Immune Gene Expression during Bovine Herpesvirus-1 Infection of MDBK Cells: Molecular Characterization of Interferon-Stimulated Gene 15
Received: 18-Dec-2017 / Accepted Date: 01-Jan-2018 / Published Date: 08-Jan-2018
Abstract
Bovine herpesvirus 1 (BoHV-1) infection extensively modulates host-cell gene expressions. To further understandings of BoHV-1 pathogenesis, Madin-Darby bovine kidney cells were infected, and RT-qPCR expression analyses were performed. Apart from classical immune genes (e.g. PKR, Mx1, IRF7, CCR4, TXNIP), EGR3, IFITM3, and ISG15 presented notably high transcripts. To gain further insight, the bovine ISG15 gene was characterized and compared against human, mouse, dog, fugu, salmon, and zebrafish homologues. In silico analysis revealed several conserved regions of interest, including a set of potential transcriptional motifs in the proximal 5’-untranslated region for all analyzed species. A protein-deduced bovine ISG15 sequence also evidenced many conserved domains in humans. Finally, in vitro infection assays with bovine viral diarrhea virus-1 or BoHV-1 both resulted in elevated blood ISG15 levels. Collectively, the obtained data indicate that ISG15 could potentially serve as an early immune marker during BoHV-1 and bovine viral diarrhea virus-1 infection.
Keywords: BoHV-1; BVDV-1; ISG15; Immune marker
Abbreviations
BoHV-1: Bovine HepaVirus 1; bp: base pairs; BVDV-1: Bovine Viral Diahrrea Virus 1; IFITM3: Interferon-Induced Transmembrane protein 3; IFN: Interferon; ISG15: Interferon- Stimulated Gene 15; MDBK: Madin-Darby Bovine Kidney; NF-kappa B: Nuclear Factor kappa-light-chain-enhancer of activated B; RT-PCR: Reverse Transcriptase-PCR
Introduction
Bovine herpesvirus 1 (BoHV-1), of the Alphaherpesvirinae subfamily, is a primary pathogen in the cattle industry. The infectious phenotypes of BoHV-1 were first reported in the USA in the 1950s, with classification as a List B-notifiable disease by the World Organization for Animal Health. Clinical symptoms of viral infection include infectious bovine rhinotracheitis, abortion, infectious pustular vulvovaginitis, and systemic infection in neonates. Just as in humans, the nervous sensory ganglia of surviving animals present a life-long latent infection, and undetermined stimuli can reactivate viral excretion. This phenomenon is responsible for BoHV-1 persistence within cattle herds. While the exact economic impacts of infection have not been quantified, estimates are high.
The BoHV-1 genome is a long, double-stranded, linear DNA molecule comprised by 135.3 kilobase pairs. A total of 73 open reading frames have been clearly identified (Genbank AJ004801). Most of the BoHV-1 gene repertoire consists of open reading frames homologous to genes found in other Alphaherpesvirinae. These homologues are generally labelled to related genes described in the prototype herpes simplex virus 1.
BoHV-1 infection of permissive cells involves virus attachment between glycoprotein B, gC, gD, gH, and gL to cell surface structures such as heparan sulfate sugar moieties [1-4]. While the virus particle is transported to the nucleus, tegument proteins are shed in the cytosol of the infected cell. These proteins might play important roles during the early infection stages, serving as a first contact with the intracellular environment and, particularly, with molecular sensors related to cellular immunity [5]. Despite these advances in knowledge, the specific impacts of BoHV-1 infection on the mechanisms of affected cell signaling pathways are not well understood. One promising step towards elucidating the entire disease process is evaluating the host transcriptome during BoHV-1 infection, with initial focus on detecting and quantifying molecules produced by the host cell as a result of viral infection. The quantification of these molecules can be used to identify potential immune biomarkers associated with specific viral families, as achieved in several prior investigations [6-8].
Independent of gene expressions, post-translational protein modifications is an essential strategy for regulating the cell proteome. Ubiquitin and ubiquitin-like modifiers covalently bind to proteins to coordinate the regulation of protein levels, signaling pathways, vesicular trafficking, and many other cellular processes. Of particular note, the interferon-stimulated gene 15 (ISG15) is an interferon (IFN)- α/β-inducible, 17 kilodalton (kDa), ubiquitin-like molecule that, in humans, is encoded by the ISG15 gene. The LRLRGG sequence is present in mature ISG15 and at the C terminus of mature ubiquitin. The main cellular function of ISG15 is ISGylation, with ISG15 existing either as a free molecule (intracellular and extracellular) or conjugated to target protein lysine residues [9]. Interestingly, ISG15 is strongly expressed in Madin-Darby bovine kidney (MDBK) cells and animals infected with bovine virus (unpublished data). Nevertheless, ISG15 in cattle remains poorly characterized.
The present study assessed the modulation of several immune markers during an infection assay of MDBK cells with BoHV-1. Following expression analyses, the bovine ISG15 gene was characterized as a potential biomarker of the immune response to viral infection.
Materials and Methods
Cell cultures and infection with BoHV-1 and BVDV-1
Madin-Darby Bovine Kidney (MDBK) cells (ATCC) were maintained in a Dulbecco’s Modified Eagle’s medium supplemented with 10% fetal horse serum at 37°C in a humidified, 5% CO2 atmosphere. Cells were confirmed free of bovine viral diarrhea virus (BVDV) by RT-PCR. Field-isolated BoHV-1 and BVDV-1 [10] were obtained from bovine blood samples and propagated in an MDBK cell culture. Endpoint viral titration was performed with six replicates in MDBK cells, and a 50% tissue culture infective dose was determined. For expression, immunofluorescence, and qPCR analyses, MDBK cells were infected with the BoHV-1 at a multiplicity of infection of 3 for 0, 6, and 24 h. Cells were then washed with fetal horse serum-free Dulbecco’s Modified Eagle’s medium and subsequently incubated in Dulbecco’s Modified Eagle’s medium supplemented with 2% fetal horse serum.
Analysis of gene expressions
To measure viral genome replication and to analyze host gene expressions, viral and total cellular RNA were extracted from cells or total blood with TRIzol (Life Technologies) and isolated using commercial viral and cellular RNA kits (Macharey-Nagel). Reverse transcription was performed using M-MLV Reverse Transcriptase (Life Technologies) and random primers or oligo d(T) for viral and cellular DNA extractions, respectively. qPCR analysis of the cDNA was carried out on 96-well plates in triplicate reactions with defined primers (Table 1) and SYBR Green Reagents (Stratagene). All qPCR reactions were performed using the Lightcycler 96 System (Roche). The mean expressions of the tested genes were normalized to housekeeping genes for validation, as based on the extent of fold-changes and/or biological significance. Fold-changes were calculated with the relative 2-(ΔΔCt) quantification method using CD81 as the housekeeping gene [11].
