Identification of RAB3IP as a Novel Oncogene Related to Ovarian Cancer
Received: 13-Feb-2024 / Manuscript No. DPO-24-127538 / Editor assigned: 15-Feb-2024 / PreQC No. DPO-24-127538 / Reviewed: 29-Feb-2024 / QC No. DPO-24-127538 / Revised: 07-Mar-2024 / Manuscript No. DPO-24-127538 / Accepted Date: 07-Mar-2024 / Published Date: 14-Mar-2024 DOI: 10.4172/2476-2024.9.1.227
Abstract
Objective: The incidence of ovarian cancer ranks third among gynecological malignancies, after cervical cancer and endometrial cancer. However, the mortality rate of ovarian cancer has always been the highest. The pathogenesis of ovarian cancer is not fully understood because the ovary is located deep in the pelvis, the onset of the disease is relatively insidious, and ovarian cancer is usually diagnosed at an advanced stage. The aim of our study was to explore new biomarkers and possible therapeutic targets for ovarian cancer.
Methods: We used public data from the cancer genome atlas The Cancer Genome Atlas (TCGA) and Genotype- Tissue Expression (GTEx) databases to explore the expression level of the RAB3IP gene in ovarian cancer patients. At the same time, we analyzed the correlation between the RAB3IP gene expression and patient survival, and use ROC curve to predict its clinical efficacy. We validated the level of RAB3IP in ovarian cancer tissue using WB and q-PCR. We used cck-8, wound-healing assay and colony formation assay to verify its potential biological function in ovarian cancer. By using molecular docking technology to predict potential drug targets. Pathway enrichment analyses was used to analyze the mechanism of the RAB3IP in the occurrence and development of ovarian cancer.
Results: In this study, we found that the expression of RAB3IP in ovarian cancer is high by bioinformatics and cell biology and verified it with tissue samples. The promotion of RAB3IP on the proliferation and migration of ovarian cancer was confirmed by cck-8, wound-healing assay and colony formation assay, and RAB3IP-related genes were enriched for their possible functions, which were believed to be involved in the immune and micro environmental regulation of ovarian cancer to some extent. The expression level of RAB3IP is also significantly correlated with the prognosis of ovarian cancer. In addition, we performed drug target prediction of RAB3IP, identifying austocystin D and belinostat as potential target drugs that may play a role, and used molecular docking for preliminary validation.
Conclusion: All these results provide preliminary evidence that RAB3IP can be used as a new ovarian cancer biomarker to develop new therapeutic targets
Keywords: Ovarian cancer; RAB3IP; Bioinformatics; Biomarkers
Introduction
RAB3IP is a major activator of RAB proteins that can affect intracellular transport processes and cytoskeletal reorganization. RAB3IP is important in the growth of malignant cells, it has been found to be up-regulated in malignant tumors and has been suggested to be a tumor-specific marker for some malignancies, such as colorectal cancer, large salivary adenocarcinomas, and gliomas. Recent studies have identified RAB3IP as a proto-oncogene and metastasis-associated protein in colorectal, large salivary adenocarcinoma and cervical cancer [1-3]. In human colorectal cancer metastasis, RAB3IP was found to be specifically activated by hypomethylation of LINE-1 [3]. Further analysis elucidated the hypomethylated state of the LINE-1 sequence in the promoter region of RAB3IP, which correlated with overexpression of RAB3IP in matched primary cancer and liver metastasis specimens. lINE-1 accompanied by downstream genes such as RAB3IP and MET may play an important role in the progression of cancer toward metastasis. In gastric cancer studies, RAB3IP has been recognized as a marker of poor prognosis because it significantly enhances cell migration, and this effect is attributed to the involvement of RAB3IP in Epithelial-Mesenchymal Transition (EMT) to perform its invasion-promoting function. In vitro studies also showed that RAB3IP overexpression induced changes in the invasive ability and EMT-like phenotype of gastric cancer cells. In addition, the high expression of RAB3IP was attributed to the aberrant activation of SSX2, while a significant increase in the invasive ability and motility of cancer cells was observed [4].
However, the expression and potential functions of RAB3IP in ovarian cancer are not yet known. In this study, we demonstrated the role of RAB3IP in ovarian cancer tissues and cell lines, revealing the potential significance of RAB3IP in participating in ovarian cancer invasion and metastasis.
Materials and Methods
Acquirement of target data
Normal tissue microarrays from the GTEx (https://xenabrowser. net/datapages/) database and tumor tissue microarrays from the TCGA (https://cancergenome.nih.gov/) database were integrated to analyze the differences in RAB3IP expression in 34 tumors. Data corresponding to TCGA for ovarian plasmacytoid cystic adenocarcinoma and corresponding normal tissue data in GTEx were extracted to determine the expression levels of RAB3IP in adjacent normal and ovarian cancer tissues.
Survival analysis
Overall survival of the RAB3IP high and low expression groups in ovarian cancer was analyzed using the Kaplan-Meier Plotter (http:// kmplot.com).
Sankey diagrams
Sankey diagrams were constructed through the ggalluvial package in the R software (4.2.2). The Sankey diagram consists mainly of edges, flows and pivots, where edges represent flowing data, flows represent specific values of the flowing data, and nodes represent different classifications. The width of the edges is shown proportionally to the flow, the wider the edge, the larger the value. The distribution of samples in different subgroups is observed through the Sankey diagram.
ROC curve
The accuracy efficacy of RAB3IP in predicting the prognosis of ovarian cancer was analyzed by ROC curve. ROC curve can reflect the relationship between sensitivity and specificity. The horizontal coordinate is 1-specificity, the vertical coordinate is sensitivity, the closer the horizontal coordinate is to the zero point, the higher the accuracy, and the farther the vertical coordinate is from the zero point, the better the accuracy. The Area Under Curve (AUC) is used to evaluate the prediction efficacy, and the AUC is usually taken as the value of 0.5~1, and the closer it is to 1, the better the prediction effect is.
Tumor stemness score
Cancer stemness score was obtained according to the One-Class Logistic Regression (OCLR) algorithm constructed by Malta, et al. [5]. RNAseq data and corresponding clinical information of 515 ovarian cancer patients were obtained from the TCGA dataset. The OCLR algorithm was used to calculate the mRNA stemness index (Stemness index, Si). In addition, Spearman correlation was used in this paper to map Si to the range (0, 1) by subtracting the minimum value and dividing by the maximum value. All analysis methods and R packages were implemented using the v4.2.2 version of the R software.
