Research Article Open Access
Identification of X. laevis Vitellogenin Peptide Biomarkers for Quantification by Liquid Chromatography Tandem Mass Spectrometry
Leah G Luna* and Katherine CoadyToxicology & Environmental Research & Consulting, The Dow Chemical Company, Midland, USA
- *Corresponding Author:
- Leah G.
Luna Toxicology, Environmental Research and Consulting
The Dow Chemical Company
Building 1803 Washington Street
Midland, MI 48674, USA
Tel: 989-636-2428
Fax: 989-638-9305
E-mail: Luna@dow.com
Received date: June 12, 2014; Accepted date: June 28, 2014; Published date: June 30, 2014
Citation: Luna LG, Coady K (2014) Identification of X. laevis Vitellogenin Peptide Biomarkers for Quantification by Liquid Chromatography Tandem Mass Spectrometry. J Anal Bioanal Tech 5:194 doi: 10.4172/2155-9872.1000194
Copyright: © 2014 Luna LG, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Abstract
Mass spectrometry (MS) offers an exciting possibility as a highly specific method for measuring vitellogenin (VTG) concentrations in toxicity tests that are aimed at evaluating perturbations in the endocrine system. Typically MS approaches to protein quantification attempt to measure intact proteins or rely on enzymatic digestion procedures to reduce the size of the protein into smaller peptide fragments which can be utilized as biomarkers for quantification. With the latter approach, the MS technique allows both the direct measurement of the VTG intact peptide biomarker mass-to-charge ratio and the generation of peptide fragmentation data for further confirmation in complex matrices. In this study we utilize trypsin digestion and a C18 Zip Tip clean-up method to generate predictably cleaved peptides to identify VTG biomarkers from small quantities of plasma collected from the African clawed frog (Xenopus laevis). From the 201,545 Da X. laevis VTG precursor protein, results indicate 2 peptides which would be excellent biomarker candidates for the quantitative measurements of VTG. This methodology may be particularly useful in the context of the Larval Amphibian Growth and Development Assay, a Tier 2 test which is currently under development as a part of the United States Environmental Protection Agency’s Endocrine Disruptor Screening Program.
Keywords
Biomarkers; Peptide; Vitellogenin;X. laevis; Mass spectrometry; Endocrine disruptors
Introduction
Since the 1980’s there has been a dramatic increase in scientific regulatory and public awareness of the presence of endocrine disrupting chemicals in the environment. In the scientific regulatory arena, the EPA’s Endocrine Disruptor Screening Program (EDSP) uses a tiered approach for determining whether a substance may have an effect that is similar to those produced by naturally occurring estrogen, androgen, or thyroid hormones. The Tier 1 screening includes a battery of eleven assays that, when interpreted in a weight of evidence approach, can assess the potential for interaction with the estrogen, androgen, or thyroid hormone systems. Chemicals that go through Tier 1 screening and are found to have the potential to interact with the estrogen, androgen, or thyroid hormone systems will potentially proceed to Tier 2 testing of the EDSP. Tier 2 testing is intended to confirm and characterize the effects observed in Tier 1 tests as well as to establish a quantitative relationship between the dose and the observed effect(s). Currently, the US EPA is still in the process of developing and validating the EDSP Tier 2 tests. One biomarker of interest in the Tier 2 Larval Amphibian Growth and Development Assay (LAGDA) is expression of vitellogenin (VTG). VTG is a yolk precursor glycolipophosphoprotein that is secreted in the liver of oviparous (egg laying) vertebrates. It is generally agreed that a change in VTG levels is a good indicator for estrogenic and potentially anti-estrogenic effects, and it is proposed as one of several endpoints in the LAGDA. Due to the small size of the developing frogs that are assessed in the LAGDA, only limited amounts of blood can be obtained from individuals (10-50 μl). As a result, sensitive and selective analytical methods are necessary.
A number of analytical methods have been developed for the quantification of VTG in plasma, liver and tissues [1-6]. These various methods differ in safety, sensitivity, specificity, and technical difficulty. Nevertheless, the most accepted approaches to measure VTG are from of an enzyme linked immunosorbent assay (ELISA) [4] or the utilization of a radioimmunoassay (RIA) [6]. However for amphibian screening the ELISA and RIA assays suffer from the lack of commercial antibodies for test species like Xenopus laevis.