| GENE | N° ACCESSION | PRIMER | PRODUCT LENGTH | |
|---|---|---|---|---|
| PKR | AB104655.1 | F | 5‘-CCG TCT GAA AAT GGC TTC TC-3' | 138 |
| R | 5‘-TGT AGG CGC CAA ACT TCT CT-3' | |||
| BCL3 | NM_001205993.1 | F | 5'-GGA GAT ACG CCA CTC CAC AT-3' | 142 |
| R | 5'-TGA TCA CAG CCA GGT GTA GC-3' | |||
| INFβ | EU276065.1 | F | 5'-ACTCCTGGGGCAGTTACCTT-3' | 139 |
| R | 5'-GTGCTGGAGCACCTCATACA-3' | |||
| MX1 | JQ766265.1 | F | 5'-TTT TTC AAC CTC CAC CGA AC-3' | 132 |
| R | 5'-GTA CAC CTG GTC CTG GCA GT-3' | |||
| MHC I | JN792884.1 | F | 5'-ACATGGAGCTTGTGGAGACC-3' | 158 |
| R | 5'-TTCCCCTCCTTTATCCCATC-3' | |||
| MHC II | NM_001034668.3 | F | 5'-CAGATCAAGGTTCGGTGGTT-3' | 103 |
| R | 5'-TCACGAGGATCTGGAAGGTC-3' | |||
| IRF7 | NM_001105040.1 | F | 5'-GCT CCA CTA CAC CGA GAA GC-3' | 196 |
| R | 5'-GAA GTC AAA GAT GGG CGT GT-3' | |||
| IER3IP1 | NM_001113320.1 | F | 5'-GAACAGACCAGGGAATTGGA-3' | 102 |
| R | 5'-TCAATGGCACTCTCATCACG-3' | |||
| IFNGR | NM_001035063.1 | F | 5'-TAAGCGAGAAGGCACCAAGT-3' | 114 |
| R | 5'-GCAATTGGCTGGTAGGTGAT-3' | |||
| NFE2L2 | NM_001011678.2 | F | 5'-CAAAATGACAAGCTGGCTGA-3' | 131 |
| R | 5'-AAATGTGGGCTGCAGTTACC-3' | |||
| ISG15 | NM_174366.1 | F | 5'-TGCAGAACTGCATCTCCATC-3' | 199 |
| R | 5'-TTCATGAGGCCGTATTCCTC-3' | |||
| RSAD2 | NM_001045941.1 | F | 5'-GATTCAACGTGGACGAGGAT-3' | 276 |
| R | 5'-TCCGCCCATTTCTACAGTTC-3' | |||
| GADD45ϒ | NM_001045901.1 | F | 5'-CAAGCGTTCTGCTGTGAGAA-3' | 133 |
| R | 5'-TGGGGTTCGAAATGAGGATA-3' | |||
| IFITM3 | NM_001078141.2 | F | 5'-GTGGCATTCGCCTACTCTGT-3' | 142 |
| R | 5'-CGATGAGGACGACAGTCAGA-3' | |||
| BST2 | AB698753.1 | F | 5'-GGCCCAGGAATTCTCACATA-3' | 154 |
| R | 5'-CAACGCGTCCTTCAGATTTT-3' | |||
| DUSP6 | XM_010804772.1 | F | 5'-CGTGGTGCTCTACGATGAGA-3' | 264 |
| R | 5'-GACTCAATGTCCGAGGAGGA-3' | |||
| MX2 | BC149724.1 | F | 5'-AGGTGTCCTGAAGGTCATGC-3' | 250 |
| R | 5'-CAAGGGGAGCGATTTATTGA-3' | |||
| NR4A1 | NM_001075911.1 | F | 5'-GGCATGGTGAAGGAAGTTGT-3' | 244 |
| R | 5'-CCGAGAGCAGGTCGTAGAAC-3' | |||
| TXNIP | NM_001101875.2 | F | 5'-CAAGTTCGGCTTTGAGCTTC-3' | 103 |
| R | 5'-GCTGGGACGATCAAGAAAAG-3' | |||
| EGR3 | NM_001289818.1 | F | 5'-CAAGTACCCCAACCGACCTA-3' | 199 |
| R | 5'-GCGGATGTGAGTGGTAAGGT-3' | |||
| CXCR4 | NM_174301.3 | F | 5'-GCCTGGTATCGTCATCCTGT-3' | 201 |
| R | 5'-TCAAACTCACACCCTTGCTG-3' | |||
| ZBP1 | XM_005194416.2 | F | 5'-GCTTCTGCTGATCCCTCAAC-3' | 213 |
| R | 5'-AATTTCGATGGAAGCGTCTG-3' | |||
| KLF4 | HQ703512.1 | F | 5'-CACTGTGACTGGGATGGTTG-3' | 146 |
| R | 5'-ATGTGTAAGGCGAGGTGGTC-3' | |||
| CSRP2 | NM_001038183.1 | F | 5'-GATCATTGGAGCTGGAAAGC-3' | 129 |
| R | 5'-TTCTTCGCGTAGCATCCTTT-3' | |||
| BCL2L11 | NM_001075310.1 | F | 5'-GCAGATACACGCCCAGAGAT-3' | 163 |
| R | 5'-ACACCAGACGCACGATGTAG-3' | |||
| TNFRSF10A | BC149759.1 | F | 5'-TTCCGGTGTACAAGGTGTGA-3' | 194 |
| R | 5'-ACAGCAGCAATGACGATGAG-3' | |||
| SAMD9L | NM_001206334.1 | F | 5'-TCTGCATCAATACCCACTGC-3' | 117 |
| R | 5'-CTTGCCCTCTGGTTTGTCAT-3' | |||
| ZC3H12A | NM_001102187.1 | F | 5'-ATCCTCCTGGCAGTGAATTG-3' | 205 |
| R | 5'-AGCGGTCGTCATAGCAGACT-3' | |||
| TNFSF10 | XM_005195915.2 | F | 5'-CAATCCCTGCTGGGAACTAA-3' | 207 |
| R | 5'-TCATTCTTGGAGCCTGGAAC-3' | |||
| PALM3 | XM_010798194.1 | F | 5'-GGCAGAGAAAACAGGAGACG-3' | 283 |
| R | 5'-TGCCTCACTCACATCCTCTG-3' | |||
| SLC7A2 | XM_005226025.2 | F | 5'-AAGGAAATGTGGCAAACTGG-3' | 101 |
| R | 5'-CCCATAGACGCTTGTTCCAT-3' | |||
| DDX58 | XM_002689480.4 | F | 5'-ACCCCAGACCGAAGAAGTTT-3' | 223 |
| R | 5'-GCAGCATCAAATGGGATCTT-3' | |||
| TAF4B | NM_001205844.1 | F | 5'-TCTTCACCTCAGCCTCACCT-3' | 195 |
| R | 5'-CTCAGCCTGGATGGAAGAAG-3' | |||
| GBP1 | BT030474.1 | F | 5'-GGATCAGAGAACACCCTGGA-3' | 291 |
| R | 5'-GTCTGGCTTCTTCGGATGAG-3' | |||
| CFOS | BT029837.1 | F | 5'-AGTGCCAACTTCATCCCAAC-3' | 190 |
| R | 5'-CTCTGCCTCCTGTCATGGTT-3' | |||
| GCH1 | XM_002690949.4 | F | 5'-TTCTTGGCCTCAGCAAACTT-3' | 176 |
| R | 5'-TTTCTGCACTCCTCGCATTA-3' | |||
| UBA7 | NM_001012284.1 | F | 5'-CCATCACCTGAAGTGGACCT-3' | 296 |
| R | 5'-TGTAGTGCAAGCGTGGAAAG-3' | |||
| IFIT3 | NM_001075414.1 | F | 5'-TGCTGACAAGGTGAAACGAG-3' | 111 |
| R | 5'-TTTTTCCCACCGCACTTTAC-3' | |||
| IFIT2 | BT025389.1 | F | 5'-ACCCCATTAACCCTTTGAGG-3' | 249 |
| R | 5'-TGTTGGGCATGCATTTTAGA-3' | |||
| MAP3K8 | BC142398.1 | F | 5'-CTGAAACACGAGGCACTGAA-3' | 108 |
| R | 5'-TTCTAACTCCTTCCGGCTCA-3' | |||
Table 1: List of primers used for RT-qPCR analyses of immune markers.
Immunofluorescence assays
MDBK cells were grown for 24 h on Labteck Chamber slides (Nunc). Cells were infected with BoHV-1 and BVDV-1 at a multiplicity of infection of 3 for either 6 or 24 h, fixed with the HistoChoice Clearing Agent (Sigma-Aldich), permeabilized with 0.1% Triton X- 100, and blocked with 1% bovine serum albumin. Cells infected by BoHV-1 were bound to the BoHV-1 conjugated monoclonal antibody against viral proteins (Cat. No. 210-69-IBR, VMRD, Inc.). Nuclei were counterstained with DAPI (Invitrogen). Chamber slides were mounted on glass slides using a fluorescence mounting medium (Dako). Images were taken using a fluorescent microscope (Model IX71, Olympus). All experiments were performed at least three times.
Western blotting
MDBK cells were lysed in an ice-cold NP-40 lysis buffer containing 1% Nonidet P-40, 25 mM Tris-HCl (pH 7.5), 150 mM sodium chloride, 1 mM EDTA, 5 mM sodium fluoride, 1 mM sodium orthovanadate, 1 mM leupeptin, and 1 mM phenylmethylsulfonyl fluoride. Western blot analysis was carried out according to a standard protocol [12]. Briefly, cells were lysed at 4°C, and the BCA Protein Assay Kit (Pierce, Thermo Scientific) was used to determine cell-lysate protein concentrations. Equal amounts of protein were loaded onto polyacrylamide gels, electrophoresed, and transferred onto nitrocellulose membranes (Schleicher and Schuell). After transfer, the membranes were blocked overnight in 5% dry milk and probed for 1 h with the anti-ISG15 rabbit (1:2500; SC-50366, Santa Cruz Biotechnology) and anti-HSP70 mouse monoclonal (1:1000; #sc-sc-24, Santa Cruz Biotechnology) antibodies. Subsequently, the membranes were probed for 1 h with an HRP-coupled secondary antibody (1:5000; Sigma-Aldrich). The reactive bands were visualized via an enhanced chemiluminescence detection system (Amersham Pharmacia Biotech) according to the manufacturer’s instructions. Signal intensity was measured with the ImageJ digital imaging system.
