Research Article Open Access
Identification of Core Microorganism in Anammox Consortia Obtained from Different Incubation Environments
Tingting Zhu1,2*, Zuotao Zhang3, Shenghua Peng1,2, Sitong Liu311State Environmental Protection, Key Laboratory of Drinking Water Source Management and Technology, Shenzhen Key Laboratory of Drinking Water Source Safety Control, Shenzhen Key Laboratory of Emerging Contaminates Detection and Control in Water Environment, Shenzhen, Guangdong, China
22Shenzhen Academy of Environmental Sciences, Shenzhen, Guangdong, China
33Department of Environmental Engineering, Peking University, Beijing, China
- *Corresponding Author:
- Zhu T
Shenzhen Academy of Environmental Sciences
Shenzhen, Guangdong, China
Tel: 008675525980292
E-mail: zttszhky@sina.com
Received date: January 30, 2017; Accepted date: February 11, 2017; Published date: February 13, 2017
Citation: Zhu T, Zhang Z, Peng S, Liu S (2017) Identification of Core Microorganism in Anammox Consortia Obtained from Different Incubation Environments. J Bioremediat Biodegrad 8:385. doi:10.4172/2155-6199.1000385
Copyright: © 2017 Zhu T, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
Anammox is a novel and sustainable wastewater treatment technology which attracts much attention. The impossible isolation of pure anammox bacteria makes the issue about which bacteria types mainly involved in the anammox consortia meaningful but still lacks of investigation nowadays. In order to elucidate the core constituent of anammox consortia, we investigated microbial communities of six anammox consortia with different origins and activities by cloning 16S rRNA gene region. It is found that the stable anammox consortia have similar community structure, even with different origins (e.g. activated sludge and ground water). The bacteria species belonging to Chlorobi, Chloroflexi, Bacteroidetes, Proteobacterium, Nitrosomonas and Armatimonadetes are the common accompanying bacteria with Plantomycetes in anammox consortia, serving as core microorganisms. The further nitrogen removal profile analysis suggested that, high nitrogen removal activity corresponded to high Plantomycetes percentage. On the contrary, the bacteria species of Rhodocyclaceae, Acidobacteria and Verrucomicrobia mainly present in anammox consortia with abnormal nitrogen removal performance, such as under conditions of bacteria being inhibited. These findings would advance the understanding of anammox microbial community, which correlates with the stability and robustness of anammox consortia.
Keywords
Anammox; Nitrogen removal; Microbial community; Core microorganism
Introduction
The anaerobic ammonium oxidation (Anammox) is an innovative nitrogen removal process to convert ammonium and nitrite or nitrate to form nitrogen gas. It needs no COD addition, reduces oxygen demand and produces low sludge comparing with the traditional way [1,2] and therefore is regarded as sustainable and energy-saving technology. Although cultivation and enrichment of anammox bacteria community have been carried out by many research groups, there are still some bottlenecks for fully understanding these bacteria community, among which is the difficult isolation of anammox bacteria [3-5].
The isolation of anammox bacteria from the consortia by the traditional spread plate method has been verified to be impossible [6,7]. The highest purity of the isolated anammox consortium is 99% obtained by the method of percoll density gradient centrifugation [8]. Clearly, there are still some other accompanying bacteria. For example, some bacteria belonging to the Chloroflexi are usually appeared in the anammox consortia [9]. Actually, no matter in the natural environments or in the engineering processes, the coupling between bacteria is very common. Some of them have been verified to have the competition relationship, but most bacteria are interacting and constituting the metabolism network to perform the substrate conversion processes. For example, some reports have demonstrated that the existence of nitrifiers in the anmmox consortia to consume trace oxygen spreading in the system, which could protect the toxic effects of oxygen on anammox bacteria [10]. The reasons for the difficult isolation of anammox bacteria have thus far remained as a question of interest lacking experimental insight. One possible explanation is that, anammox bacteria only express activity at conditions that the cell concentration is higher than 1010-1011 cell/mL [8].