Target gene prediction
Based on differentially expressed genes mined from the Linked Omics (https://www.linkedomics.org/admin.php) database, the R packages cluster profiler and ggplot2 were used for functional enrichment and visualization analysis.
Mutation analysis
Use the cBioPortal (http://www.cbioportal.org/) website to learn about the frequency of alterations, mutation types, and copy number aberration results for RAB3IP in TCGA across all cancer types. Obtain somatic mutation data for ovarian cancer patients from the TCGA dataset. Mutation data were downloaded and extracted using the maftools package in R software, and then somatic nuclear mutation data from patients in the high and low gene expression groups were visualized and analyzed.
Immunoassay
The relationship between RAB3IP expression levels and immune infiltration of all types of cancers in TCGA was explored using TIMER2 (http://timer.cistrome.org/). The TIDE algorithm was used to find out the correlation between RAB3IP expression levels and tumor immune infiltration. p-values and partial correlation core values were obtained by purity-adjusted Spearman rank correlation test. The results were visualized as heatmaps.
Drug target prediction
The Gene Set Cancer Analysis (GSCA) is a knowledge base that integrates multiple chemicals, genes, functional phenotypes, and disease-dependent interactions to help study disease-associated environmental exposures and potential drug mechanisms of action, as well as to provide information on oncology drug response data and genome-sensitive markers. The structures of small molecule compounds are available through the National Library of Medicine online at https:// www.nlm.nih.gov/medline/index.html. And the interactions between small molecule compounds and genes are simulated by molecular docking computers.
Enrichment analysis
R package GSVA was used to analyze the correlation between genes and pathway scores by selecting the parameter method="sgsea" and analyzed by Spearman correlation. p<0.05 was considered statistically significant.
miRNA prediction
miRNA prediction was performed based on the miRDB online database, which is used for miRNA target prediction and functional annotation by analyzing miRNA target interactions in high-throughput sequencing experiments. In addition, visual analysis was performed by Cytoscape.
Cell culture
Human epithelial ovarian cancer cells A2780 were purchased from the Cell Resource Center of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. All cells were grown in an incubator at 37°C, 5% CO2 .
Western blotting
In this study, cells were lysed in RIPA buffer (#P0013B; Beyotime, Shanghai, China) containing a protease inhibitor (#K1007; APExBIO, Houston, USA) and a phosphatase inhibitor (#K1015; APExBIO) for 20 min at 4°C, and the lysate was centrifuged for 20 min at 4°C at 12,000 rpm/min. Centrifugation was performed for 20 min to obtain the supernatant. Proteins were quantified by BCA assay kit (#P0010S, Beyotime), 5 × loading and 1 × loading were added to ensure the consistency of protein concentration, and protein samples were boiled at 100°C for 5 min to prepare the proteins to be detected. And 20 μg per lane was separated by SDS-PAGE and transferred to PVDF membrane (#IPVH00010, Millipore, USA). Then, the membrane was incubated with RAB3IP antibody (PA5-96927, ThermoFisher) at 4°C overnight, and on the following day, the PVDF membrane was washed three times with TBST for 5 min each time, incubated with rabbit secondary antibody (A7016, Beyotime) for 2 hr at room temperature, and finally developed using ECL reagent (#180-506, Tanon).
Quantitative Polymerase Chain Reaction (qPCR)
Total RNA was extracted using the TransZol Up Plus RNA kit (Ambion, USA), and cDNA was synthesized using the reverse transcriptase kit (Vazyme, Nanjing, China) in addition, the AceQ qPCR SYBR Green Master Mix kit (Vazyme, Nanjing, China) was used for the qPCR reaction. Meanwhile, the data were normalized to GAPDH mRNA, and the qPCR primers for RAB3IP were as follows: forward 5′-GTGTCATCTACCGGCCACAC-3′ and reverse 5′-CCCTCTGAGCTTGCGAGT-3′. Cell Counting Kit-8 (CCK-8) for cell viability assay, cells from different groups were incubated for 2 h according to four different time points 0 h, 24 h, 48 h, 72 h by adding 100 μL of CCK-8 reagent (Vazyme, Nanjing, China) diluted with RPIM 1640. The results were detected at 450 nm on an enzyme meter and the data were collected.
Wound-healing assay
Different groups of cells were planted in 6-well plates, and when the cells were in the exponential growth period and the cell density reached 80%, a line was drawn in the 6-well plate with a 10-μL lance tip straight, and the initial image of the cell scratch was collected under the microscope, and then the image was taken for the second time under the same field of view after 24 h.
Cell cloning experiment
Cells from different groups were planted in 6-well plates and incubated in the incubator for 14 days. The cells were fixed with 4% paraformaldehyde for 10 min, stained with crystal violet solution and washed with water. The colonies were photographed and quantified after drying at room temperature.
Statistical analysis
Data were expressed as mean ± Standard Deviation (SD). Statistical analysis was performed using SPSS 25.0 software, and graphs of difference statistics were plotted using Graphpad Prism 8 software. Differences were detected by independent samples t-test or paired samples test. In addition, overall survival analysis was performed using the Kaplan-Meier method and Log-rank test. P<0.05 was considered statistically significant difference. * indicates P<0.05, ** indicates P<0.01, and *** indicates P<0.001.