In silico analysis
Potential motifs and domains were searched for using Tfsitescan (http://www.ifti.org/cgi-bin/ifti/Tfsitescan.pl). Subsequent comparisons were performed against human (Homo sapiens), mouse (Mus musculus), dog (Canis lupus familiaris), Atlantic salmon (Salmo salar), fugu (Takifugu rubripes), and zebrafish (Danio rerio) ISG15, available from the ENSEMBL database (http://www.ensembl.org/ index.html). The MEME/MAST v3 system (http://meme-suite.org/) for motif discovery and searching was used to predict conserved sequence domains in the promoter region of the genes. Known protein domains were defined using Interpro (http://www.ebi.ac.uk/interpro/), SMART (http://smart.emblheidelberg.de/), and Chimera v1.8 (http://www. cgl.ucsf.edu/chimera/). For molecular phylogenetic analyses, protein sequences were first aligned using ClustalW [13]. phylogenetic analyses were performed using the neighbor-joining method within the Mega v4.0 software [14]. Data were analyzed using Poisson correction, and gaps were removed by pairwise deletion. The bootstrap values of the branches were obtained by testing the tree 10,000 times.
Sample collection and processing
A total of 21 blood samples were collected from cows suspected of infection and stored in EDTA-evacuated tubes (Saarstedt, Nümbrecht, Germany). The total blood was frozen at -80°C until analyses of BVDV- 1/BoHV-1 infection and ISG15 expression.
Statistical analysis
All data are shown as means ± standard error (SE). Differences were evaluated using ANOVA followed by the Student’s t-test. Statistical significance was defined as P<0.05.
Results
Viral infection
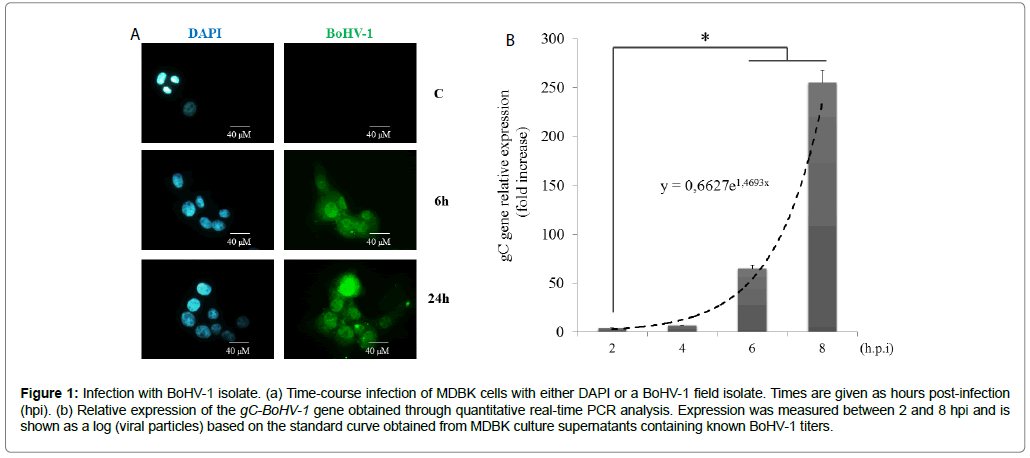
To examine if BoHV-1 isolates induced transcriptomic responses, MDBK cells were infected with a field isolate at a multiplicity of infection of 3, thus ensuring a high viral particle to cell ratio. Immunofluorescence staining of the virus confirmed that>98% of the cells were positive for BoHV-1 at 6 h post-infection (Figure 1a). There was a slight decrease in the number of viable cells at 24 hpi, which could indicate onset of the cytopathic effect. This decrease was significant 2 days post-infection (data not shown). Viral gC gene levels also rapidly increased during infection, as confirmed by real-time qPCR (Figure 1b).
Figure 1: Infection with BoHV-1 isolate. (a) Time-course infection of MDBK cells with either DAPI or a BoHV-1 field isolate. Times are given as hours post-infection (hpi). (b) Relative expression of the gC-BoHV-1 gene obtained through quantitative real-time PCR analysis. Expression was measured between 2 and 8 hpi and is shown as a log (viral particles) based on the standard curve obtained from MDBK culture supernatants containing known BoHV-1 titers.
Expression analyses
The principal aim of this study was to elucidate the diverse processes of morbidity triggered during the course of BoVH-1 infection. Since these processes begin from virus entrance to the host (i.e. early infection stages), a short infection period was used to characterize transcript modulations. In particular, transcripts related to key regulators in immune processes and in signaling pathways responsible for immune modulation and cellular processes were assessed. Markers potentially necessary for or present within the host at virus-infection onset were identified through transcriptomic analysis. For this, 40 randomly selected immune markers (Table 1) were detected by RT-qPCR, as based on prior research regarding immune responses to BVDV-1 [10,11,15].
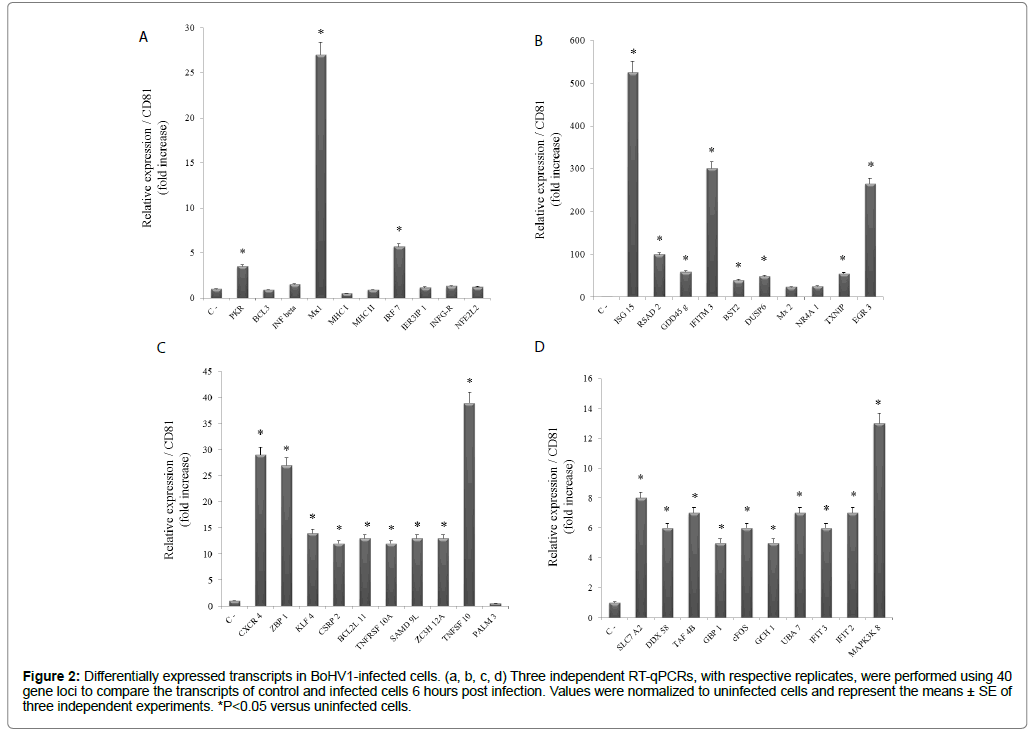
Alpha-herpesviruses can cause several clinical manifestations, including neurological diseases in cattle. As with other alphaherpesviruses, BoHV-1 establishes a lifelong latent infection, primarily in the neural ganglia of animals that survive acute infection [16,17]. The mechanisms underlying the clinical manifestations of BoHV-1 infection are not well-defined. However, it is likely that the immune system plays an important role in the development of a pathological condition [18]. In this context, the presently performed analyses identified transcriptional modulations for a number of important immunological markers involved in the antiviral state (e.g. PKR, INF beta, Mx1) and antigen presentation (e.g. MHCI, MHC II), among others (Figure 2a). Furthermore, an antiviral response was evidenced through very strong expression levels of eight immune genes (Figure 2b).
Figure 2: Differentially expressed transcripts in BoHV1-infected cells. (a, b, c, d) Three independent RT-qPCRs, with respective replicates, were performed using 40 gene loci to compare the transcripts of control and infected cells 6 hours post infection. Values were normalized to uninfected cells and represent the means ± SE of three independent experiments. *P<0.05 versus uninfected cells.
The strongly activated immune genes included ISG15, an ubiquitinlike protein with powerful antiviral activity against influenza A and B, the Sindbis virus, HIV-1, herpes simplex-1, and the murine herpesvirus [19,20]. The expression of ISG15 increased more than 500 times that of the control. A similar phenomenon was observed for transcripts of the IFN-induced transmembrane protein 3 (IFITM3) and the early growth response protein 3, which increased ≈ 300 and 250 times, respectively. While other immune markers had lower expression levels, all were significantly increased in infected cells (Figure 2b).
A molecular marker modulated by BoHV-1 infection was the DNA-dependent activator of IFN regulatory factor, also referred to as the Z-DNA binding protein 1. This marker, which acts as a cytosolic B-form DNA sensor that induces type I IFNs, was initially identified as a highly upregulated protein in mouse tumor stromal cells and in macrophages treated by gamma IFN or lipopolysaccharides [21]. Recently, the Z-DNA binding protein 1 was reported to limit herpes simplex virus 1growth through mechanisms involving the suppression of viral genomes [22]. In the presently assessed infection model, Z-DNA binding protein 1 transcripts interestingly increased more than 25 times that of the control (Figure 2c). Further research should be conducted to determine the biological consequence of this phenomenon.