Besides that, there is another question, which kinds of bacteria are usually associated with the anammox bacteria in different consortia and why they persistently existed with anammox bacteria. The elucidation of these questions is critical not only for better understanding anammox bacteria, but also for keeping the successful operation of anammox process. Bacteria composition is strongly dependent on anammox consortia obtained by different incubating environments according to the previous reports [9]. However, some bacteria strains are the common ones in the anammox consortia, namely core microorganism. The concept of core microorganism is defined as that, although the change of microbiological community is unavoidable, in the gene level, the identified microorganism existing here to exhibit special functions. The microbes that appear in all hosts are thought to fulfill a functional niche within the community and as such may provide valuable information for the ‘normal’ state of the community [11].
In this work, we analyzed six different anammox consortia obtained in different niches. Bacteria composition of the consortia was investigated by molecular microbiological technologies with the final aim to clarify the core microorganism of anammox consortia and the significant accompanying bacteria with anammox bacteria. The relationships of microbial communities with the nitrogen removal activities and nitrogen removal profiles were also investigated to further elucidate the anammox consortia behavior.
Material and Methods
Sampling
In this study, six anammox bacterial consortia were collected from different incubation environments. Sample 1 (S1) stands for bacterial consortium obtained from anammox pilot-scale plant [12]. S2 parameters bacterial consortium got from activated sludge and cultivated in anaerobic fermenter for more than one year. S3 is anammox consortium is also from activated sludge, but was seriously damaged by overheating. S4 denotes the anammox consortium obtained from upper part of percoll density centrifugation from S3 and anaerobically cultured for several months. S5 represents the anammox consortium being inhibited by high nitrite. S6 marks anammox consortium from ground water and anaerobically cultured for more than ten years in the lab [13].
Percoll density gradient centrifugation
The anammox consortium for percoll density gradient centrifugation (PDGC) was firstly washed by phosphate buffer solution (PBS) for three times. After centrifugation for 5 min at 5000 rpm to remove supernatant, the mixture of buffer and percoll solution with volume ratio of 3.1:6.9 was added. The dispersed cells were suspended in the mixture by vortex and then performed PDGC. The centrifugation time and speed were set up at 30 min and 8000 rpm. The anammox cells then appeared as a red band at the lower part of the gradient. After carefully extracted this red band by syringe needle, the purified anammox consortium in the bottom part of percoll density centrifugation was obtained after being washed in PBS. The residual bacteria in the upper part of the percoll solution were also collected for bacteria community analysis.
DNA extraction and cloning library construction
We analyzed the bacterial communities of the collected six different bacterial consortia through 16S rRNA clone library. Samples were taken in sterile polyethylene bottles and kept in an ice box immediately following collection. Then these samples were stored at -80°Cuntil the DNA extraction. After thawing, each sample was pipetted 3 mL mixedliquor into sterile eppendorf tubes and centrifuged at 14,000 xg for 10 min. The supernatant was decanted and the pellets of activated sludge samples were washed three times by centrifugation using sterile highpurify water for 5 min at 15,000 xg. DNA extraction was then performed using a Fast DNA SPIN Kit for Soil (MP Biotechnology, USA) according to the manufacturer’s protocol. DNA integrity was checked by 0.8% agarose gel electrophoresis and stored at -20°C.
PCR was firstly carried out by a universal bacterial forward primer 27F:5’- AGAGTT TGA TCC TGG CTC AG-3’ and reverse primer 1492R:5’-ACG GCT ACC TTG TTA CGA CTT-3’ to amplify the 16S rRNA genes [14]. For each sample, products of three replicate PCRs were pooled to minimize PCR artifacts. All PCR amplifications were performed using a GeneAmp PCR system 9700 thermocycler (Applied Biosystems). Each 50 μL PCR mixture contained 34.5 μL of sterile Milli-Q water, 5 μL of 10x rTaq PCR Buffer, 200 μM each deoxyribonucleoside triphosphate, 0.2 μM of each primer, 1.25 U of rTaq DNA polymerase (TaKaRa), and 20-50 ng of genomic DNA. PCR amplifications program was set up as previously described. The purified product was then ligated into the PCR-®4-TOPO ® plasmid vector and transformed into competent E. coli TOP 10 cells by using TOPO-TA cloning kit according to the manufacturer’s instructions (Invitrogen, USA). After cultivation at 37°C for 1 h, 100 colonies were randomly selected. Sequencing was performed by an auto sequencer (ABI PRISMTM3730 XL DNA Sequencer) using the universal bacteria primers M13F and M13R. The obtained DNA sequences were compared with the reference microorganisms available in the Gen Bank by BLAST search [15].