Results
High expression of RAB3IP in ovarian cancer
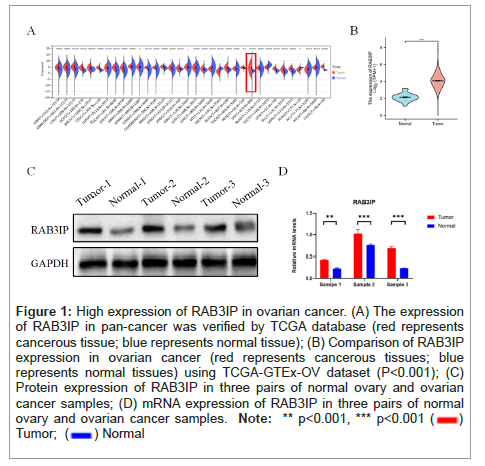
In order to investigate the role of RAB3IP in tumors, the mRNA expression levels of RAB3IP in pan-cancers in the TCGA database were first analyzed by bioinformatics. Not surprisingly, the expression of RAB3IP was significantly different in each tumor (Figure 1A). To validate the expression of RAB3IP transcriptome level in ovarian cancer, we compared the mRNA levels of RAB3IP in 88 normal ovarian samples and 427 ovarian cancer samples using the TCGA-GTEx- OV dataset in the XENA database, and the expression of RAB3IP in ovarian cancer was much higher than that in the normal control group (P<0.001) (Figure 1B). Subsequently, further by verifying whether the results of clinical samples were consistent with the database, tissue samples from three pairs of ovarian cancer patients were collected, and Western Blot experiments confirmed that the protein expression level of RAB3IP in ovarian cancer was higher than that of normal controls (Figure 1C). In addition, significantly high mRNA level of RAB3IP expression in ovarian cancer was also confirmed by real-time quantitative PCR experiments (Figure 1D).
Figure 1: High expression of RAB3IP in ovarian cancer. (A) The expression of RAB3IP in pan-cancer was verified by TCGA database (red represents cancerous tissue; blue represents normal tissue); (B) Comparison of RAB3IP expression in ovarian cancer (red represents cancerous tissues; blue represents normal tissues) using TCGA-GTEx-OV dataset (P< 0.001); (C) Protein expression of RAB3IP in three pairs of normal ovary and ovarian cancer samples; (D) mRNA expression of RAB3IP in three pairs of normal ovary and ovarian cancer samples.

Relationship between the expression level of RAB3IP and the prognosis of ovarian cancer patients
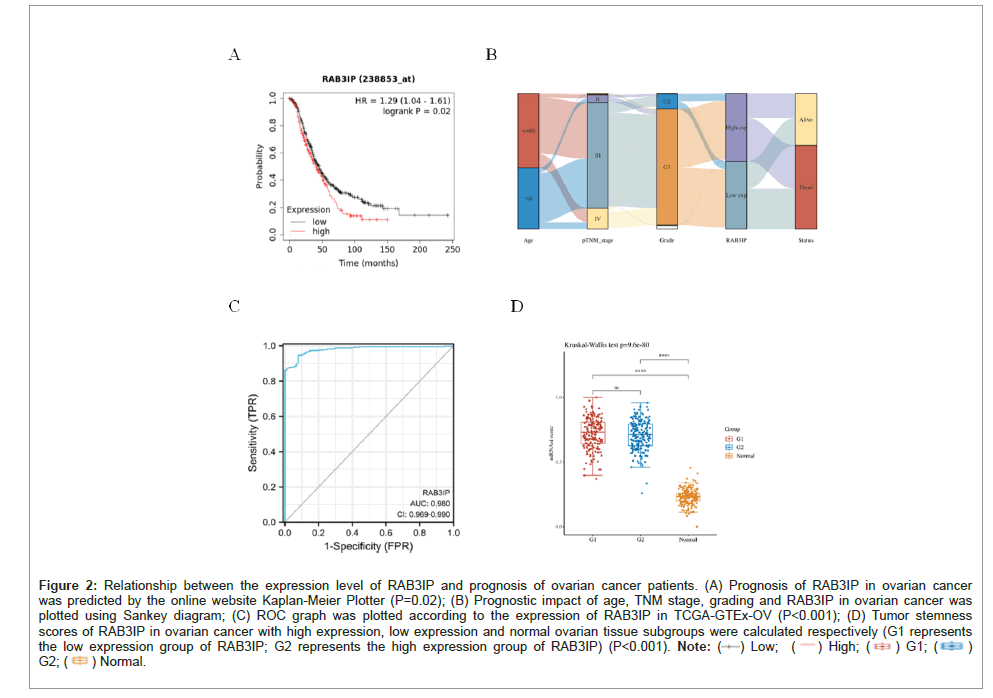
Based on the above results that RAB3IP is highly expressed in ovarian cancer, we hypothesized that RAB3IP promotes the progression of ovarian cancer and is closely related to the prognosis of ovarian cancer patients. Thus, the effect of RAB3IP on the overall survival of ovarian cancer patients in the TCGA database was analyzed, and the results suggested that ovarian cancer patients with high expression of RAB3IP had a shorter overall survival (HR=1.29, 95% CI=1.04-1.61, P=0.02) and poorer prognosis than those with low expression (Figure 2A). Immediately after that, based on the TCGA dataset to obtain the RNAseq data and corresponding clinical information of ovarian cancer, the distribution trend of age, TNM stage, grade, high and low expression of RAB3IP, and patient's life and death status in ovarian cancer samples was demonstrated by Sankey diagram, and the results suggested that the branch width of high expression of RAB3IP had more data flow volume tending to the patient's death status (Figure 2B), which suggests that the patients with high expression of RAB3IP had a poorer prognosis. The ROC plot is a curve reflecting the relationship between sensitivity and specificity, by which we verified the prognostic ability of RAB3IP in the TCGA-GTEx-OV cohort, and the AUC area of RAB3IP was 0.980 (P<0.001) (Figure 2C). Cancer progression involves the progressive loss of differentiated phenotype and the acquisition of stem cell-like features; therefore, we demonstrated that the RAB3IP high-expression group had better tumor stem cell features compared to the normal control group by calculating the Tumor Stemness Score (P<0.001).
Figure 2: Relationship between the expression level of RAB3IP and prognosis of ovarian cancer patients. (A) Prognosis of RAB3IP in ovarian cancer was predicted by the online website Kaplan-Meier Plotter (P=0.02); (B) Prognostic impact of age, TNM stage, grading and RAB3IP in ovarian cancer was plotted using Sankey diagram; (C) ROC graph was plotted according to the expression of RAB3IP in TCGA-GTEx-OV (P<0.001); (D) Tumor stemness scores of RAB3IP in ovarian cancer with high expression, low expression and normal ovarian tissue subgroups were calculated respectively (G1 represents the low expression group of RAB3IP; G2 represents the high expression group of RAB3IP) (P<0.001). 