Also highly regulated were transcripts of the tumor necrosis factor receptor superfamily member 10A (>13 times control) and tumor necrosis factor (ligand) superfamily, member 10 (>35 times control) (Figure 2c). These increases are supported by the documented ability of BoHV-1 to evade adaptive immune cells by inducing apoptosis in CD4+ cells during replication in epithelial respiratory-tract cells [23,24]. Also concerning apoptosis, the Bcl-2-like protein 11 and zinc finger CCCH-type containing 12A were strongly activated by the effects of viral infection (Figure 2c). These respectively fulfill roles as an anti-/ pro-apoptotic regulator involved in a wide variety of cellular activities [25] and as a protein acting in transcriptional activation and cell death in Myasthenia Gravis patients, specifically via the induction of genes associated with apoptosis [26]. Interestingly, neither gene has been previously implicated in apoptotic processes linked to viral infections.
DEAD box protein 58, or the retinoic acid-inducible gene 1 protein, is an innate immune receptor that acts as a cytoplasmic sensor of viral nucleic acids. This is in addition to playing major roles in sensing viral infection and in activating a cascade of antiviral responses, including the induction of type I INFs and proinflammatory cytokines, particularly in Herpesviridae infections [27]. This viral sensor robustly responded in MDBK cells infected by BoHV-1 (Figure 2d). A similar finding was obtained for the IFN-induced guanylate-binding protein 1, which belongs to the GTPase superfamily. Notably, this superfamily has described antiviral effects [28]. In general, the activated immune markers were associated with the detection of viral components, as well as with the activation of apoptotic processes. Considering this, the strong activation (>14 times the control) of the mitogen-activated protein kinase 8 gene is interesting. This gene is involved in the nuclear production of nuclear factor kappa-light-chain-enhancer of activated B (NF-κB) cells, which play a key role in regulating the immune response to infection.
Motifs and domains of bovine ISG15
During a viral infection, several molecules are activated and produced. This includes IFN-stimulated genes (ISG) [29]. Specifically, ISG15 is a type I IFN-inducible gene encoding a protein with pleiotropic functions. ISG15 has been extensively characterized in human and mouse models, under both healthy and disease conditions [30].
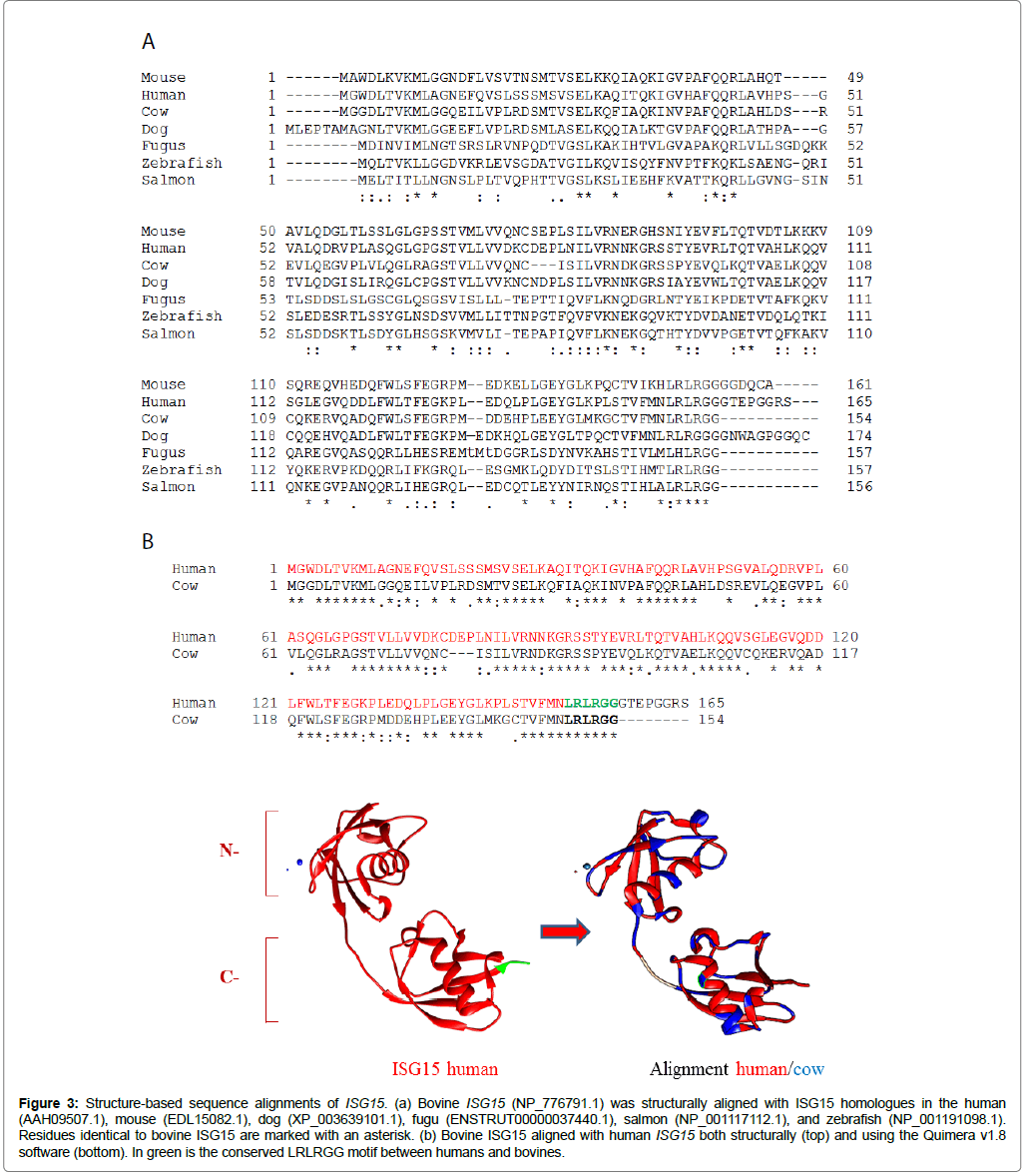
The deduced amino acid sequence of bovine ISG15 (NP_776791.1) was compared with those of vertebrate species, including human, mouse, dog, salmon, fugu, and zebrafish sequences (Figure 3a). A relatively low level of identity was found between bovine ISG15 and respective counterparts of the assessed vertebrates. The highest identity was with the dog (71%), while the lowest identity was with the salmon (32%) (Table 2). Bovine ISG15 (NM_174366.1) contained a 465 base pair (bp) open reading frame translating for a 154 amino acid product with a predicted molecular mass of 17.310 kDa and pI of 6.73. Homology comparisons revealed a structural domain in the ISG15 C-terminal corresponding to the LRLRGG sequence, which is required for conjugation to the lysine residues of target proteins (Figure 3b). Finally, structural alignment, using the human crystal of the protein as a mold, strongly suggested a conserved spatial structural conformation between human and bovine ISG15 (Figure 3b) [31].
Figure 3: Structure-based sequence alignments of ISG15. (a) Bovine ISG15 (NP_776791.1) was structurally aligned with ISG15 homologues in the human (AAH09507.1), mouse (EDL15082.1), dog (XP_003639101.1), fugu (ENSTRUT00000037440.1), salmon (NP_001117112.1), and zebrafish (NP_001191098.1). Residues identical to bovine ISG15 are marked with an asterisk. (b) Bovine ISG15 aligned with human ISG15 both structurally (top) and using the Quimera v1.8 software (bottom). In green is the conserved LRLRGG motif between humans and bovines.
| Isg15 | dog | mouse | Zebrafish | salmon | fugu | human |
|---|---|---|---|---|---|---|
| cow | 71/80 | 62/74 | 37/56 | 32/53 | 34/49 | 65/71 |
| dog | 66/78 | 34/53 | 30/49 | 30/45 | 68/73 | |
| mouse | 39/56 | 34/52 | 35/48 | 65/77 | ||
| Zebrafish | 55/67 | 40/59 | 36/56 | |||
| salmon | 51/66 | 34/55 | ||||
| fugu | 33/50 |
Table 2: Identity/similarity (%) of Isg15 homologues.