In contrast to utilizing antibodies for quantitative measurements, mass spectrometry (MS) presents an exciting possibility for becoming a reference method for quantifying VTG in amphibian assays. Typically MS approaches to protein quantification, like VTG, attempt to measure the intact protein directly or rely on enzymatic digestion procedures to reduce the size of the protein into smaller fragments called peptides [7,8]. With the enzymatic digestion approach, the MS technique allows both the direct measurement of the VTG intact peptide biomarker mass-to-charge ratio and the generation of peptide fragmentation data for further protein confirmation in complex matrices. Quantitative measurements utilizing a signature peptide approach with liquid chromatography tandem mass spectrometry (LC/MS/MS) has been developed for the fathead minnow VTG protein [9].
In order to implement quantitative measurements for amphibians utilizing VTG peptide biomarkers by LC/MS/MS, identification of X. laevis VTG peptide biomarkers is required. Therefore, in this study we screened for potentialX. laevis VTG peptide biomarker candidates by employing a targeted in silico enzymatic tryptic digestion approach and utilized synthetic standards for each candidate VTG peptide biomarker. Multiple Reaction Monitoring (MRM) transitions for candidate VTG peptide biomarkers, which are employed for quantitative measurements by LC/MS/MS, were developed and optimized by utilization of synthetic standards to maximize sensitivity. Synthetic standards were also used to verify retention times. Mass spectral confirmation transitions were verified utilizing fragmentation data from enhanced product ion (EPI) trap scans. We validated the identified VTG peptide biomarkers by tryptic digestion of 10 μL of adult femaleX. laevis plasma followed by enzymatic digest and clean up with C18 Zip-Tips for subsequent LC/MS/MS analysis.
Materials and Methods
in silico targeted VTG biomarker screening
An in silico trypsin digestion was performed on the 201,545 Da X. laevis vitellogenin precursor protein P18709 (VITA2_XENLA) utilizing the non-redundant protein sequence database UniProtKB/ Swiss-Prot peptide mass cutter to produce predictably cleaved tryptic peptides. Protein Prospector MS Pattern search [10], a proteomics tool for mining sequence databases in conjunction with Mass Spectrometry experiments, was utilized to search the non-redundant protein NCBI database to ensure selectivity of the identified VTG peptides for X. laevis.
Targeted Mass Spectrometry screening of candidate VTG peptide biomarkers
Synthesis of VTG target peptides: Unlabeled X. laevis VTG peptides identified from the in silico trypsin digestion were synthesized by Celtek Peptides (Franklin, TN), purity for all peptides were >97% via HPLC method. One mg/mL stock solutions of each candidate VTG peptides were prepared in ultrapure water.
Test species: Sexually mature X. laevis females were removed from their husbandry tanks and anesthetized in a solution of MS- 222 buffered to a pH of approximately 7 with sodium bicarbonate. Following anesthesia, blood was collected via cardiac puncture using heparin-coated 26 ½ gauge needles attached to a syringe. After a sufficient amount of blood was collected (approximately 1 mL), frogs were euthanized (while still anesthetized) by severing the spinal cord. All housing and treatment of the frogs was performed under the oversight of the Institutional Animal Care and Use Committee at an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) certified laboratory. Blood was immediately placed on ice and then centrifuged (735×g for 10 minutes) to remove red blood cells from the plasma. Plasma was stored at -80°C prior to its use for biomarker identification experiments by mass spectrometry.
X. laevis plasma trypsin digestion: Adult femaleX. laevis plasma was removed from the -80°C freezer, brought to room temperature and subsequently vortex-mixed. A 10 μL aliquot of the adult femaleX. laevis plasma was added to a clean microfuge tube. A 10 μL aliquot of 0.1% Rapigest (Waters Corporation, Bedford, MA) in 50 mM ammonium bicarbonate was added to the microcentrifuge tube containing the plasma and vortex-mixed. The microcentrifuge tube containing the X. laevis plasma and Rapigest was incubated at 100°C on a heated block for 5 minutes to solubilize proteins and to enhance susceptibility to enzymatic cleavage without inhibiting enzyme activity. Samples were removed from the heat block, briefly microfuged and allowed to cool to room temperature. A 20 μL aliquot of sequencing grade modified trypsin (Promega Corporation, Madison, WI, ~1:40 protease: protein ratio) was added to the microcentrifuge tube, vortex-mixed and incubated overnight at 37°C.
C18 Zip-Tip cleanup of trypsin digestion plasma: TheC18 Zip- Tip bed was activated with two 10 μL aliquots of acetonitrile and subsequently equilibrated with two 10 μL aliquots of ultrapure water with 0.1% trifluoracetic acid (TFA). TheX. laevis trypsin digested plasma sample was removed from the incubator, allowed to come to room temperature, and then applied to the Zip-Tip by drawing up and releasing the digested sample ten times through the Zip-Tip bed. The Zip-Tip bed was subsequently washed with five 10 μL aliquots of ultrapure water with 0.1% TFA. The peptides from the digested sample were eluted from the Zip-Tip bed with three 10 μL aliquots of 50:50 ultrapure water: methanol with 0.1% formic acid. This was performed three times on the same trypsin digested sample and subsequently analyzed by liquid chromatography mass spectrometry for presence of the targeted VTG peptide biomarkers.