Malignant biological behavior of RAB3IP in ovarian cancer
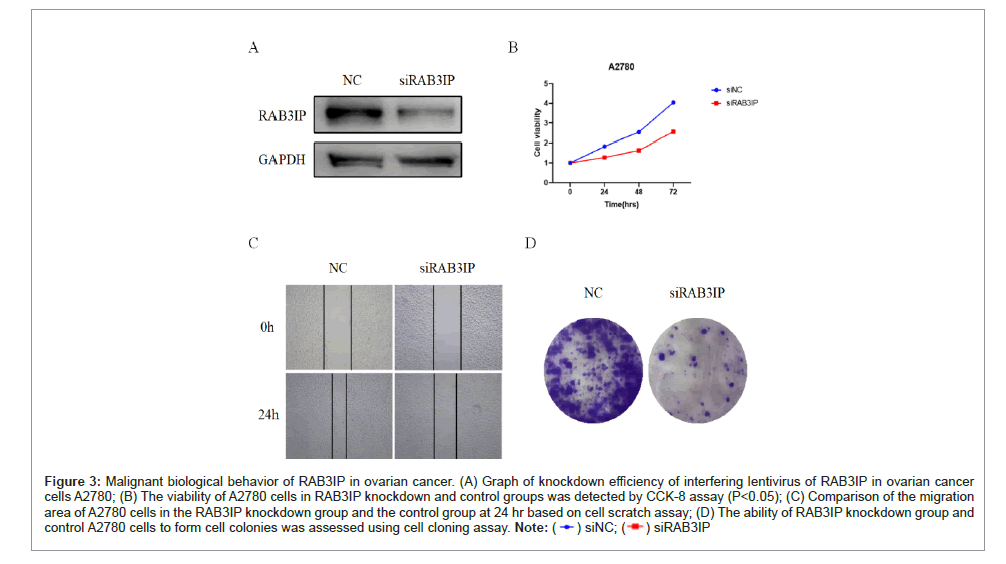
In view of the high expression of RAB3IP in ovarian cancer and its impact on the prognosis of ovarian cancer patients, we further explored its effect on the biological behavior of ovarian cancer cells. First, lentiviral siRAB3IP was transfected in ovarian cancer cells A2780 for knockdown, and the knockdown efficiency of RAB3IP was verified by Western Blot experiments, as shown in Figure 3A, the knockdown lentivirus of RAB3IP demonstrated effective interference efficiency. To further investigate whether RAB3IP had any effect on the proliferation and migration of ovarian cancer cells, cell survival viability was detected by CCK-8 assay, and the results showed that ovarian cancer cells A2780 with siRAB3IP could significantly inhibit cell viability with a significant temporal trend at the three time points of 24 h, 48 h, and 72 h, as compared with the control cells (Figure 3B) (P<0.05 ). The 24 h cell migration area was compared by cell scratch assay, and it was found that the siRAB3IP group was able to significantly inhibit the rate of cell migration (Figure 3C). Consistent with the above results, the number of A2780 cell colonies in the siRAB3IP group was significantly reduced compared with that in the control group (Figure 3D), and the above results indicated that RAB3IP was obviously able to promote the proliferation and migration of ovarian cancer cells.
Figure 3: Malignant biological behavior of RAB3IP in ovarian cancer. (A) Graph of knockdown efficiency of interfering lentivirus of RAB3IP in ovarian cancer cells A2780; (B) The viability of A2780 cells in RAB3IP knockdown and control groups was detected by CCK-8 assay (P<0.05); (C) Comparison of the migration area of A2780 cells in the RAB3IP knockdown group and the control group at 24 hr based on cell scratch assay; (D) The ability of RAB3IP knockdown group and control A2780 cells to form cell colonies was assessed using cell cloning assay. 
Exploring genes associated with RAB3IP in ovarian cancer
The Linked Omics database was used to assess the genes associated with RAB3IP in ovarian cancer. The volcano plot in Figure 4A shows that genes positively associated with RAB3IP are clustered on the right side of the 0.0 value of the abscissa, while genes negatively associated with it are clustered on the left side. The heatmaps of Figures 4B and 4C show the top 50 positively and negatively correlated genes extracted and analyzed according to the Spearman test. In addition, these positively and negatively correlated genes were analyzed for two-by-two correlation in ovarian cancer, respectively, and visualized by heatmaps (Figures 4A and 4C). Immediately after that, the above 100 correlated genes were included in the GO and KEGG pathway enrichment analysis, and it was found that these 100 correlated genes were closely associated with nuclear membrane, nuclear chromatin, mitophagy and cell cycle progression (Figures 4A-4C).
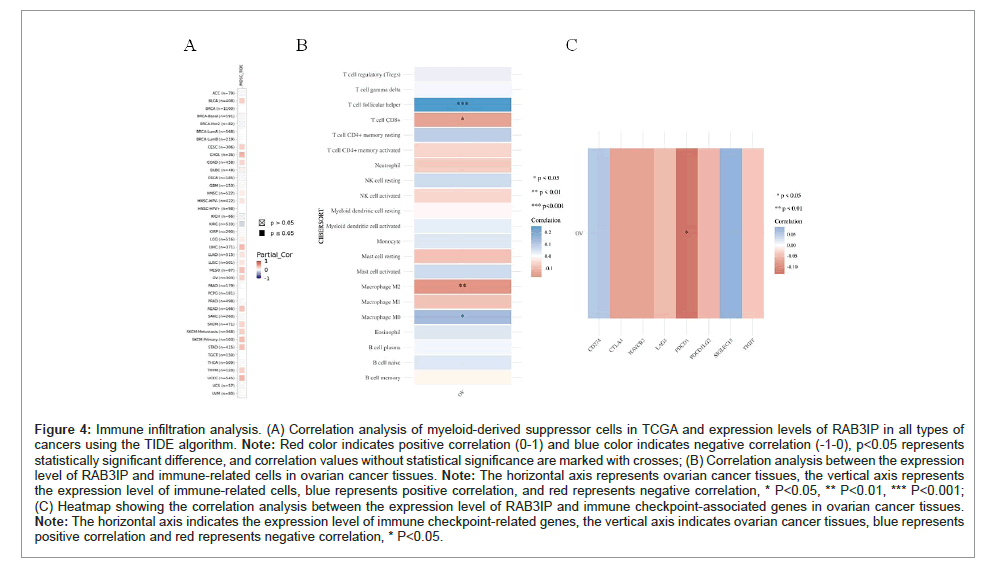
Figure 4: Immune infiltration analysis. (A) Correlation analysis of myeloid-derived suppressor cells in TCGA and expression levels of RAB3IP in all types of cancers using the TIDE algorithm. Note: Red color indicates positive correlation (0-1) and blue color indicates negative correlation (-1-0), p<0.05 represents statistically significant difference, and correlation values without statistical significance are marked with crosses; (B) Correlation analysis between the expression level of RAB3IP and immune-related cells in ovarian cancer tissues. Note: The horizontal axis represents ovarian cancer tissues, the vertical axis represents the expression level of immune-related cells, blue represents positive correlation, and red represents negative correlation, * P<0.05, ** P<0.01, *** P<0.001; (C) Heatmap showing the correlation analysis between the expression level of RAB3IP and immune checkpoint-associated genes in ovarian cancer tissues. Note: The horizontal axis indicates the expression level of immune checkpoint-related genes, the vertical axis indicates ovarian cancer tissues, blue represents positive correlation and red represents negative correlation, * P<0.05.