Clustering analysis
The shared bacterial community at different taxa levels was clustered by PCORD-5 software according to PCORD-5 tutorial. The detail data processing procedure is as follows. Firstly the bacterial community data matrix transforms to present-absent data matrix on PCORD-5 software. Then the processed data matrix was clustered by Sorensen (Bray-Curtis) distance.
Batch test for testing anammox activity
The ammonium and nitrite removal profile of the anammox consortia were determined in the anaerobic serum bottles. The consortium was washed and resuspended in the cultivation medium as described previously [8]. pH was adjusted to 7-8 with 0.1M NaOH or 0.1M HCl. After this, the headspace and liquid phase were sparged with nitrogen gas to keep dissolved oxygen (DO)<0.5 mg/L. After an experiment cycle, which last for 24-72 h depending on the bacterial activities, the supernatant was poured and fresh medium was added. A syringe needle was inserted to the cover of the vessels to measure the gas volume produced in a cycle. The reactions were conducted at 35°C. Samples were taken at the beginning and the end of the cycles to measure nitrogen compounds concentrations and Volatile Suspended Solids (VSS).
Chemical analysis
The concentrations of ammonium, nitrite, nitrate and VSS were measured according to standard methods, as set out by the American Publish Health Association [16]. Total nitrogen (TN) was measured by using TOC analyzer (ASI-V, Germany). DO was measured by DO analyzers (Mettler, Switzerland). pH and Oxidation-reduction potential (ORP) were obtained by pH meter (Mettler, Switzerland).
Results and Discussion
Bacterial diversity of anammox consortia
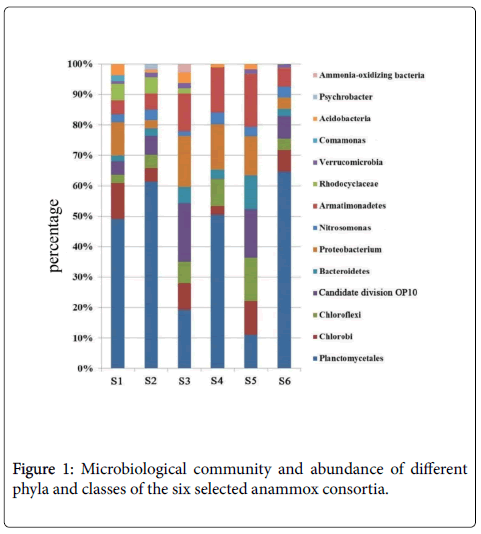
In this study, to investigate the phylogenetic diversity of bacteria communities of anammox consortia, six clone libraries data of 16S rRNA genes from different incubation environments were analyzed. In total, 500 sequences were obtained from the six libraries and they were identified as 16S rRNA gene sequences using the NCBI blast. Figure 1 showed that nearly all the bacteria consortia contain the bacteria strains belonging to Planctomycetes, Chorobi, Chloroflexi, Bacteroidetes, Proteobacterium, Nitrosomonas and Armatimonadetes . It is easily to be discovered that, the Planctomycetes percentages are distinct for these anammox consortia, the values of which were identified as 49%, 62%, 19%, 51%, 11% and 65% for S1, S2, S3, S4, S5 and S6, respectively. It could be well understood that the obvious higher Planctomycetes percentage for S2 than S1, due to the effective cultivation and enrichment of anammox bacteria in anaerobic conditions.
S3 was the inhibited anammox consortium by over-heating, and this process inevitably damaged the bacteria and decreased the percentage of functional Planctomycetes . In this part, the pretreatment by ultrasound as proposed previously [8] was ignored, inducing the incomplete separation of the anammox bacteria. Even though, the increased purity of Planctomycetes was observed at the bottom of the percoll solution (S4). As mentioned in the methods, S6 was the one anaerobically cultured in the lab for more than ten years and thus the bacteria community seems to be stable and the Planctomycetes accounts for as high as 65%. For the steady operation of the reactors, we could regard that S2 and S6 all have steady bacteria community with the Planctomycetes percentage of around 60-70%.