VTG unlabeled standards: Unlabeled synthetic standard peptide stocks of GEGIEEFLR and QVLQVTPFAER (Celtek Biosciences, Franklin, TN) were prepared in ultrapure water to provide a concentration of 1 mg/mL. The 1 mg/mL solutions were further diluted 1:100, 1:1000, 1:10,000 and 1:100,000 in ultrapure water to provide working stocks of 10 μg/mL, 1 μg/mL, 0.1 μg/mL and 0.01 μg/mL, respectively, for liquid chromatography mass spectrometry experiments. All peptide stocks solutions and dilutions thereof were stored at -80°C until needed.
VTG labeled internal standards: 13C and 15N labeled synthetic standard peptide stocks of GEGIEEFLR(+10) and QVLQVTPFAER(+10) (New England Peptide, Garnder, MA, purity for ISTDs were >95% via HPLC method) were prepared in ultrapure water to provide a concentration of 1 mg/mL. The 1 mg/mL solutions were further diluted 1:100 and 1:1000 in ultrapure water to provide working stocks of 10 μg/mL and 1 μg/mL respectively. The stocks solutions were stored at -80°C until needed.
Analysis: High Performance Liquid Chromatography and AB/ Sciex 4000 QTRAP Mass Spectrometry:
The analytical column utilized was an AdvanceBio Peptide Map, 2.1×150 mm, 2.7 μm particle size (part number 653750-902, Agilent, Santa Clara, CA). The aqueous mobile phase (A) consisted of Ultrapure (18 MΩ) water with 0.1% formic acid, while the organic phase (B) was ACN with 0.1% formic acid. The autosampler was programmed to deliver a 20 μL injection. A gradient profile was utilized at a flow rate of 350 μL/min. Initially, the mobile phase consisted of 88% A and 12% B and held for 2 minutes. A 3.25% change per minute was utilized over the next 8 minutes where the mobile phases had a final composition of 62% A and 38% B, respectively. At 10 minutes, the gradient was stepped to 98% B for 5 minutes to clean the column and then stepped to 88% A and 12% B for the next 10 minutes to re-equilibrate the column to initial conditions. The total run time was 25 minutes per sample. The column eluent was introduced into a 4000 QTRAP (AB/Sciex, Foster City, CA) mass spectrometer with a turbo ion spray interface.
The MRM initiated detection, and sequencing workflow was utilized to create in silico multiple reaction monitoring transitions which optimized collision energies for trypsin cleaved peptides from theX. laevis VTG protein. Peptide transitions were generated by selecting peptide precursor ions (Q1) with masses greater than 400 Da in the +2 charge state. The y fragmentation ions (Q3) were selected based on y ions that were greater than the peptide precursor mass-to-charge ratio. Information dependent acquisition (IDA) was set to trigger EPI trap scans for MS/MS data when the MRM scan reached 500 counts per second (cps) for surveying precursor peptides for fragmentation. All trap scans were performed at 4000 Da/sec. The 4000 QTRAP instrument (AB/Sciex, Foster City, CA) was operated in positive ion mode. Instrument parameters were as follows: curtain gas=10, collision gas=high, ion spray voltage=5500 V, source temperature=500°C, source gas 1=20 psi, source gas 2=20 psi, cell entrance potential=10 V, collision cell exit potential=10 V, declustering potential=70 V and dwell time=50 ms per MRM transition. Q0 trapping was utilized with a fixed fill time of 20 ms for EPI scans
Results and Discussion
Targeted in silicoX. laevis VTG biomarker screening
Selection of the targetX. laevis VTG peptide biomarkers were based on several factors including host specificity of the peptide and a sequence of 7-12 amino acids following digestion. Other factors included the absence of: 1) Methionine, cysteine or tryptophan amino acids in the sequences which can be oxidized either in vivo or during sample processing; 2) Three or more repeating residues; and 3) A peptide that contained acid labile aspartic acid (D) and proline (P) pairs. Furthermore, theoretical percent cleavage probabilities of less than 100% with the digestion enzyme trypsin were discarded as biomarker candidates in order to maintain target biomarker stability and detection [11]. Utilizing these criteria, resultingX. laevis VTG peptide candidates from the in silico trypsin digest are listed in Table 1.