Phylogenetic relationships of the ISG15 gene
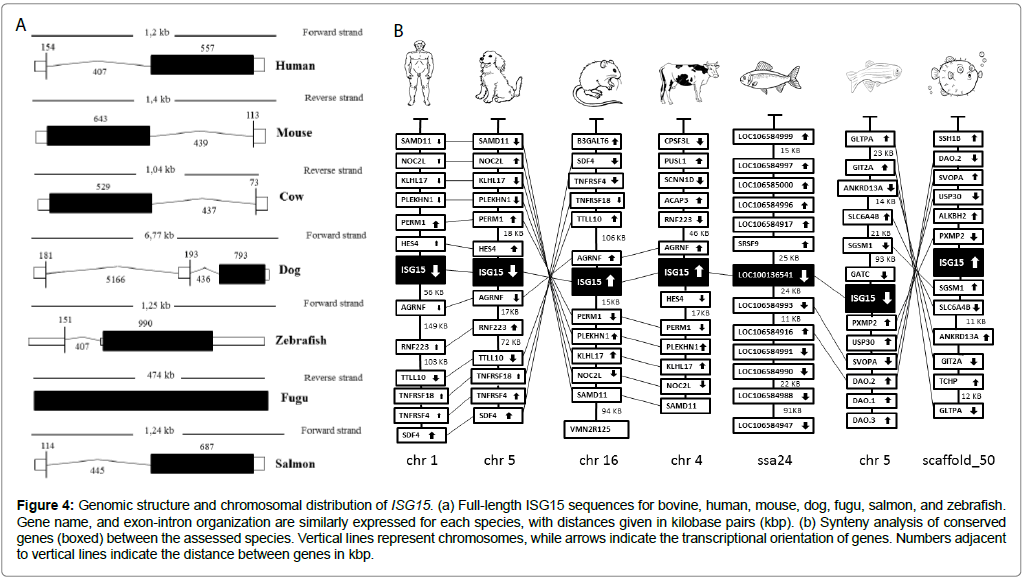
The structure of the ISG15 gene was phylogenetically characterized through comparisons against the dog, human, fugu, mouse, salmon, and zebrafish sequences, resulting in the genomic organization of ISG15 between these vertebrates (Figure 4a). The size of the genomic structure for ISG15 varied from 0.474 to 6.77 kilobase pairs, with the longest observed in the dog and shortest in the fugu (Figure 4a). In all cases, ISG15 had only one exon in the genome.
Figure 4: Genomic structure and chromosomal distribution of ISG15. (a) Full-length ISG15 sequences for bovine, human, mouse, dog, fugu, salmon, and zebrafish. Gene name, and exon-intron organization are similarly expressed for each species, with distances given in kilobase pairs (kbp). (b) Synteny analysis of conserved genes (boxed) between the assessed species. Vertical lines represent chromosomes, while arrows indicate the transcriptional orientation of genes. Numbers adjacent to vertical lines indicate the distance between genes in kbp.
Considering the gained genomic perspectives, the distribution of ISG15 was then evaluated in each species through synteny analysis. The ISG15 genes were located by using the BLAST algorithm against chromosomes (Figure 4b). Transcription orientation was conserved for the mouse and fugu genes, but the human, dog, salmon, and zebrafish genes were transcribed in the opposite direction (Figure 4b). There was a certain degree of conservation in the ISG15 loci of the studied vertebrates (Figure 4b). Moreover, genes adjacent to ISG15 were comparable, with largely conserved arrangements between the cow, human, mouse, and dog (Figure 4b).
In vitro and in vivo expression of ISG15
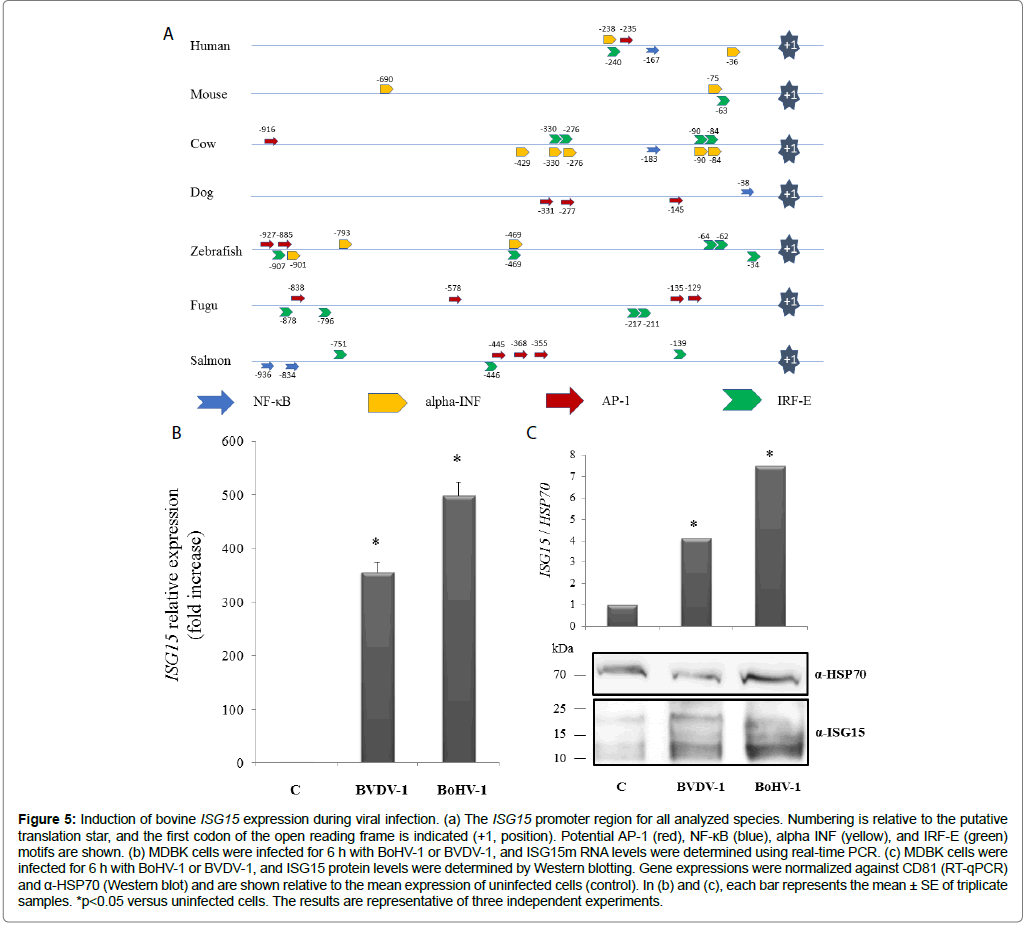
The robust increases observed for various molecular markers as a result of BoHV-1 infection naturally raise new questions requiring further experimental study. One point of interest chosen for further exploration was to more precisely characterize the expression profile of ISG15 in the context of viral infection. As a starting point, in silico analyses were performed for the 5 ‘UTR of the ISG15 gene in bovine, human, dog, mouse, fugu, salmon, and zebrafish models, all of which evidenced a potential to respond to classical immune pathways (e.g. NF- κB, AP-1, IRFs, among others) (Table 3 and Figure 5a). Interestingly, the bovine ISG15 regulatory region had a high density of NF-κB and IRF response elements as compared to the other assessed species. Nevertheless, the participation of these transcriptional regulators remains to be confirmed and will be a subject of future research.
Figure 5: Induction of bovine ISG15 expression during viral infection. (a) The ISG15 promoter region for all analyzed species. Numbering is relative to the putative translation star, and the first codon of the open reading frame is indicated (+1, position). Potential AP-1 (red), NF-κB (blue), alpha INF (yellow), and IRF-E (green) motifs are shown. (b) MDBK cells were infected for 6 h with BoHV-1 or BVDV-1, and ISG15m RNA levels were determined using real-time PCR. (c) MDBK cells were infected for 6 h with BoHV-1 or BVDV-1, and ISG15 protein levels were determined by Western blotting. Gene expressions were normalized against CD81 (RT-qPCR) and α-HSP70 (Western blot) and are shown relative to the mean expression of uninfected cells (control). In (b) and (c), each bar represents the mean ± SE of triplicate samples. *p<0.05 versus uninfected cells. The results are representative of three independent experiments.