The genetic alterations of RAB3IP in TCGA
We explored the genetic changes of RAB3IP in ovarian cancer through cBioPortal based on the TCGA cohort. As shown in Figure 5A, the frequency of RAB3IP changes in ovarian cancer was higher than 4% compared to various tumors. Moreover, the majority of RAB3IP alteration frequencies in ovarian cancer were of the "amplification" type, followed by the "mutation" type, and a small number of "multiple variants". Figure 5B shows the types and locations of mutations in the RAB3IP gene. Based on the mutation map, ovarian cancer patients in the TCGA cohort were categorized into high and low expression groups according to the median expression value of RAB3IP. In the RAB3IP high-expression group, the RYR2, CMYA5, ABCA1, PAPPA2, and LAMC1 genes were more inclined to be mutated, whereas in the RAB3IP low-expression group, significant mutations were observed in the RYR2, NF1, ATRX, NLRP3, and VPS13B genes (Figures 5A-5E).
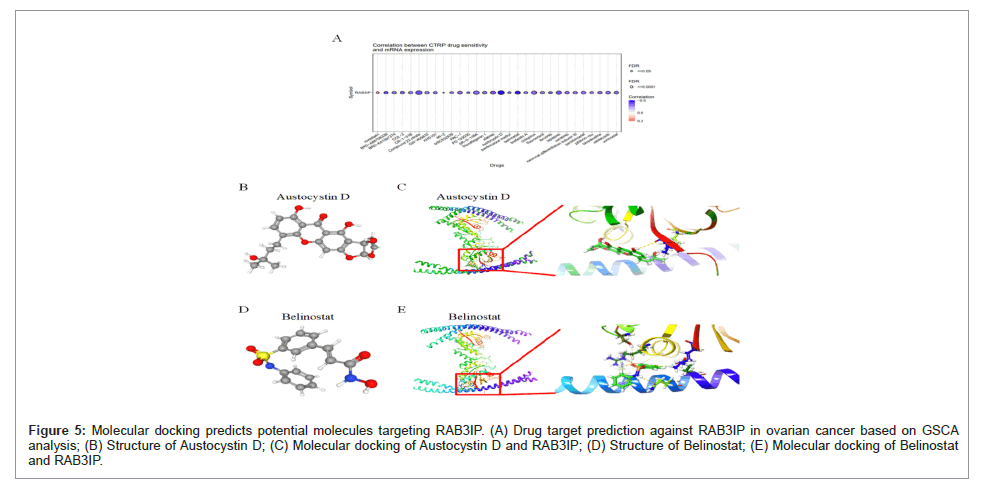
Figure 5: Molecular docking predicts potential molecules targeting RAB3IP. (A) Drug target prediction against RAB3IP in ovarian cancer based on GSCA analysis; (B) Structure of Austocystin D; (C) Molecular docking of Austocystin D and RAB3IP; (D) Structure of Belinostat; (E) Molecular docking of Belinostat and RAB3IP.
Immune infiltration analysis
Myeloid-derived suppressor cells are a heterogeneous population of immature myeloid cells with immunosuppressive activity (it blocks the proliferation and activity of T cells and natural killer cells). In addition to their role in suppressing immune responses, myeloid-derived suppressor cells directly stimulate tumor cell proliferation, metastasis, and angiogenesis [6]. Therefore, we used TIDE in TIMER2 to explore the correlation between the level of myeloid-derived suppressor cell infiltration and the level of RAB3IP expression in multiple types of cancers. Importantly, we found that the expression level of RAB3IP was positively correlated with the infiltration of myeloid-derived suppressor cells and negatively correlated with KIRC in CHOL, LIHC, MESO, OV, and UCEC cancer types (Figure 6A). According to the immune correlation analysis, in ovarian cancer, the expression level of RAB3IP was positively correlated with follicular helper T cells (P<0.001) and M0 macrophages (P<0.05) and negatively correlated with CD8-positive T cells (P<0.05) and M2 macrophages (P<0.01) (Figure 6B). Immune checkpoint-related genes have attracted the attention of scholars in cancer therapy. Therefore, we explored the correlation between the expression levels of RAB3IP and the expression levels of immune checkpoint-related genes by co-expression analysis. We found that in ovarian cancer, the expression level of RAB3IP was negatively correlated with that of the immune checkpoint-related gene PDCD1 (P<0.05) (Figure 6C).
Molecular docking predicts potential molecules targeting RAB3IP
With the above results we demonstrated the pro-carcinogenic role of RAB3IP in ovarian cancer, and immediately after, in order to explore molecular compounds that could be used as targeting RAB3IP, we analyzed the drug sensitivity of RAB3IP. The two molecular compounds most relevant to RAB3IP were selected, and Austocystin D and Belinostat were identified as potential targeting agents that could play a role (Figure 7A). And based on Spearman correlation analysis, the structures of the potential target drugs were identified (Figures 7B- 7D). To further validate the binding activity of RAB3IP and these two drugs, we predicted whether the small molecules had the potential to become drug candidates by computer simulation of molecular docking, and the results showed that both Austocystin D and Belinostat could interact with RAB3IP (Figures 7C-7D).