Similarity analysis of the six anammox consortia
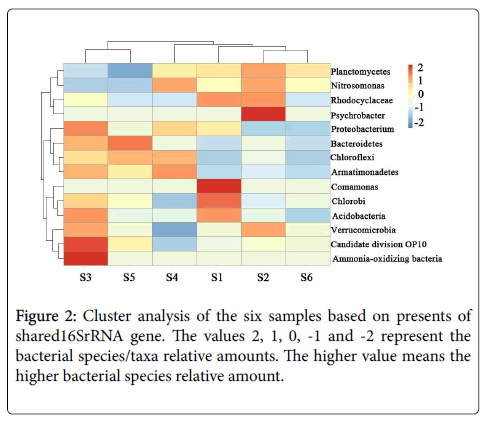
The similarity of the six samples was evaluated using clustering analysis. As we can see from Figure 2, the clustering analysis based on present-absent of shared orders in samples revealed that bacterial communities in the six samples could be clustered into two groups: group I includes samples S3 and S5. These two samples have a low Planctomycetes percentage with high bacteria diversity. Group II contains samples S1, S2, S4 and S6, among which samples S2 and S6 show high similarities. This finding was also consistent with the result of Planctomycetes percentage analysis and obviously suggested that the stable anammox consortia have similar community structure, even with different origins (one from activated sludge and one from ground water). These two samples have comparatively high Planctomycetes percentage and low bacteria diversity, since they were collected from the steady anaerobically operated container. Therefore, we can draw the conclusion that the anammox bacteria communities are similar when they were cultivated to stable stage, even with different origins.
Identification of core microorganism in the anammox consortia
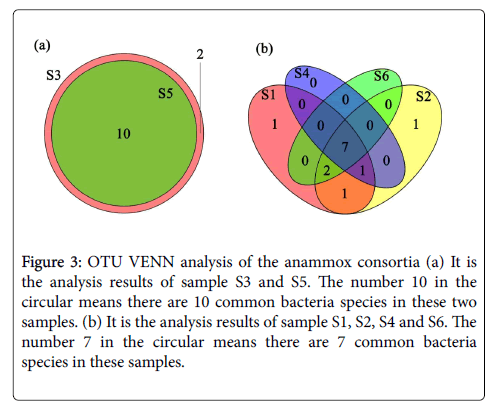
Although the consortia were from different incubation environments, they have some other common bacteria strains besides Planctomycetes . The relationship of these bacteria strains and the common part has been identified and shown in Figures 3a and 3b. Thes dominant common bacteria in the these consortia were identified as Planctomycetes, Chlorobi, Chloroflexi, Bacteroidetes, Proteobacterium, Nitrosomonas and Armatimonadetes linkages, the clones of which were in range of 10-30% for all these members. We then deduce that Chlorobi, Chloroflexi, Bacteroidetes, Proteobacterium, Nitrosomonas and Armatimonadetes may be the bacteria species which are easily accompanied with Planctomycetes (special functional bacteria for anammox reaction) in anammox consortia.
Figure 3: OTU VENN analysis of the anammox consortia (a) It is the analysis results of sample S3 and S5. The number 10 in the circular means there are 10 common bacteria species in these two samples. (b) It is the analysis results of sample S1, S2, S4 and S6. The number 7 in the circular means there are 7 common bacteria species in these samples.
Besides, Acidobacteria, Verrucomicrobia and Rhodocyclaleae were detected in the damaged or inhibited anammox consortia (samples S3 and S5). Bacteroidetes and Proteobacterium are considered to be responsible for the degradation of protein and the fermentation of acetate acid. The species in the genus Chorobi and Chloroflexi are facultative anaerobic bacteria and play important roles in fixing CO2 for growth [17]. This study verified the concomitant existence of these bacteria with Planctomycete in anammox consortium under anaerobic condition. Nitrosomonas are recognized to have functions to anaerobically oxidize ammonium [18,19].