| Peptide # | Position of Cleavage Site | Peptide Sequence | # Amino Acids in Peptide | Peptide MW | Percent Cleavage Probability |
|---|---|---|---|---|---|
| 1 | 66 | VEISAYAQR | 9 | 1036.2 | 100 |
| 2 | 115 | FEYSNGR | 7 | 871.9 | 100 |
| 3 | 223 | VIQGAATYTYK | 11 | 1214.4 | 100 |
| 4 | 250 | QVLQVTPFAER | 11 | 1287.5 | 100 |
| 5 | 280 | SGQLTPPQIQLK | 12 | 1309.5 | 100 |
| 6 | 694 | GEGIEEFLR | 9 | 1049.2 | 100 |
| 7 | 883 | VHAHLPAK | 8 | 872.1 | 100 |
| 8 | 1100 | DTNETALYR | 9 | 1082.1 | 100 |
Table 1: Potential peptide biomarker candidates from the in silico trypsin digestion of the X. laevis Vitellogenin protein.
Analysis of candidateX. laevis VTG peptide biomarkers
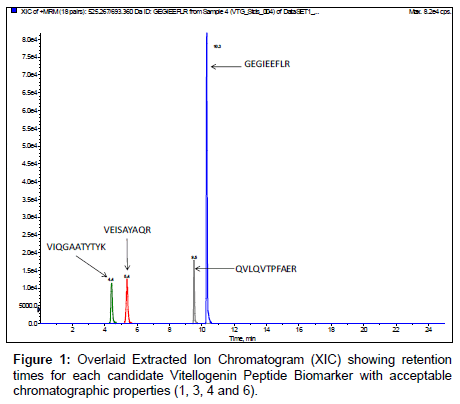
Candidate unlabeledX. laevis VTG peptide biomarkers identified from the in silico trypsin digestion were synthesized to determine retention times and verify amino acid sequences by MRM initiated IDA EPI trap scans for MS/MS fragmentation data. MRM transitions of the monitoredX. laevis VTG peptide biomarkers are listed in Table 2 along with their optimized collision energies. Results demonstrate that of the eight VTG peptide biomarker candidates initially monitored, four of the peptides had very hydrophilic retention times (peptides 2, 5, 7 and 8 in Table 1) eliminating them as good candidates for biomonitoring. In addition, utilization of hydrophilic peptides could be easily lost due to washing steps required during plasma sample clean-up with an aqueous media containing 0.1 % TFA which is a necessary step with the use of C18 ZipTips. Figure 1 shows overlaid chromatograms for each of the 4 remaining VTG peptide biomarkers candidates. The chromatography for the 4 remaining VTG peptide biomarkers candidates (1, 3, 4 and 6 in Table 1) showed resolution >1.5 with retention times of 4.4 ± 0.1, 5.4 ± 0.1, 9.5 ± 0.2 and 10.3 ± 0.2 minutes respectively. Peak symmetry demonstrated diminutive tailing. The candidate VTG peptide biomarker peak widths averaged approximately 9 seconds.
| Peptide | Q1 Precursor Ion | Precusor m/z Ratio | Q3 Product Ion | Optimized Collision Energy (V) |
|---|---|---|---|---|
| VEISAYAQR | 518.8 | +2 | 808.4 | 27.8 |
| VEISAYAQR | 518.8 | +2 | 695.3 | 27.8 |
| VEISAYAQR | 518.8 | +2 | 537.3 | 27.8 |
| FEYSNGR | 436.7 | +2 | 596.4 | 24.2 |
| FEYSNGR | 436.7 | +2 | 725.3 | 24.2 |
| VIQGAATYTYK | 607.8 | +2 | 1002.5 | 31.7 |
| VIQGAATYTYK | 607.8 | +2 | 874.4 | 31.7 |
| VIQGAATYTYK | 607.8 | +2 | 675.3 | 31.7 |
| SGQLTPPQIQLK | 655.4 | +2 | 1037.6 | 33.8 |
| SGQLTPPQIQLK | 655.4 | +2 | 924.6 | 33.8 |
| SGQLTPPQIQLK | 655.4 | +2 | 823.5 | 33.8 |
| GEGIEEFLR | 525.3 | +2 | 863.5 | 28.1 |
| GEGIEEFLR | 525.3 | +2 | 806.1 | 28.1 |
| GEGIEEFLR | 525.3 | +2 | 693.4 | 28.1 |
| VHAHLPAK | 436.8 | +2 | 872.5 | 24.2 |
| VHAHLPAK | 436.8 | +2 | 773.4 | 24.2 |
| VHAHLPAK | 436.8 | +2 | 636.4 | 24.2 |
| DTNETALYR | 541.8 | +2 | 1082.5 | 28.8 |
| DTNETALYR | 541.8 | +2 | 967.5 | 28.8 |
| DTNETALYR | 541.8 | +2 | 866.4 | 28.8 |
| QVLQVTPFAER | 644.4 | +2 | 1060.6 | 33.4 |
| QVLQVTPFAER | 644.4 | +2 | 947.5 | 33.4 |
| QVLQVTPFAER | 644.4 | +2 | 720.4 | 33.4 |
Table 2: Multiple Reaction Monitoring Transitions for Targeted Candidate Vitellogenin Peptide Biomarkers.