| Species | Motif | Length | Position | Score | Occurrence |
|---|---|---|---|---|---|
| Cow | NF-kB-mIg-kappa' | 11 | -183 | 8 | 1 |
| NF-kB-vimentin | 10 | -183 | 10 | 1 | |
| NF-kappaB-TNF-a' | 11 | -183 | 11 | 1 | |
| NFkB-HIV-1.2' | 11 | -183 | 8 | 1 | |
| NFkB-HIgk' | 11 | -183 | 8 | 1 | |
| NFkB-IgkLC' | 11 | -183 | 8 | 1 | |
| NFkB-TNFa-K.1' | 10 | -183 | 10 | 1 | |
| NFkB-vimentin' | 10 | -183 | 10 | 1 | |
| NFkB_CS1' | 10 | -183 | 10 | 1 | |
| NFkB_CS2' | 11 | -183 | 11 | 1 | |
| NFkB_CS4' | 9 | -182 | 9 | 1 | |
| AP-1-thrombomod' | 7 | -916 | 7 | 1 | |
| AP-1_CS | 7 | -916 | 7 | 1 | |
| AP-1-cyclinD2' | 7 | -822 | 7 | 1 | |
| AP-1_CS3' | 7 | -822 | 6 | 1 | |
| AP1-TRE-1/T' | 9 | -568 | 9 | 1 | |
| alpha-INF.2' | 6 | -887 | 6 | 2 | |
| -818 | 6 | 5 | |||
| -732 | 6 | 4 | |||
| -656 | 6 | 3 | |||
| -522 | 6 | 2 | |||
| -496 | 6 | 1 | |||
| alpha-INF.2 | 6 | -429 | 6 | 4 | |
| -330 | 6 | 3 | |||
| -276 | 6 | 2 | |||
| -90 | 6 | 1 | |||
| IRF-2_RS' | 8 | -682 | 6 | 3 | |
| -122 | 6 | 1 | |||
| -524 | 6 | 2 | |||
| IRF-2_RS | 8 | -336 | 6 | 4 | |
| -330 | 6 | 3 | |||
| -90 | 6 | 2 | |||
| -84 | 6 | 1 | |||
| IRF-7_CS' | 12 | -526 | 10 | 1 | |
| IRF-7_CS | 12 | -332 | 10 | 2 | |
| -86 | 10 | 1 | |||
| IRF-3_CS | 12 | -86 | 11 | 1 | |
| Human | NFkB_CS4' | 9 | -574 | 8 | 1 |
| NF-kB-FasL-kB1 | 11 | -167 | 8 | 1 | |
| AP-1-involucrin | 7 | -235 | 7 | 1 | |
| IRF-2_RS' | 8 | -759 | 6 | 1 | |
| -30 | 6 | 1 | |||
| IRF-2_RS | -244 | 6 | 4 | ||
| -238 | 6 | 3 | |||
| -36 | 6 | 2 | |||
| IRF-7_CS | 12 | -240 | 10 | 2 | |
| -32 | 10 | 1 | |||
| IRF-3_CS | 12 | -38 | 11 | 2 | |
| -32 | 11 | 1 | |||
| alpha-INF.2' | 6 | -950 | 6 | 4 | |
| -757 | 6 | 3 | |||
| -731 | 6 | 3 | |||
| -617 | 6 | 1 | |||
| alpha-INF.2 | 6 | -238 | 6 | 2 | |
| Mouse | -36 | 6 | 1 | ||
| AP-1-BTEB | 8 | -755 | 8 | 1 | |
| AP-1-PAI-2 | 7 | -754 | 7 | 1 | |
| AP-1_CS3 | 7 | -754 | 6 | 2 | |
| -630 | 6 | 1 | |||
| AP-1_CS3' | 7 | -754 | 6 | 2 | |
| -630 | 6 | 1 | |||
| AP-1_CS2' | 8 | -631 | 8 | 1 | |
| AP-1_CS4' | 7 | -630 | 7 | 1 | |
| AP-1_CS4 | 7 | -630 | 7 | 1 | |
| AP1-TRE-4/T' | 9 | -631 | 9 | 1 | |
| AP-1-gamma-INF | 7 | -135 | 7 | 1 | |
| alpha-INF.2 | 6 | -690 | 6 | 2 | |
| -75 | 6 | 1 | |||
| alpha-INF.2' | -149 | 6 | 2 | ||
| -97 | 6 | 1 | |||
| IRF-2_RS' | 8 | -263 | 6 | 2 | |
| -127 | 6 | 1 | |||
| IRF-2_RS | 8 | -75 | 6 | 3 | |
| -69 | 6 | 2 | |||
| -63 | 6 | 1 | |||
| Zebrafish | AP-1-cyclinD2 | 7 | -927 | 7 | 1 |
| -885 | 7 | 1 | |||
| AP-1_CS3 | 7 | -927 | 6 | 1 | |
| -885 | 6 | 1 | |||
| IRF-2_RS | 8 | -907 | 6 | 5 | |
| -469 | 6 | 4 | |||
| -68 | 6 | 3 | |||
| -62 | 6 | 2 | |||
| -34 | 6 | 1 | |||
| IRF-2_RS' | 8 | -554 | 6 | 1 | |
| IRF-3_CS | 12 | -64 | 11 | 1 | |
| IRF-7_CS | 12 | -64 | 10 | 1 | |
| alpha-INF.2 | 6 | -901 | 6 | 3 | |
| -793 | 6 | 2 | |||
| -469 | 6 | 1 | |||
| alpha-INF.2' | -552 | 6 | 2 | ||
| -495 | 6 | 1 | |||
| Dog | NF-kB-TNF-alpha | 11 | -38 | 8 | 1 |
| AP-1-IL-6' | 11 | -332 | 8 | 1 | |
| AP-1-IL-6 | 11 | -279 | 11 | 1 | |
| AP-1_CS1 | 8 | -331 | 8 | 1 | |
| AP-1_CS1' | 8 | -277 | 8 | 1 | |
| AP-1-TBXAS1' | 7 | -330 | 7 | 1 | |
| AP-1_CS3 | 7 | -330 | 7 | 2 | |
| -277 | 7 | 1 | |||
| AP-1_CS3' | 7 | -277 | 6 | 1 | |
| AP-1_CS4 | 7 | -330 | 7 | 2 | |
| 7 | 1 | ||||
| AP-1_CS4' | 7 | -277 | 6 | 1 | |
| AP-1-IL-3 | 7 | -277 | 7 | 1 | |
| AP-1-involucrin' | 7 | -145 | 7 | 1 | |
| alpha-INF.2' | 6 | -577 | 6 | 2 | |
| -482 | 6 | 1 | |||
| alpha-INF.2 | 6 | -333 | 6 | 2 | |
| -251 | 6 | 1 | |||
| Salmon | NFkB_CS2 | 11 | -935 | 8 | 1 |
| NFkB_CS1 | 10 | -934 | 10 | 1 | |
| NFkB_CS2' | 11 | -934 | 8 | 1 | |
| NF-kB-TNF-alpha | 11 | -834 | 8 | 1 | |
| AP-1-involucrin | 7 | -445 | 7 | 1 | |
| AP-1-gamma-INF' | 7 | -368 | 7 | 1 | |
| AP-1-TBXAS1 | 7 | -355 | 7 | 1 | |
| AP-1_CS1' | 8 | -355 | 8 | 1 | |
| AP-1_CS3' | 7 | -355 | 6 | 1 | |
| AP-1_CS4' | 7 | -355 | 7 | 1 | |
| AP1-ET-I' | 8 | -355 | 8 | 1 | |
| AP1-SV40.2' | 7 | -355 | 7 | 1 | |
| IRF-2_RS' | 8 | -979 | 6 | 4 | |
| -973 | 6 | 3 | |||
| -700 | 6 | 2 | |||
| -363 | 6 | 1 | |||
| IRF-2_RS | 8 | -751 | 6 | 2 | |
| -139 | 6 | 1 | |||
| IRF-1-DE1' | 7 | -446 | 7 | 1 | |
| Fugu | AP-1-PAI-2' | 7 | -838 | 7 | 1 |
| AP-1_CS3' | 7 | -838 | 6 | 2 | |
| -129 | 6 | 1 | |||
| AP-1_CS3 | 7 | -838 | 6 | 2 | |
| AP-1-IL-3_(2)' | 7 | -578 | 7 | 2 | |
| -135 | 7 | 1 | |||
| AP-1-IHABP' | 7 | -129 | 7 | 1 | |
| AP-1_CS4' | 7 | -129 | 7 | 1 | |
| m4-AP-1_site' | 8 | -129 | 8 | 1 | |
| IRF-2_RS' | 8 | -929 | 6 | 2 | |
| -356 | 6 | 1 | |||
| IRF-2_RS | 8 | -878 | 6 | 5 | |
| -796 | 6 | 4 | |||
| -379 | 6 | 3 | |||
| -217 | 6 | 2 | |||
| -211 | 6 | 1 | |||
| IRF-3_CS | 12 | -213 | 11 | 1 | |
| IRF-7_CS | 12 | -213 | 10 | 1 |
Table 3: List of putative cis elements in the 5’ UTR of the ISG15 gene for the assessed species.
For the current study, focus was given to evaluating ISG15 expression under conditions of controlled in vitro, as well as natural, infection with BoHV-1 and BVDV-1. Specifically, MDBK cells were infected with the BoHV-1 and BVDV-1 viruses for 24 h. At the end of the infection period, ISG15 expression levels were evaluated through Western blotting and RT-qPCR analysis (Figures 5b and 5c). The expression of ISG15 strongly increased as a consequence of infection with either virus, with levels comparable between BoHV-1 and BVDV- 1. Therefore, ISG15 apparently responds powerfully to the presence of either RNA or DNA genomic viruses. This is an interesting result for a bovine viral infection model. However, these results were obtained in vitro and require further corroboration.