Single-sample gene enrichment analysis
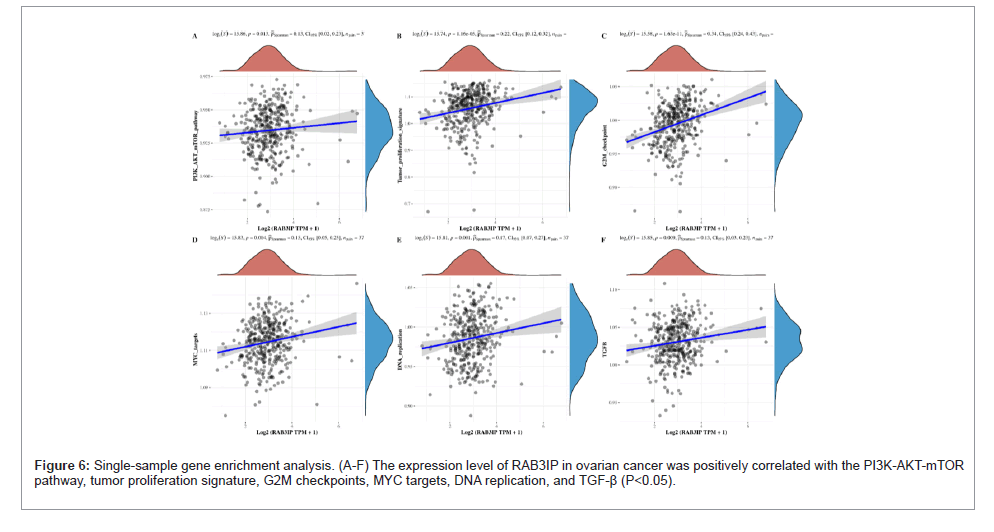
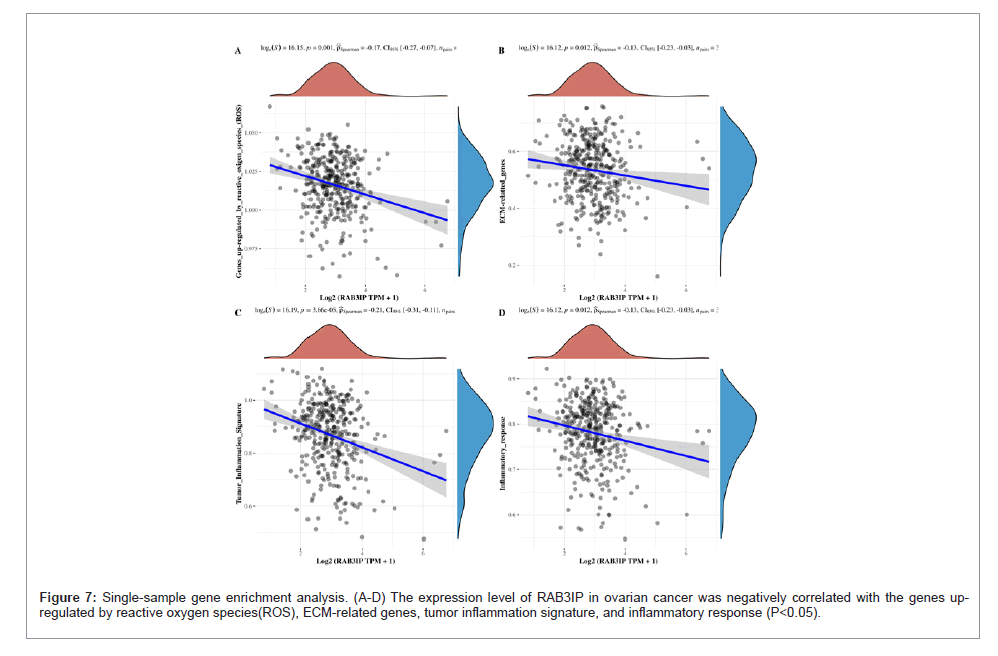
Using the collection of genes contained in the relevant pathways in the TCGA database, the enrichment scores of each sample on each pathway were sequentially calculated according to the single- sample gene enrichment analysis algorithm, so as to obtain the connection between the samples and the pathways, and by calculating the correlation between the gene expression and the pathway scores, the relationship between the gene and the pathway can be obtained. Based on this, we found that in ovarian cancer, the expression level of RAB3IP was positively correlated with the PI3K-AKT-mTOR pathway, tumor proliferation signature, G2M checkpoints, MYC targets, DNA replication, and TGF-β (P<0.05) (Figure 8), whereas it was negatively correlated with the genes up-regulated by reactive oxygen species(ROS), ECM-related genes, tumor inflammation signature, and inflammatory response (P<0.05).
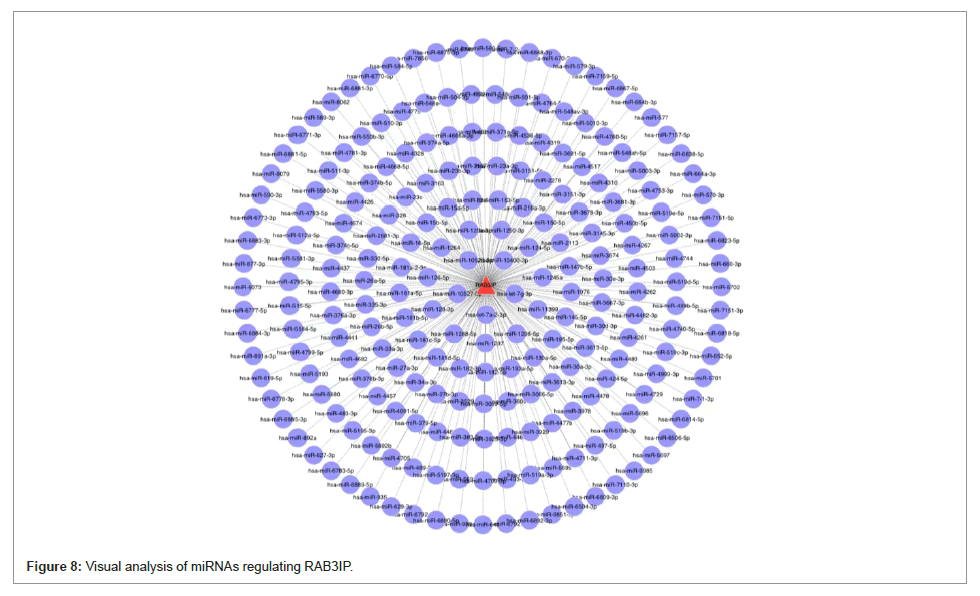
Prediction of miRNAs regulating RAB3IP
MiRNAs are small non-coding RNAs, typically 18-25 nucleotides long that inhibit protein translation by binding to complementary target mRNAs. miRNAs regulate many biological processes, including cell cycle regulation, cell growth, proliferation, differentiation, apoptosis, metabolism, neuronal patterns, and aging [7]. In this study, we also predicted 224 miRNAs that may target RAB3IP (Figure 8 ). It can provide information about miRNAs as biomarkers and therapeutic targets in ovarian cancer.
