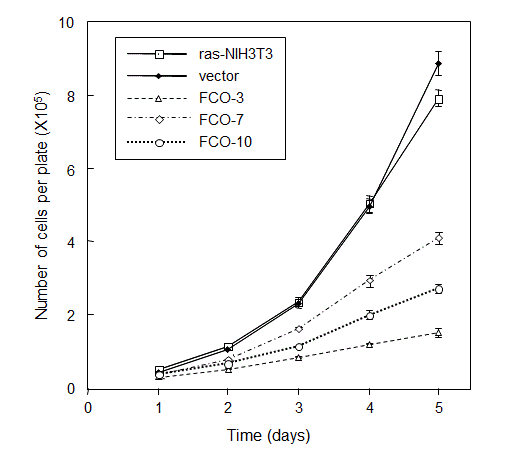
When the proliferation rates were examined by direct counting of cell numbers, all of COUP-TFI-transfected cells grew much more slowly than the parental and vector-transfected control cells (Figure 2). The apparent inhibitory effect of COUP-TFI on cell proliferation prompted us to investigate changes in cell cycle distribution by the flow cytometric analysis.
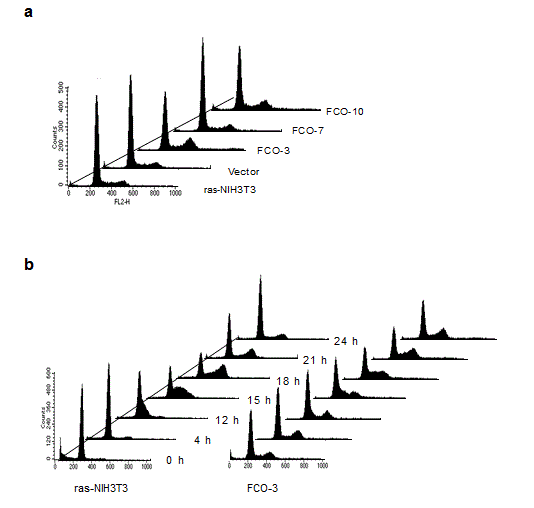
As shown in Figure 3a, a different cell cycle distribution was observed between COUP-TFI-transfected cells and control cells: COUP-TFI transfectants with the exception of one clone (FCO-7) showed accumulation in late S to G2/M phase of the cell cycle. A further analysis of cell cycle distribution with semi-synchronized cells at various times after release from serum starvation was performed (Figure 3b). Serum starvation for two days led to an increase in G0/G1 population and a concomitant decrease in S and G2/M populations in parental ras-NIH3T3 cells. A small but apparent portion of hypodiploid apoptotic cells was also detected in the parental cells. On the other hand, a substantial amount of the G2/M population was still detectable in FCO-3 cells. Although the parental cells entered S phase around 12 h after serum stimulation, an apparent delay was observed in progression of FCO-3 cells from S phase to the sequential phase, suggesting that COUP-TFI repressed mid-S to G2/M progression. The extent of prolongation in mid-S to G2/M progression correlated well with that of decreases in cell proliferation rates (Figure 2). Taken together, we concluded that a delay of progression through mid-S to G2/M phase is responsible for growth inhibition in COUP-TFI-transfected cells. Reduced anchorage-independent growth in COUP-TFI transfectants
Colony-forming ability in soft agar medium is well established as a useful mean to assess the transformed state of a given cell population, and was examined on COUP-TFI transfectants.
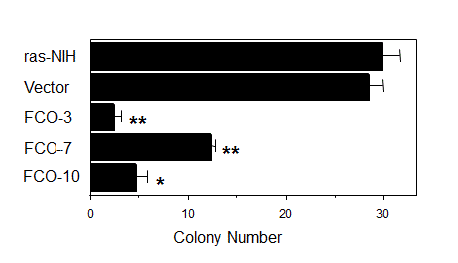
As shown in Figure 4, the number of colonies formed in soft agar decreased significantly in COUP-TFI transfectants compared with parental cells and a vector-transfected clone, suggesting that COUP-TFI may have a potential to suppress a transformed phenotype of tumor cells. cDNA microarray and northern blot analysis
cDNA microarray analysis was performed to elucidate candidates for genes that are targets of COUP-TFI, and that may account for the growth-inhibitory effects. Total RNAs prepared from three COUP-TFI-transfected clones, FCO-3, 7 and 10, were each mixed before poly(A)+ RNA isolation, in order to avoid detecting non-specific alterations in mRNA levels fluctuating from one clone to another. For the control, a mixture of total RNAs prepared from parental cells harvested at two different times was used. Possible difference of labeling efficiency of Cy3 and Cy5 was controlled by interconverting the two dyes in cDNA synthesis. Genes whose expressions were altered by transfection of COUP-TFI are listed in Table 1.