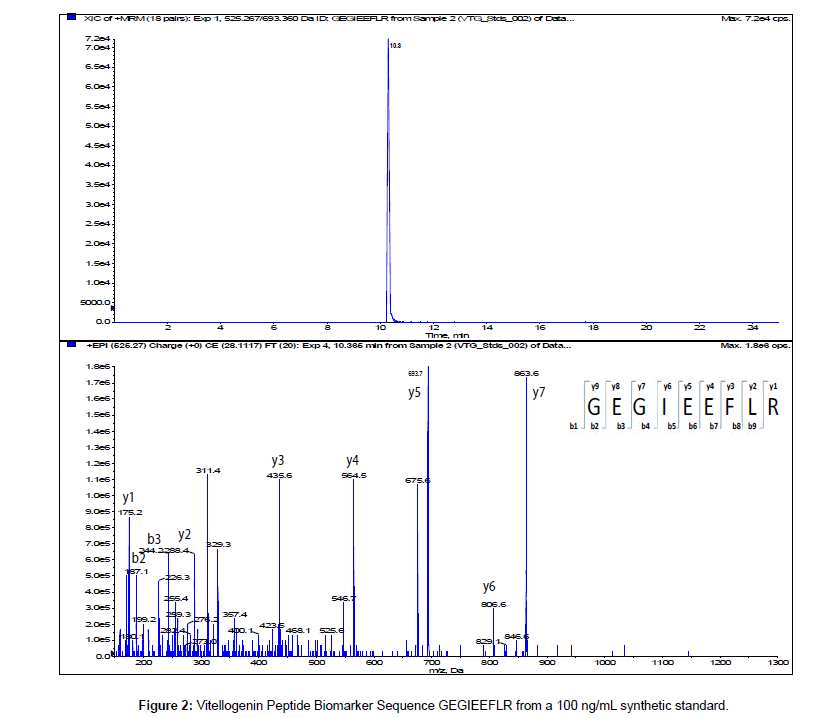
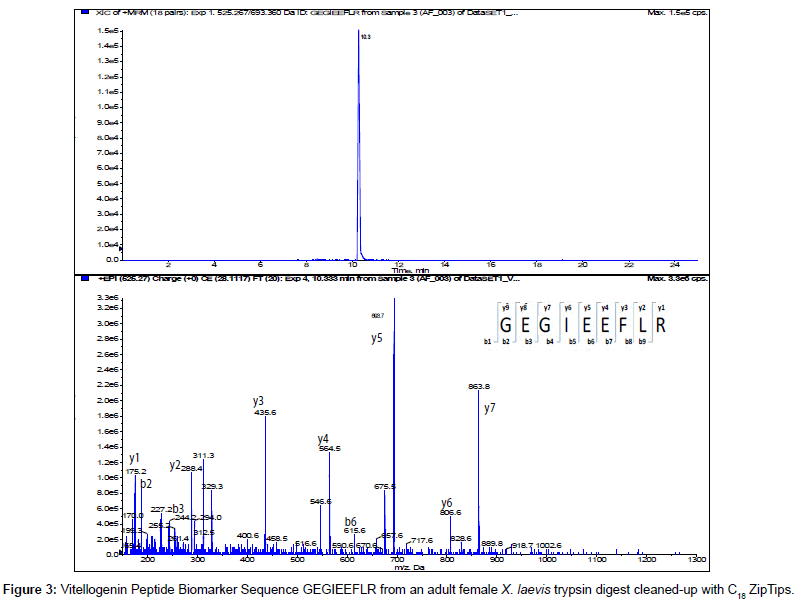
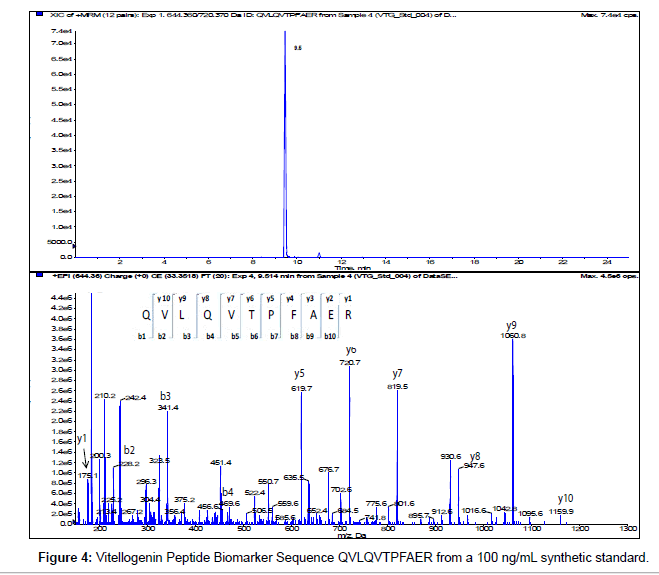
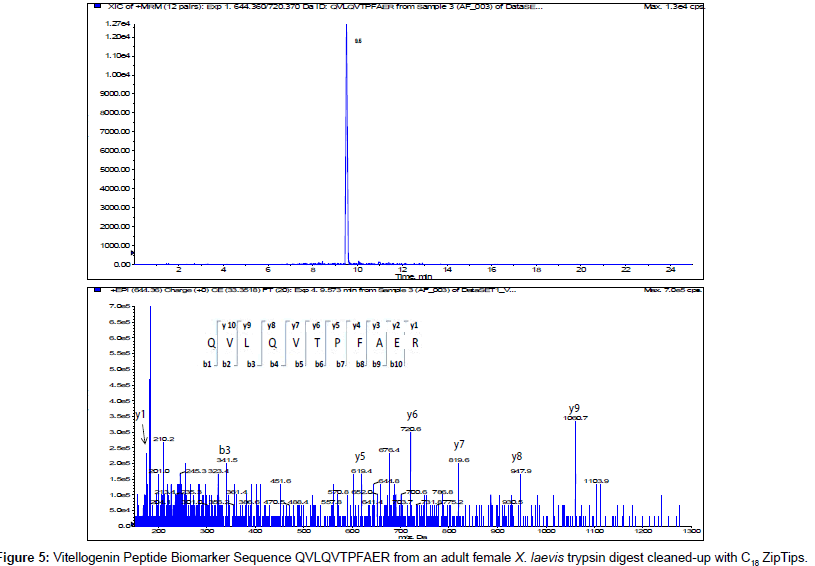
The MS/MS Fragment Ion Calculator (Institute of Systems Biology) was utilized to generate theoretical b and y ions (peptide fragment ions that extend from the N-terminus and C-terminus respectively) from the intact precursor ion for each candidate VTG peptide biomarker. The amino acid sequence tag was determined from fragmentation ions generated from the EPI trap scans. The difference between specific fragmentation y and b ions represents the masses of amino acid residues. Prior to LC/MS/MS analysis,X. laevis plasma digests were cleaned up to remove components such as unwanted matrix from the plasma, sample buffers, salts and other additives added during the sample preparation of the trypsin digest. A C18 ZipTip, 10 μl pipette tip with a standard 0.6 μL bed of C18 silica based medium fixed at its tip, was utilized to desalt, enrich and purify theX. laevis plasma trypsin digests. Typically purification of tryptic peptides with the C18 ZipTip results in high recovery, however the capacity of the C18 ZipTip is limited as only up to 5 μg digested protein can be loaded per ZipTip without losses. Therefore five C18 ZipTips were utilized on the sameX. laevis plasma trypsin digest and eluates combined to confirm the VTG peptide biomarkers. A representative extracted ion chromatogram (top) and an EPI trap scan is presented in Figure 2 for the VTG peptide biomarker synthetic standard GEGIEEFLR (bottom), while Figure 3 shows a representative extracted ion chromatogram (top) of the VTG peptide biomarker candidate GEGIEEFLR from an adult femaleX. laevis trypsin digest cleaned-up with C18 ZipTips (bottom). From the fragmentation ions generated we can validate that the peptide monitored is GEGIEEFLR from the adultX. laevis tryptic peptide digest. We also validate the peptide biomarker QVLQVTPFAER (Figures 4 and 5). The VTG candidate biomarkers VIQGAATYTYK and VGISAYQR were detectable in the adult femaleX. laevis trypsin digest, but qualitatively at much lower levels than GEGIEEFLR or QVLQVTPFAER peptide biomarkers. Thus Table 3 lists the precursor ion as well as the three most intense fragmentation (product) ions results for the two most intense VTG peptide biomarkers.