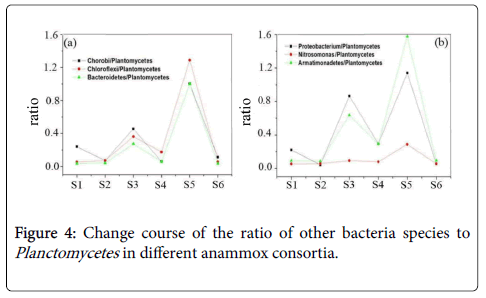
Members of Acidobacteria are fermentative and could degrade small molecular acids [5]. The species of Verrucomicrobia can oxidize methane and play important roles in the anaerobic fermentation of organic compounds [20]. For the inhibited anammox consortia, the increase of these bacteria could be well understood for the existence of many dead bacteria or cell fractions with potential release of protein and organic compounds. In addition to the observation of the core and accompanying bacteria, we have explored the composition of the bacterial community of anammox consortia, in aspect of the proportional relationship of different bacteria species to Planctomycetes . Corresponding ratios of Chorobi/Planctomycetes, Chloroflexi/Planctomycetes, Bacteroidetes/Planctomycetes,Proteobacterium/Planctomycetes, Armatimonadetes/Planctomycetes and Nitrosomonas/Planctomycetes have been presented in Figure 4.
By the statistical analysis, note that almost the seven-key species has the same variation tendency with Planctomycetes, except for the Nitrosomonas species. These data once again verified the status as the significant accompanying microorganisms of Chorobi, Chloroflexi, Bacteroidetes, Proteobacterium and Armatimonadetes in the anmmox consortia. However, the ratio of Nitrosomonas/Planctomycetes was kept at a constant value. This result reflects that different from other species, Nitrosomonas decreased in bacteria consortium with low Plantomycetes percentage or anammox activity.
From Figures 4(a) and 4(b), we can also see that after long time culturing, sample S2 and sample S6, the ratios of Chorobi/ Plantomycetes, Chloroflexi/Plantomycetes, Bacteroidetes/ Plantomycetes, Proteobacterium/Plantomycetes, Nitrosomonas/ Plantomycetes and Armatimonadetes/Plantomycetes were lower than other bacterial consortia. In sample S2 and S6, these ratios are almost the same, which illustrates that the ratios of the above bacteria species and Plantomycetes tend to be stable after long time culturing. This further illustrates these bacteria are the bacteria species which are easily attached to Planctomycetes to constitute the anammox consortia. They could exist under the anaerobic condition with Planctomycetes together for long time. As previously reported, the bacteria species such as Verrucomicrobia, Proteobacterium and Bacteroidetes have the important functions for the degradation of proteins, polysaccharide and organic compounds [21-23]. The Extracellular Polymeric Substances (EPS) including proteins polysaccharide and organic compounds, released by the involved bacteria then could be used as the necessary energy source for these bacteria and ensure their existence in the anammox consortia. This result also confirmed that these bacteria directly and also inevitably emerged to be the accompanying bacteria with Planctomycetes species under the anaerobic and autotrophic conditions for long time.