Discussion
Ovarian cancer is a fatal gynecological cancer with diverse biology at the molecular, cellular and clinical levels [8]. It is the eighth most common cause of death leading to 140,000 women's deaths [9]. It has the worst prognosis and high mortality rate due to lack of proper screening, poor diagnosis and late detection of the disease, hence it has been called the "silent killer" [10]. Most women diagnosed at advanced stages have a 5-year survival rate of only 46% after standard treatment [11]. CA125 is a useful biomarker for detecting the progression of ovarian cancer, but it lacks sensitivity and specificity because it is increased in benign conditions, such as ovarian cysts, uterine fibroids, and infections [12]. Therefore, there is a need to identify new biomarkers to screen for ovarian cancer.
Comprehensive bioinformatics analysis of microarray data is a powerful tool for analyzing gene expression, protein function, and signaling pathways under various physiological and pathological conditions [13]. It provides a new approach to explore disease- associated genes and predict new molecular interactions, key regulatory molecules, and therapeutic drug targets [14]. Recent studies using bioinformatics analysis based on the TCGA database demonstrated that pyroptosis plays a non-negligible role in the prognostic value, clinicopathological features, and tumor immune infiltration microenvironment of ovarian cancer, which provides new insights into identifying and characterizing the tumor immune microenvironmental landscape, thus guiding a more effective assessment of the prognosis and tailoring of immunotherapeutic strategies for ovarian cancer [15]. However, public databases of extensive genetic profiling data from ovarian cancer patients have not been thoroughly analyzed. RAB3IP, a newly discovered member of the RAB protein family in recent years, acts as an activator protein of the RAB proteins, which is involved in the disease mainly by affecting intracellular transport processes and cytoskeletal remodeling.RAB3IP is closely associated with cancer development and disease progression, and it has been suggested that RAB3IP may play an important role in malignant cell growth, as it was found to be upregulated and considered as a tumor-specific marker for some malignant tumors (e.g., colorectal cancer, macrosomal adenocarcinoma and gliomas) [1-3]. In this study, the expression of RAB3IP in ovarian cancer tissues was detected for the first time by Western Blot and qPCR experiments, and the results showed that the expression of RAB3IP in ovarian cancer tissues was significantly higher than that in normal controls, suggesting that the high expression of RAB3IP may be involved in the development of ovarian cancer. The role of RAB in tumors has not been well studied, and some researchers have suggested that the regulatory effect of RAB3IP on vesicle transport may be through natural immune regulation and thus affect tumor progression [16]. In addition, RAB proteins promote signaling between tumor cells and stromal cells, providing a favorable microenvironment for tumor cell proliferation and invasion [17].
The interaction between RAB3IP and SSX2 was confirmed by de Bruijn, et al. In their study, the co-localization of these two proteins in the nucleus was observed, revealing for the first time that RAB3IP is a protein that interacts with SSX2. In addition, the interaction between these two molecules was revealed by glutathione chain transferase pull-down assay, which determined that they were co-expressed [18]. SSX2 is a cancer-associated protein that interacts with RAB3IP, which was also found to enhance the invasiveness of breast cancer cells by inhibiting Erα signaling [19]. In gastric cancer studies, RAB3IP has been recognized as a marker of poor prognosis because it significantly enhances cell migration, and this effect has been attributed to the involvement of RAB3IP in EMT to perform its invasion-promoting function. In vitro studies also showed that RAB3IP overexpression induced changes in the invasive capacity and EMT-like phenotype of gastric cancer cells. In addition, the high expression of RAB3IP was attributed to the aberrant activation of SSX2, while a significant increase in the invasive ability and motility of cancer cells was observed [4]. In this study, we further analyzed the relationship between RAB3IP expression and the survival prognosis of ovarian cancer patients, and the results showed that RAB3IP expression was an independent risk factor affecting the overall survival of ovarian cancer patients, and the overall survival time of ovarian cancer patients with high expression of RAB3IP was significantly shorter, which indicated that the expression of RAB3IP could be used as an evaluation index for the survival prognosis of ovarian cancer patients.
In this study, RAB3IP was knocked down for the first time using siRNA in ovarian cancer cells A2780, and it was found that silencing of RAB3IP significantly inhibited the viability, proliferation and migration ability of ovarian cancer cells A2780 (P<0.05). It further indicated that RAB3IP was involved in ovarian cancer progression.
In addition, bioinformatics analysis was performed to screen 50 genes positively and negatively associated with RAB3IP in ovarian cancer, and found that these 100 genes were closely associated with the nuclear membrane, nuclear chromatin, mitophagy, and cell cycle progression. The frequency of RAB3IP alterations was high, and most of the alterations were of the "amplification" type ; in the group with high RAB3IP expression, RAB3IP was highly expressed. In the RAB3IP high-expression group, RYR2, CMYA5, ABCA1, PAPPA2, and LAMC1 genes were more likely to be mutated, while in the RAB3IP low-expression group, significant mutations were observed in RYR2, NF1, ATRX, NLRP3, and VPS13B genes. According to the immune correlation analysis, the expression level of RAB3IP was positively correlated with the infiltration of myeloid-derived suppressor cells, follicular helper T cells, and M0 macrophages (P<0.05), whereas it was negatively correlated with KIRC, CD8-positive T cells, and M2 macrophages (P<0.05). In addition, in ovarian cancer, the expression level of RAB3IP was negatively correlated with that of the immune checkpoint-related gene PDCD1 (P<0.05). Molecular docking was simulated by computer and predicted that Austocystin D and Belinostat could be used as drugs targeting RAB3IP in ovarian cancer. By single- sample gene enrichment analysis, the expression level of RAB3IP in ovarian cancer was positively correlated with the PI3K-AKT-mTOR pathway, tumor proliferation signature, G2M checkpoints, MYC targets, DNA replication, and TGF-β (P<0.05) ; and negatively correlated with the genes up-regulated by Reactive Oxygen Species (ROS), ECM-related genes, tumor inflammation signature, and inflammatory response (P<0.05). Finally, we predicted 224 miRNAs that might target RAB3IP. However, there are still some unanswered questions in this study, such as what is the specific mechanism by which RAB3IP promotes ovarian cancer progression, and what genes regulate RAB3IP to exert its pro- cancer effects. We will continue to explore these questions in depth in the next step.