Considering the high prevalence rates of BoHV-1 and BVDV-1 in cattle farms, the following question was proposed: Is there any potential relationship between the presence of these viruses and ISG15 expression? To approximate an answer, the number of amplicon copies for BoHV-1, BVDV-1, and ISG15 were obtained via absolute quantification through qPCR assays. Interestingly, notable ISG15 levels/200 μL of whole blood were recorded in the presence of these viral pathogens for 21 animals from 8 different locations. Evidently, the evaluated animals might be exposed to other viruses, pathogens, or nutritional conditions that could trigger an increase of ISG15. However, the performed assessments describe a possible pattern of ISG15 expression in animals infected with BoHV-1 and BVDV-1. Interestingly, animals infected with BVDV-1 had higher ISG15 levels than animals infected with BoHV-1 (Table 4).
| Bovine | BoHV-1 copies | BVDV-1 copies | ISG15 copies |
|---|---|---|---|
| 10.5.02.0029.08 | nct | 3977 | 2535 |
| 10.5.02.0029.11 | nct | 747 | 2130 |
| 10.5.02.0029.12 | nct | 2395 | 1674 |
| 10.5.02.0029.13 | nct | 1469 | 2247 |
| 10.5.02.0499.14 | nct | 974 | 2692 |
| 10.5.02.0082.22 | nct | 1161 | 1742 |
| 10.5.02.0082.23 | nct | 2409 | 1504 |
| 10.5.02.0089.01 | nct | 2268 | 2765 |
| 10.5.02.0089.02 | nct | 1004 | 1351 |
| 10.5.02.0089.03 | nct | 573 | 4819 |
| 10.5.02.0089.04 | nct | 784 | 2087 |
| 10.5.02.0029.05 | nct | 533 | 3225 |
| 14.1.06.0148.02 | 39 | 26 | 1098 |
| 14.1.06.0148.14 | 54 | 33 | 290 |
| 14.1.06.1386.06 | nct | 298 | 669 |
| 14.1.06.1386.10 | 77 | 3633 | 2264 |
| 14.1.06.1386.12 | 30 | 84 | 266 |
| 14.1.06.1386.13 | 125 | 21 | 479 |
| 14.1.06.1386.14 | 70 | 33 | 548 |
| 10.5.09.0318.01 | nct | nct | nct |
| 14.1.06.1462.08 | nct | nct | 108 |
Table 4: Absolute quantification of ISG15, BoHV-1, and BVDV-1 copies from bovine blood samples.
Discussion
Due to the high impact that BoHV outbreaks have on the global bovine industry, many studies have sought to understand the infective process of this virus, as well as the immune responses of the host. To this end, the discovery of virus-regulated genes has been aided by transcriptomic analysis, a powerful tool for identifying candidate marker genes of infection.
In the current study, IFITM3 transcripts were among the most abundant, reaching expression levels >300-fold relative to uninfected cells. In recent years, the crucial role of this protein has been studied mostly in the context of influenza A infection. Mice lacking the IFITM3 gene display fulminant viral pneumonia on infection, even with a low-pathogenicity influenza virus [32]. The majority of the viruses restricted by IFITMs are enveloped RNA viruses; however, the precise mechanism that allows IFITMs to inhibit virus replication remains unclear [33]. Cells infected by DNA viruses, such as herpes simplex virus and cytomegalovirus, can also induce the expression of ISGs, including IFITMs. Nevertheless, the role of IFITMs in the lifecycle of DNA viruses is largely unknown [34]. Recently, IFITM3 knockdown by RNA interference reduced the titer of the human cytomegalovirus ≈ 100-fold by 8 days post-infection [35]. This finding suggests that herpes viruses, which would include BoHV, somehow require high IFITM levels to form new viral progeny. These prior results, together with the data obtained in the presents study, lay the groundwork for an interesting line of research related to the replication mechanism of BoHV.
The thioredoxin system is a key antioxidant system that protects cells from oxidative stress through disulfide reductase activity. Thioredoxins are highly conserved in many organisms, including from bacterial organisms to plants and mammals. This range indicates that the thioredoxin system is cellular and essential for cell survival and functioning. Interestingly, viral infection induced strong transcript expression for the thioredoxin-interacting protein at 6 hpi (Figure 4b). While this phenomenon has been reported for various single-stranded RNA viruses [36,37], the same does not hold true for double-stranded DNA viruses.
The virus inhibitory protein, endoplasmic reticulum-associated, INF-inducible, also known as radical SAM domain-containing 2, is a multifunctional protein in viral processes. Indeed, this protein can be induced in a variety of cell types by different cellular factors, such as type I, II, and III INFs, DNA, and viral RNA [38]. Furthermore, genic expression was recently associated with the persistence of viral infection [39]. These prior reports are consistent with the classic phenotype of this viral family. In the presently assessed model, the radical SAM domain-containing 2 gene was robustly activated by BoHV-1. This activation has not been previously assigned to the immune response against BoHV-1 infection. Nevertheless, whole transcript expression suggests that, from an immunological perspective, the host tries to protect against viral infection by expressing radical SAM domaincontaining 2 in the immediate hours after pathogen infection.
The reactivation of a latent infection may occur under certain natural or induced stimuli, and this phenomenon provides adequate means for virus transmission and expansion [40]. A number of immune markers were found modulated as a consequence of BoHV-1 infection. These included tetherin, also known as bone marrow stromal antigen 2, a lipid raft associated protein that, in humans, is encoded by the gene [41]. Tetherin has also been termed CD317. This gene is only expressed as a response to stimuli from the IFN pathway; howeverm, tetherin is constitutively expressed in mature B cells, plasma cells, and plasmacytoid dendritic cells. In the current assays, BST2 transcripts strongly increased (≈ 37 times the control) at 6 hpi (Figure 4b). The presences of a virus and viral components are detected in humans by recognition molecules such as the retinoic acid-inducible gene 1 protein. Cascades of interactions occur between signaling molecules, and eventually the signal reaches the nucleus, resulting in the upregulated expression of ISGs. In turn, this up-regulation activates the IFN-α pathway to send the signal to neighboring cells, which causes an up-regulation in the expression of other ISGs and many viral restriction factors, such as tetherin [42,43]. No information currently links BoHV-1 infection and increased transcripts of the BST2 mRNA gene. However, the present results support an apparent link between BTS2 gene activation and a sharp increase in ISG15 transcripts (Figure 2b). In fact, viral infection triggered ISG15 expression >500-times that of the control at 6 hpi (Figure 2b). Several studies have found that ISG15 can impact the viral lifecycle at the virus-release stage. For example, ISG15 expression inhibits ubiquitination of the HIV-1 Gag protein and interaction between Gag and Tsg101, both of which are events important in mediating HIV-1 budding and release [44].
Worth noting, the present study is the first to provide a general vision of the ISG15 gene based on the respective sequence (conserved protein motif) and degree of in vitro and in vivo expressions. While there is compelling information related to this protein in human and mouse models, little to no information is available on bovine ISG15. This protein should be of particular interest in the context of bovine health due to evidencing increased expression as a result of BoHV or BVDV-1 infection, two important bovine pathogens connected to significant economic losses in the cattle industry. Although genic/ proteic similarities and differences exist, the bovine homologue of ISG15 was generally consistent in responses to cellular stimuli and should, therefore, preserve the same function described in humans [45].
In the assessed bovine models, ISG15 expression strongly increased as a consequence of BoHV infection. However, an RNA virus model (i.e. BVDV-1) also potentiated the synthesis of ISG15 in vitro (Figures 5b and 5c). A similar result was obtained by checking the presence of this immune marker in different bovines infected with both viruses. Interestingly, ISG15 tended to increase more in BVDV-infected animals. This phenomenon could be explained by the viral load detected for BoHV, which, in all cases, was lower than that detected for BVDV (Table 4).
The current study is the first to identify and confirm, at a transcriptomic level, the potential participation of cellular proteins in diverse processes activated during the course of BoHV-1 infection. Assessments were conducted using a molecularly characterized and infectious particle of BoHV-1. Notwithstanding, additional experimental strategies should be used to confirm the participation of these identified proteins. Such information would provide valuable insight towards comprehending the infection mechanism of BoHV-1in bovines.
In conclusion, specific transcriptomic analyses of MDBK cells infected with BoHV-1 produced high-quality marker genes of infection. Infected cells showed notable modulation of numerous genes from diverse functional classes, including of apoptosis, immunity, and cell signaling. These findings establish the foundation for interesting research lines aiming to expand on understandings of the infection mechanism employed by this pathogen.
Acknowledgments
This work was supported by the Direccion de Investigacion, Universidad Austral de Chile and the FONDEF I+T IT15i10016 Grant.
References
- Gerdts V, Beyer J, Lomniczi B, Mettenleiter TC (2000) Pseudorabies virus expressing bovine herpesvirus 1 glycoprotein B exhibits altered neurotropism and increased neurovirulence. J Virol 74: 817-827.
- Liang X, Pyne C, Li Y, Babiuk LA, Kowalski J (1995) Delineation of the essential function of bovine herpesvirus 1 gD: an indication for the modulatory role of gD in virus entry. Virology 207: 429-441.
- Ligas MW, Johnson DC (1988) A herpes simplex virus mutant in which glycoprotein D sequences are replaced by beta-galactosidase sequences binds to but is unable to penetrate into cells. J Virol 62: 1486-1494.