| Peptide | Q1 Precursor Ion | Precusor m/z Ratio | Q3 Product Ion | Optimized Collision Energy (V) | Retention Time (min) |
|---|---|---|---|---|---|
| QVLQVTPFAER | 644.4 | +2 | 1060.4 (y9)* | 33.4 | 9.5 |
| QVLQVTPFAER | 644.4 | +2 | 819.5 (y7) | 33.4 | 9.5 |
| QVLQVTPFAER | 644.4 | +2 | 720.5 (y6) | 33.4 | 9.5 |
| GEGIEEFLR | 525.3 | +2 | 863.5 (y7) | 28.1 | 10.3 |
| GEGIEEFLR | 525.3 | +2 | 693.4 (y5)* | 28.1 | 10.3 |
| GEGIEEFLR | 525.3 | +2 | 564.5 (y4) | 28.1 | 10.3 |
*denotes most intense fragment ion for the VTG peptide biomarker candidate
Table 3: Optimized Fragmentation (Product) Ions for the two X. laevisVitellogenin Peptide Biomarkers.
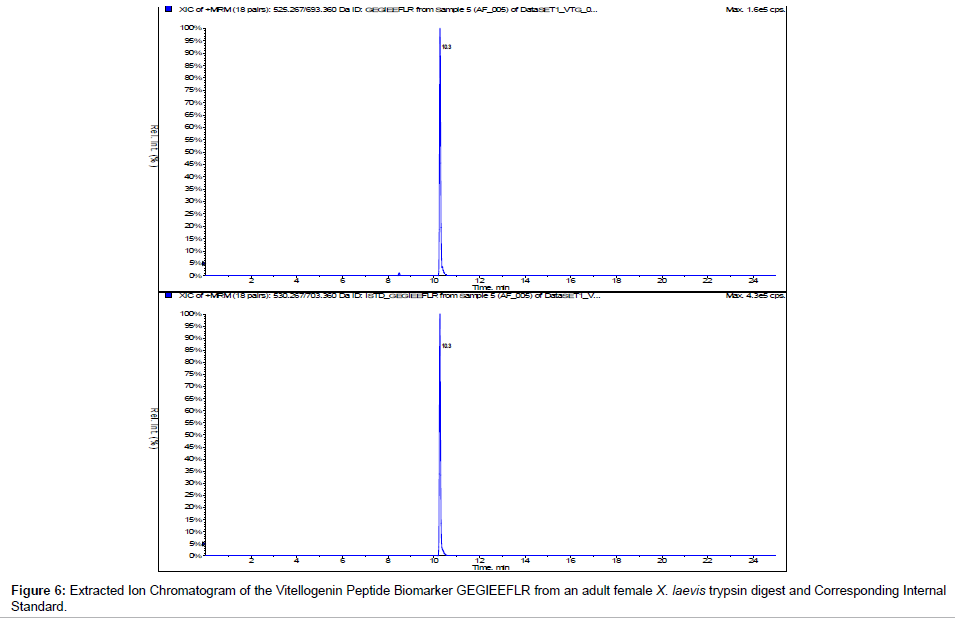
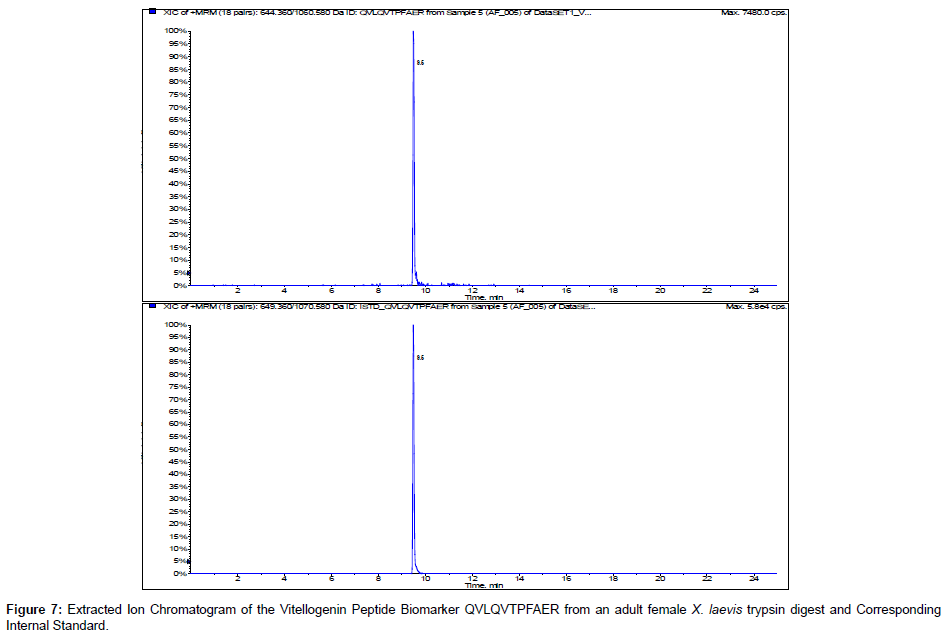
We incorporated 13C 15N internal standards for multiple reactions monitoring of the peptide biomarkers GEGIEEFLR and QVLQVTPFAER. Figures 6 and 7 show the representative chromatograms of the peptide biomarkers and corresponding internal standards for the VTG peptide biomarkers GEGIEEFLR and QVLQVTPFAER respectively. Future investigation will include determining trypsin optimization, sensitivity limits, stability, recovery and quantification of the VTG peptide biomarkers in adult female, adult male and juvenileX. laevis samples.