Conclusion
Overall, in the present study, we demonstrated that RAB3IP is highly expressed in ovarian cancer and promotes ovarian cancer progression, which is positively correlated with the proliferation and migration of ovarian cancer cells. The high expression of RAB3IP is significantly associated with poor prognosis of ovarian cancer patients, and it can be used as a new biomarker for ovarian cancer prognosis.
Statements and Declarations
Funding
This work was supported by Nantong Science and Technology Bureau(MS22022007).
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Author contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Xiaohao Li. The first draft of the manuscript was written by Qian He. Aiqin He conducted supervision and quality control. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
- Fischer U, Keller A, Leidinger P, Deutscher S, Heisel S, et al. (2008) A different view on DNA amplifications indicates frequent, highly complex, and stable amplicons on 12q13-21 in glioma. Mol Cancer Res 6:576-84.
[Crossref] [Google Scholar] [PubMed]
- Zhang L, Mitani Y, Caulin C, Rao PH, Kies MS, et al. (2013) Detailed genome-wide SNP analysis of major salivary carcinomas localizes subtype-specific chromosome sites and oncogenes of potential clinical significance. Am J Pathol 182:2048-57.
[Crossref] [Google Scholar] [PubMed]
- Hur K, Cejas P, Feliu J, Moreno-Rubio J, Burgos E, et al. (2014) Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut 3:635-46.
[Crossref] [Google Scholar] [PubMed]
- Ren H, Xu Z, Guo W, Deng Z, Yu X (2018) Rab3IP interacts with SSX2 and enhances the invasiveness of gastric cancer cells. Biochem Biophys Res Commun 503:2563-2568.
[Crossref] [Google Scholar] [PubMed]
- Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, et al. (2018) Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell 173:338-354.e15.
[Crossref] [Google Scholar] [PubMed]
- Mabuchi S, Yokoi E, Komura N, Kimura T (2018) Myeloid-derived suppressor cells and their role in gynecological malignancies. Tumour Biol 40:1010428318776485.
[Crossref] [Google Scholar] [PubMed]
- Uddin A, Chakraborty S (2018) Role of miRNAs in lung cancer. J Cell Physiol 20.
[Crossref] [Google Scholar] [PubMed]
- Matulonis UA, Sood AK, Fallowfield L, Howitt BE, Sehouli J, et al. (2016) Ovarian cancer. Nat Rev Dis Primers 2:16061.
[Crossref] [Google Scholar] [PubMed]
- Mahmood RD, Morgan RD, Edmondson RJ, Clamp AR, Jayson GC (2020) First-line management of advanced high-grade serous ovarian cancer. Curr Oncol Rep 22:64.
[Crossref] [Google Scholar] [PubMed]
- Bast RC Jr, Hennessy B, Mills GB (2009) The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer 9:415-28.
[Crossref] [Google Scholar] [PubMed]
- Jones HM, Fang Z, Sun W, Clark LH, Stine JE, et al. (2017) Atorvastatin exhibits anti-tumorigenic and anti-metastatic effects in ovarian cancer in vitro. Am J Cancer Res 7:2478-2490.
[Crossref] [Google Scholar] [PubMed]
- Duffy MJ, Bonfrer JM, Kulpa J, Rustin GJ, Soletormos G, et al. (2005) CA125 in ovarian cancer: European group on tumor markers guidelines for clinical use. Int J Gynecol Cancer 15:679-91.
[Crossref] [Google Scholar] [PubMed]
- Huang S, Sun C, Hou Y, Tang Y, Zhu Z, et al. (2018) A comprehensive bioinformatics analysis on multiple gene expression omnibus datasets of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Sci Rep 8:7630.
[Crossref] [Google Scholar] [PubMed]
- Petryszak R, Burdett T, Fiorelli B, Fonseca NA, Gonzalez-Porta M, et al. (2014) Expression Atlas update--a database of gene and transcript expression from microarray- and sequencing-based functional genomics experiments. Nucleic Acids Res 42:D926-32.
[Crossref] [Google Scholar] [PubMed]
- Gao L, Ying F, Cai J, Peng M, Xiao M, et al. (2023) Identification and validation of pyroptosis-related gene landscape in prognosis and immunotherapy of ovarian cancer. J Ovarian Res 16:27.
[Crossref] [Google Scholar] [PubMed]
- Pfeffer SR (2017) Rab GTPases: master regulators that establish the secretory and endocytic pathways. Mol Biol Cell 28:712-715.
[Crossref] [Google Scholar] [PubMed]
- Vieira OV (2018) Rab3a and Rab10 are regulators of lysosome exocytosis and plasma membrane repair. Small GTPases 9:349-351.
[Crossref] [Google Scholar] [PubMed]
- de Bruijn DR, dos Santos NR, Kater-Baats E, Thijssen J, van den Berk L, et al. (2002) The cancer-related protein SSX2 interacts with the human homologue of a Ras-like GTPase interactor, RAB3IP, and a novel nuclear protein, SSX2IP. Genes Chromosomes Cancer 34:285-98.
[Crossref] [Google Scholar] [PubMed]
- Chen L, Zhou WB, Zhao Y, Liu XA, Ding Q, et al. (2012) Cancer/testis antigen SSX2 enhances invasiveness in MCF-7 cells by repressing ERα signaling. Int J Oncol 40:1986-94.
[Crossref] [Google Scholar] [PubMed]
Citation: Li X, He, He A (2024) Identification of RAB3IP as a Novel Oncogene Related to Ovarian Cancer. Diagnos Pathol Open 9: 227. DOI: 10.4172/2476-2024.9.1.227
Copyright: © 2024 Li X, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.