Levels of mRNAs for six genes were significantly elevated in COUP-TFI-transfected cells. Among these genes, the decorin and cyclin G genes were especially noteworthy because they are involved in the control of cell proliferation [35,36]. Contrary to our expectations, none was listed as the gene down-regulated by introduction of COUP-TFI, which often works as a transcriptional repressor in a reporter transfection assay [6-10]. Elevated mRNA expression levels of decorin and cyclin G in COUP-TFI-transfectants
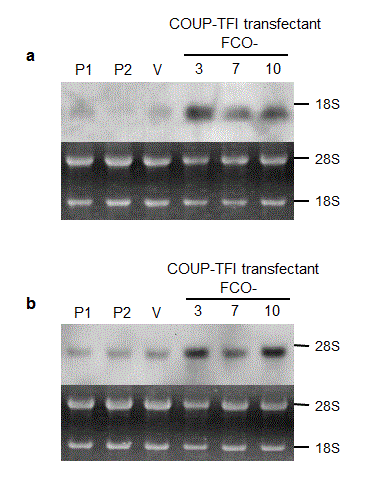
To confirm changes in decorin and cyclin G mRNA levels, we performed Northern blot analysis (Figure 5).
Decorin mRNA levels were elevated in all three clones of COUP-TFI transfectants as compared to those of parental cells and a control vector-transfected clone. Cyclin G mRNA levels were elevated in at least two of COUP-TFI transfectants. Interestingly, the extents of elevation in decorin and cyclin G mRNA levels in COUP-TFI transfectants (Figure 5) seem to correlate roughly with degrees of cell growth inhibition (Figure 2), changes in cell cycle distribution (Figure 3a) and suppression of anchorage-independent growth (Figure 4). Activation of decorin promoter by COUP-TFI
To examine whether decorin is a target of COUP-TFI, luciferase reporter assay was performed using reporter plasmids containing the promoter region of the decorin gene. The regions between –252 and +42 and between –204 and +42 were activated approximately 30-fold by COUP-TFI as compared to the empty vector (Figure 6).
Among several constructs, the region between –252 and -205 was most activated by COUP-TFI, although the ratio of activation by COUP-TFI versus activation by control vector was also 30 fold. Further dividing this region into two portions indicated COUP-TFI-responsive domains are located in both regions between –252 and –231 and between –231 and –205. Activation of the region between –252 and +42 was lower than that between –252 and –205, suggesting the presence of cis-acting suppressive element(s) in the region between –204 and +42. Thus, multiple COUP-TFI-responsive elements may locate in the promoter region of the decorin gene. Discussion
In this study, we examined the effects of COUP-TFI on cell growth, and observed that COUP-TFI transfection resulted in reduction of proliferation rates and anchorage-independent growth. This growth-inhibitory activity of COUP-TFI is well consistent with the recent report that COUP-TFI is down-regulated in bladder transitional cell carcinoma [37]. Cell cycle progression of COUP-TFI-transfected cells was mainly blocked around G2/M phase and the transition from mid-S to G2/M phase was also delayed. A previous study using breast cancer cell lines has demonstrated that COUP-TFII inhibits cell growth and cell cycle progression by modulating the expression of p21Cip1, cyclin D1, and the activity of cdk2 [21]. In the present study, we could not detect any elevation of the p21Cip1 level in COUP-TFI transfectants (data not shown). In addition, cDNA microarray analysis and succeeding Northern blot analysis revealed elevation of mRNA levels for decorin and cyclin G in COUP-TFI transfectants.