Nitrogen removal profile of the six anammox consortia
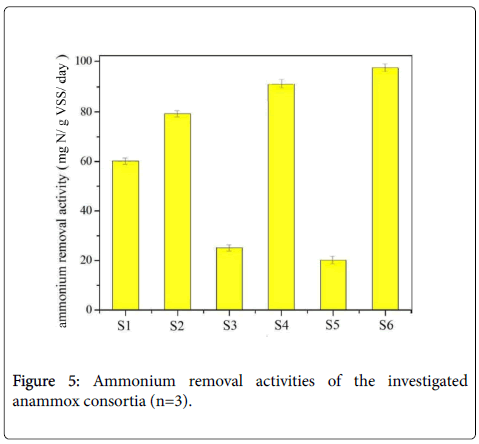
Microbiological community analysis revealed the significant difference of anammox consortia obtained from different incubation environments. Now we turn to the nitrogen removal profile of the six anammox consortia. The primary concern becomes whether the anammox activities were different and whether the nitrogen removal profiles have direct relationship with the microbiological community composition. We then investigated batch experiment to survey the ammonium removal activity and nitrogen removal profile of the anammox consortia provided in this study. This activity tests were performed for three times for getting the average value, and the six bacteria consortia have different nitrogen removal activities. The ammonium removal activities of these consortia were finally identified as 59 NH4 +-N gVSS-1 day-1, 78 NH4+-N gVSS-1 day-1, 24 NH4+-N gVSS-1 day-189 NH4+-N gVSS-1 day-1, 18 NH4+-N gVSS-1 day-1and 95 NH4+-N gVSS-1 day-1, respectively for samples 1, 2, 3, 4, 5 and 6. The relative results are displayed in Figure 5. Generally, there is a trend that the high ammonium removal activity responded to the high Planctomycetes percentage, the values of which have been verified as 49%, 62%, 19%, 51%, 11% and 65% for samples S1, S2, S3, S4, S5 and S6, respectively. However, not only the Plantomycetes percentage decided the ammonium removal activity. For sample S4, the bottom part of percoll density centrifugation contained about 51% Plantomycetes percentage, but it has a higher ammonium removal activity (89 NH4+-N gVSS-1 day-1) than sample S2, containing about 61% Plantomycetes (about 78 NH4+-N gVSS-1day-1). Bacteria construction may cause this. The bottom part of percoll density centrifugation has less bacteria species, and it contains the necessity species Chorobi, Chloroflexi, Bacteroidetes, Proteobacterium, Nitrosomonas and Armatimonadetes in the anammox consortia. The other main finding here was that, although absolute anaerobic condition was maintained, typical nitrogen removal characteristics of anammox reaction (the ratio of ammonium and nitrite was defined as 1:1.32) were not observed in some samples. The abnormal nitrogen removal profile with the overconsumption of ammonium to nitrite was specially found in samples S3 and S5. They converted ammonium and nitrite with the ratio of 1:0.51 as illustrated from the batch experiment. This ammonium removal related to the high inhibition levels of anammox consortia and the low Planctomycetes percentage. Cloning analysis addressed the existence of Rhodocyclaceae, Armatimonadetes, Acidobacteria and Verrucomicrobia in the consortia at the beginning with the over-consumption of ammonium, but declined in other consortia. The appearance of these species indicated the declination of the anammox consortia and the corresponding decreased ammonium removal activities.
Conclusions
In this study, we explored the 16S rRNA gene analysis to analyze the community composition and core microorganism of the six anammox consortia obtained from different incubation environments. The results show that the stable anammox consortia have similar community structure, even with different origins (e.g. activated sludge and ground water). The bacteria species belonging to Chlorobi, Chloroflexi, Bacteroidetes, Proteobacterium, Nitrosomonas and Armatimonadetes are the common accompanying bacteria with Plantomycetes in anammox consortia. They could exist with Plantomycetes in the anaerobic conditions for a long period, the amount of which varied simultaneously for anammox consortia. The bacteria species belonging to Rhodocyclaceae, Armatimonadetes, Acidobacteria and Verrucomicrobia emerged as the declination of anammox consortia, suggesting the decrease of ammonium nitrogen activity and the abnormal nitrogen removal profile. These findings are useful to advance the understanding of anammox microbial community, which correlates with the stability and robustness of anammox consortia.
Acknowledgements
The authors are grateful to the foundation (No. JCYJ20130402094730839 and No. CXZZ20130517152035590) for financial support.
References
- Kuenen JG (2008) Anammox bacteria: from discovery to application. Nature Reviews Microbiology 6: 320-326
- Abma WR, Driessen W, Haarhuis R, van Loosdrecht MC (2010) Upgrading of sewage treatment plant by sustainable and cost-effective separate treatment of industrial wastewater. Water Science and Technology 61: 1715-1722.
- Jetten M, Schmid M, Schoonen KVP, SinningheDamste J, Strous M (2005) Anammox organisms: enrichment, cultivation and environmental analysis. Methods in enzymology 397: 34-57.
- Banihani Q, Hadadin N, Jamrah A (2012) Start-up of anaerobic ammonium oxidation (anammox) from conventional return activated sludge in up-flow anaerobic sludge blanket reactor for autotrophic nitrogen removal from wastewater. Jordan Journal of Civil Engineering 6: 17-27.