Conclusion
From the 201,545 DaX. laevis VTG precursor protein, results showed 2 VTG peptides which are excellent biomarkers that can be utilized for quantitative measurements of VTG from small volumes ofX. laevis plasma. We utilized EPI trap scans to confirm the amino acid sequence of the peptide biomarkers and compared them to synthetic standards. The three most intense fragmentation ions appear to be y4, y5 and y7 for the VTG peptide biomarker GEGIEEFLR, while the three most intense fragmentation ions appear to be y6, y7 and y9 for QVLQVTPFAER and these six fragmentation ions are expected to be the most sensitive to monitor for quantification and confirmation ofX. laevis VTG. Typically one transition (precursor ion/ fragment ion pair) is utilized for quantification and the other two are used for confirmation of the peptide biomarker. This methodology of quantifying ofX. laevis VTG may be particularly useful to screen for endocrine active chemicals inX. laevis, or may specifically be useful in the context of the Larval Amphibian Growth and Development Assay, a Tier 2 test which is currently under development as a part of the United States Environmental Protection Agency’s Endocrine Disruptor Screening Program.
Acknowledgements
We thank The Dow Chemical Company for funding this research
References
- Oka T, Mitsui N, Hinago M, Miyahara M, Fujii T, et al. (2006) All ZZ male Xenopus laevis provides a clear sex-reversal test for feminizing endocrine disruptors. Ecotoxicol Environ Saf 63: 236-243.
- Knechtges PL, Sprando RL, Porter KL, Brennan LM, Miller MF, et al. (2007) A novel amphibian tier 2 testing protocol: a 30-week exposure of Xenopus tropicalis to the antiandrogen flutamide. Environ Toxicol Chem 26: 555-564.
- Matsumura N, Ishibashi H, Hirano M, Nagao Y, Watanabe N, et al. (2005) Effects of nonylphenol and triclosan on production of plasma vitellogenin and testosterone in male South African clawed frogs (Xenopus laevis). Biol Pharm Bull 28: 1748-1751.
- Mitsui N, Tooi O, Kawahara A (2003) Sandwich ELISAs for quantification of Xenopus laevis vitellogenin and albumin and their application to measurement of estradiol-17 beta effects on whole animals and primary-cultured hepatocytes. Comp Biochem Physiol C Toxicol Pharmacol 135: 305-313.
- Oka T, Tooi O, Mitsui N, Miyahara M, Ohnishi Y, et al. (2008) Effect of atrazine on metamorphosis and sexual differentiation in Xenopus laevis. Aquat Toxicol 87: 215-226.
- Sumpter JP (1985) The purification, radioimmunoassay and plasma levels of vitellogenin from the rainbow trout. Lofts B, Holmes WN, Current Trends in Comparative Endocrinology, University Press, Hong Kong 355-357.
- Luna LG, Williams TL, Pirkle JL, Barr JR (2008) Ultra performance liquid chromatography isotope dilution tandem mass spectrometry for the absolute quantification of proteins and peptides. Anal Chem 80: 2688-2693.
- Wahl KL, Wunschel SC, Jarman KH, Valentine NB, Petersen CE, et al. (2002) Analysis of microbial mixtures by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Chem 74: 6191-6199.
- Zhang F, Bartels MJ, Brodeur JC, Woodburn KB (2004) Quantitative measurement of fathead minnow vitellogenin by liquid chromatography combined with tandem mass spectrometry using a signature peptide of vitellogenin. Environ Toxicol Chem 23: 1408-1415.
- Clauser KR, Baker P, Burlingame AL (1999) Role of accurate mass measurement (+/- 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal Chem 71: 2871-2882.
- Keil B (1992) Specificity of proteolysis. Springer-Verlag Berlin-Heidelberg, NewYork, 335.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 14601
- [From(publication date):
July-2014 - Jul 03, 2025] - Breakdown by view type
- HTML page views : 9995
- PDF downloads : 4606