Cyclin G was initially identified as a target of the c-src family kinase pathway in rat fibroblasts [38]. It has attracted attention because the cyclin G gene is one of the target genes of p53 tumor suppressor protein: two distinct high-affinity p53-binding sites were found in the promoter region of cyclin G, and its expression is induced after DNA damage via p53 [39,40]. In addition, it was reported that cyclin G is involved in G2/M checkpoint control [36]. Therefore, cyclin G might cause G2/M arrest through its direct induction by p53. However, in other studies, ectopic overexpression of cyclin G in human RKO colon carcinoma cells accelerated cell growth [41], and cyclin G1 was essential for the growth of human osteosarcoma cells [42]. This contradiction may be attributable to difference in cell types and/or in the expression levels of cyclin G (ectopic enforced expression etc.). We also identified decorin as a candidate for the cell growth inhibitor under the control of COUP-TFI. Decorin is a member of small leucine-rich proteoglycan gene family [43]. It has been reported that decorin inhibits cell proliferation by neutralizing transforming growth factor β (TGFβ) activity [35,44] and/or by inducing p21Cip1 through epidermal growth factor receptor (EGFR) activation [45-47]. The latter mechanism may not account for the growth inhibition in the present study because EGFR is not expressed in ras-NIH3T3 cells (data not shown) and also because p21 was not induced in COUP-TFI transfectants (data not shown). On the other hand, it is plausible that suppression of TGFβ activity by decorin is the cause of growth inhibition in response to COUP-TFI introduction, because TGFβ stimulates the proliferation of NIH3T3 cells [48]. It is noteworthy that decorin-overexpressing COUP-TFI transfectants showed apparent reduction of anchorage-independent growth (Figure 4). This observation is concordant with the notion that decorin plays a role in suppression of some transformed phenotypes of tumor cells. Indeed, it has been demonstrated that de novo decorin gene expression suppressed the malignant phenotype in human colon cancer cells [49]. We also isolated stable decorin transfectants, which showed similar cell cycle distribution to that of COUP-TFI transfectants (Supplemental Figure S1). The accumulation at mid-S to G2/M phase was more prominent than COUP-TFI-tranfectants, possibly due to the enforced overexpression of decorin. COUP-TFs belongs to the superfamily of the steroid/thyroid hormone-activated nuclear receptors [1], which binds to direct repeat of the hexanucleotide 5’-AGGTCA-3’ motif (DR1 motif) [50,51]. Our present study showed that the regions between –252 and –205 bp and between -204 and +42 in the decorin promoter are responsive to COUP-TFI (Figure 6). The 30-fold transactivation of the reporter gene suggests the direct activation of the decorin promoter by COUP-TFI. However, this region contains no similar DR1 consensus sequence. Therefore, we cannot rule out the possibility that the decorin promoter was indirectly activated by COUP-TF1 via some other transcription factor(s). We also failed to find any DR1-like motif in the promoter region of the cyclin G. However, the potent growth-inhibition and the specific mid-S to G2/M arrest of cell cycle progression by both decorin and cyclin G suggest that these two molecules have a key role in the growth-inhibitory activity of COUP-TFI. Acknowledgements
We are grateful to T. Sekiya for Ha-ras-transformed NIH3T3 cells, Tatsuya Kimura and Shoaib Chowdhury (Kumamoto University) for the COUP-TFI plasmid and Ming-Jer Tsai (Baylor College of Medicine) for the COUP-TFI antiserum. We also thank our colleague for suggestions, help, and discussion.
Grant sponsor: Grant in Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Competing interests: The authors declare that they have no competing interests. References
|
Tables and Figures at a glance
| Table 1 |
Figures at a glance
 |
 |
 |
 |
| Figure 1 | Figure 2 | Figure 3 | Figure 4 |
 |
 |
 |
|
| Figure 5 | Figure 6 | Supplemental Figure S1 |
Relevant Topics
- Analytical Biochemistry
- Applied Biochemistry
- Carbohydrate Biochemistry
- Cellular Biochemistry
- Clinical_Biochemistry
- Comparative Biochemistry
- Environmental Biochemistry
- Forensic Biochemistry
- Lipid Biochemistry
- Medical_Biochemistry
- Metabolomics
- Nutritional Biochemistry
- Pesticide Biochemistry
- Process Biochemistry
- Protein_Biochemistry
- Single-Cell Biochemistry
- Soil_Biochemistry
Recommended Journals
- Biosensor Journals
- Cellular Biology Journal
- Journal of Biochemistry and Microbial Toxicology
- Journal of Biochemistry and Cell Biology
- Journal of Biological and Medical Sciences
- Journal of Cell Biology & Immunology
- Journal of Cellular and Molecular Pharmacology
- Journal of Chemical Biology & Therapeutics
- Journal of Phytochemicistry And Biochemistry
Article Tools
Article Usage
- Total views: 13750
- [From(publication date):
September-2015 - Nov 21, 2024] - Breakdown by view type
- HTML page views : 9331
- PDF downloads : 4419