- Costas AMG, Liu Z, Tomsho LP, Schuster SC, Ward DM, et al. (2012) Complete genome of CandidatusChloracido bacterium the thermophilum, achlorophy II- based photoheterotroph belonging to the phylum Acidobacteria. Environmental microbiology 14: 177-190.
- Strous M, Kuenen JG, Jetten MSM (1999) Key physiology of anaerobic ammonium oxidation. Applied and Environmental Microbiology 65: 3248-3250.
- Kartal B, vanNiftrik L, Keltjens JT, OpdenCamp HJ, Jetten MS (2012) Anammox-growth physiology, cellbiology and metabolism. AdvMicrobPhysiol 60: 211-262.
- Strous M, Fuerst JA, Kramer EHM, Logemann S, Muyzer G, et al. (1999) Missing lithotroph identified as a new planctomycete. Nature 400: 446-449.
- Kindaichi T, Yuri S, Ozaki N, Ohashi A (2012) Ecophysiological role and function of uncultured Chloroflexi in ananammox reactor. Water Science and Technology 66: 2556-2561.
- Liu ST, Yang FL, Xue Y, Gong Z, Chen HH, et al. (2008) Evaluation of oxygen adaptation and identification of functional bacteria composition for anammox consortium in non-woven biological rotating contactor. Bioresource Technology 99: 8273-8279.
- Meriweather M, Matthews S, Rio R, Baucom RS (2013) A 454 Survey Reveals the Community Composition and Core Microbiome of the Common Bed Bug (Cimexlectularius) across an Urban Landscape. PLoSONE 8: e61465
- Liu ST, Mueller E, Horn H (2013) A systematic insight into a single-stage deammonification process operated in granular sludge reactor with high-loaded reject-water: characterization and quantification of microbiological community.Journal of Applied Microbiology 114: 339-351.
- Liu ST, Zhang ZT, Ni JR (2013) Effects of Ca2+ on activity restoration of the damaged anammox consortium. Bioresource Technology 143:315-321.
- Lane DJ (1991) Nucleic acid techniques in bacterial systematic. John Wiley & Sons, New Jersey, USA 5: 205-248.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. Journal of Molecular Biology 215: 403-410.
- APHA (1998) Standard methods for the examination of water and wastewater. American Public Health Association, Washington, USA.
- Hug LA, Castelle CJ, Wrighton KC, Thomas BC, Sharon I, et al. (2013) Community genomic analyses constrain the distribution of metabolictraits across the Choloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome 1: 22
- Poth M, Focht DD (1985) 15N kinetic analysis of N2O production by Nitrosomonaseuropaea: an examination of nitrifierdenitrification. Applied and Environmental Microbiology 49: 1134-1141.
- Zart D, Bock E (1998) High rate of aerobic nitrification and denitrification by Nitrosomonaseutropha grown in a fermentor with complete biomass retention in the presence of gaseous NO2 or NO. Archives of Microbiology 169: 282-286
- Lee KC, Webb RI, Janssen PH, Sangwan P, Romeo T, et al. (2009) Phylum Verruco microbial representatives share a compartmentalized cell plan with members of bacterial phylum Planctomycetes. BMC Microbiology 9:5
- LakshmiKV, SasikalaCh, RamanaChV (2009) Rhodoplanespokkaliisoli sp. nov., a phototrophic alphaproteo bacterium isolated from a waterlogged brackish paddy soil. International Journal of Systematic and Evolutionary Microbiology 59: 2153-2157.
- Martinez-Garcia DM, Swan BK, Arnosti C, Chain PSG, Reitenga KG, et al. (2012) Capturing Single Cell Genomes of Active Polysaccharide Degraders: An Unexpected Contribution of Verrucomicrobia. PLoSONE 7: e35314
- Kabisch A, Otto A, Konig S, Becher D, Albrecht D, et al. (2014) Functional characterization of polysaccharide utilization loci in the marine Bacteroidetes âÂ?Â?GramellaforsetiiâÂ?Â? KT0803. The ISME Journal 8: 1492-1502
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 3942
- [From(publication date):
March-2017 - Apr 02, 2025] - Breakdown by view type
- HTML page views : 3032
- PDF downloads : 910