- Meyer G, Hanon E, Georlette D, Pastoret PP, Thiry E (1998) Bovine herpesvirus type 1 glycoprotein H is essential for penetration and propagation in cell culture. J Gen Virol 79: 1983-1987.
- Carpenter DE, Misra V (1991) The most abundant protein in bovine herpes 1 virions is a homologue of herpes simplex virus type 1 UL47. J Gen Virol 72: 3077-3084.
- Beer M, König P, Schielke G, Trapp S (2002) Diagnostic markers in the prevention of bovine herpesvirus type 1: possibilities and limitations. Berl Munch Tierarztl Wochenschr 116: 183-191.
- Leite F, Kuckleburg C, Atapattu D, Schultz R, Czuprynski CJ (2004) BHV-1 infection and inflammatory cytokines amplify the interaction of Mannheimia haemolytica leukotoxin with bovine peripheral blood mononuclear cells in vitro. Vet Immunol Immunopathol 99: 193-202.
- Rajput MKS, Darweesh MF, Park K, Braun LJ, Mwangi W, et al. (2014) The effect of bovine viral diarrhea virus (BVDV) strains on bovine monocyte-derived dendritic cells (Mo-DC) phenotype and capacity to produce BVDV. J Virol 11: 44.
- Scagnolari C, Monteleone K, Selvaggi C, Pierangeli A, D’Ettorre G, et al. (2016) ISG15 expression correlates with HIV-1 viral load and with factors regulating T cell response. Immunobiol 221: 282-290.
- Villalba M, Fredericksen F, Otth C, Olavarría V (2016) Transcriptomic analysis of responses to cytopathic bovine viral diarrhea virus-1 (BVDV-1) infection in MDBK cells. Mol Immunol 71: 92-202.
- Fredericksen F, Delgado F, Cabrera C, Yáñez A, Gonzalo C, et al. (2015b) The effects of reference genes in qRT-PCR assays for determining the immune response of bovine cells (MDBK) infected with the Bovine Viral Diarrhea Virus 1 (BVDV-1). Gene 569: 95-103.
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold spring harbor laboratory press.
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673-4680.
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol Biol Evol 24: 1596-1599.
- Fredericksen F, Villalba M, Olavarría VH (2016) Characterization of bovine A20 gene: Expression mediated by NF-κB pathway in MDBK cells infected with bovine viral diarrhea virus-1. Gene 581: 117-129.
- Fredericksen F, Carrasco G, Villalba M, Olavarría VH (2015a) Cytopathic BVDV-1 strain induces immune marker production in bovine cells through the NF-κB signaling pathway. Mol Immunol 68: 213-222.
- Meyer G, Lemaire M, Ros C, Belak K, Gabriel A, et al. (2001) Comparative pathogenesis of acute and latent infections of calves with bovine herpesvirus types 1 and 5. Arch Virol 146: 633-652.
- Perez SE, Bretschneider G, Leunda MR, Osorio FA, Flores EF, et al. (2002) Primary infection, latency, and reactivation of bovine herpesvirus type 5 in the bovine nervous system. Vet Pathol 39: 437-444.
- Abril C, Engels M, Liman A, Hilbe M, Albini S, et al. (2004) Both viral and host factors contribute to neurovirulence of bovine herpesviruses 1 and 5 in interferon receptor-deficient mice. J Virol 78: 3644-3653.
- Lenschow DJ, Lai C, Frias-Staheli N, Giannakopoulos NV, Lutz A, et al. (2007) IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc Natl Acad Sci 104: 1371-1376.
- Pincetic A, Kuang Z, Seo EJ, Leis J (2010) The interferon-induced gene ISG15 blocks retrovirus release from cells late in the budding process. J Virol 84: 4725-4736.
- Fu Y, Comella N, Tognazzi K, Brown LF, Dvorak HF, et al. (1999) Cloning of DLM-1, a novel gene that is up-regulated in activated macrophages, using RNA differential display. Gene 240: 157-163.
- Pham TH, Kwon KM, Kim YE, Kim KK, Ahn JH (2013) DNA sensing-independent inhibition of herpes simplex virus 1 replication by DAI/ZBP1. J Virol 87: 3076-3086.
- Muylkens B, Thiry J, Kirten P, Schynts F, Thiry E (2007) Bovine herpesvirus 1 infection and infectious bovine rhinotracheitis. Vet Res 38: 181-209.
- Nandi S, Kumar M, Manohar M, Chauhan RS (2009) Bovine herpes virus infections in cattle. Anim Heal Res Rev 10: 85-98.
- Hsu SY, Lin P, Hsueh AJW (1998) BOD (Bcl-2-related ovarian death gene) is an ovarian BH3 domain-containing proapoptotic Bcl-2 protein capable of dimerization with diverse antiapoptotic Bcl-2 members. Mol Endocrinol 12: 1432-1440.
- Park KH, Jung J, Lee JH, Hong YH (2016) Blood transcriptome profiling in myasthenia gravis patients to assess disease activity: a pilot RNA-seq study. Exp Neurobiol 25: 40-47.
- da Silva LF, Jones C (2013) Small non-coding RNAs encoded within the herpes simplex virus type 1 latency associated transcript (LAT) cooperate with the retinoic acid inducible gene I (RIG-I) to induce beta-interferon promoter activity and promote cell survival. Virus Res 175: 101-109.
- Niu P, Shabir N, Khatun A, Seo BJ, Gu S, et al. (2016) Effect of polymorphisms in the GBP1, Mx1 and CD163 genes on host responses to PRRSV infection in pigs. Vet Microbiol 182: 187-195.
- Schneider WM, Chevillotte MD, Rice CM (2014) Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32: 513-545.
- Hermann M, Bogunovic D (2017) ISG15: In Sickness and in Health. Trends Immunol 38: 79-93.
- Malakhov MP, Malakhova OA, Kim KIl, Ritchie KJ, Zhang DE (2002) UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J Biol Chem 277: 9976-9981.
- Everitt AR, Clare S, Pertel T, John SP, Wash RS, et al. (2012) IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484: 519-523.
- Perreira JM, Chin CR, Feeley EM, Brass AL (2013) IFITMs restrict the replication of multiple pathogenic viruses. J Mol Biol 425: 4937-4955.
- Nicholl MJ, Robinson LH, Preston CM (2000) Activation of cellular interferon-responsive genes after infection of human cells with herpes simplex virus type 1. J Gen Virol 81: 2215-2218.
- Xie M, Xuan B, Shan J, Pan D, Sun Y, et al. (2015) Human cytomegalovirus exploits interferon-induced transmembrane proteins to facilitate morphogenesis of the virion assembly compartment. J Virol 89: 3049-3061.
- Li S, Wang J, He WR, Feng S, Li Y, et al. (2015) Thioredoxin 2 is a novel E2-interacting protein that inhibits the replication of classical swine fever virus. J Virol 89: 8510-8524.
- Taylor DW, Zhu Y, Staals RHJ, Kornfeld JE, Shinkai A, et al. (2015) Structures of the CRISPR-Cmr complex reveal mode of RNA target positioning. Science 348: 581-585.
- Seo JY, Yaneva R, Cresswell P (2011) Viperin: a multifunctional, interferon-inducible protein that regulates virus replication. Cell Host Microbe 10: 534-539.
- Moal V, Textoris J, Ben AA, Mehraj V, Berland Y, et al. (2012) Chronic hepatitis E virus infection is specifically associated with an interferon-related transcriptional program. J Infect Dis 207: 125-132.
- Zajac MPDM, Ladelfa MF, Kotsias F, Muylkens B, Thiry J, et al. (2010) Biology of bovine herpesvirus 5. Vet J 184: 138-145.
- Rollason R, Korolchuk V, Hamilton C, Schu P, Banting G (2007) Clathrin-mediated endocytosis of a lipid-raft-associated protein is mediated through a dual tyrosine motif. J Cell Sci 120: 3850-3858.
- Douglas JL, Gustin JK, Viswanathan K, Mansouri M, Moses AV, et al. (2010) The great escape: viral strategies to counter BST-2/tetherin. PLoS Pathog 6: e1000913.
- Kühl A, Pöhlmann S (2012) How Ebola virus counters the interferon system. Zoonoses Public Health 59: 116-131.
- Pincetic A, Leis J (2009) The mechanism of budding of retroviruses from cell membranes. Adv Virol.
Citation: Stepke C, Fredericksen F, Arriagada V, Oltra J, Toledo C, et al. (2018) Immune Gene Expression during Bovine Herpesvirus-1 Infection of MDBK Cells: Molecular Characterization of Interferon-Stimulated Gene 15. J Mol Immunol 3: 117.
Copyright: © 2017 Stepke C, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited
Share This Article
Open Access Journals
Article Usage
- Total views: 3809
- [From(publication date): 0-2018 - Apr 04, 2025]
- Breakdown by view type
- HTML page views: 2984
- PDF downloads: 825