Identification and Comparison of Venom Toxin Proteins of Puff Adder and African Bush Viper by Mass Spectrometrum
Received: 20-Jul-2022 / Manuscript No. ECR-22-69662 / Editor assigned: 23-Jul-2022 / PreQC No. ECR-22-69662 / Reviewed: 06-Aug-2022 / QC No. ECR-22-69662 / Revised: 10-Aug-2022 / Manuscript No. ECR-22-69662 / Published Date: 17-Aug-2022
Abstract
Tropical and equatorial Africa is a paradise for the herpetologist. Many species of reptiles are present, but often difficult to discover and observe [1]. With an area of 2,345,095 km2 and crossed by lakes, rivers, forests (equatorial and tropical) and savannas, the Democratic Republic of Congo has several geozoological zones that are near 168 species of snakes. In addition to these reptiles, Congolese forests and savannas are home to many other poisonous animals such as scorpions, ants, bees, etc. The venoms of all these animals are rich sources of biomolecules. Currently, only 0.01% of the 40 million toxins estimated in nature are characterized [2]. But it is paradoxical to note that despite its rich biodiversity in poisonous animals, the Democratic Republic of Congo has no internal researchers interested in work on venoms. However, this sector is booming in the 21st century and offers particularly fertile opportunities for medical research and modern therapeutics. It is in this context that we are interested in studying the biochemical composition of the venoms of certain snakes, in this case Puff Adder and African Bush Viper. These two vipers are typically African [3, 4] and populate the Congolese ophidian fauna. The first is terricolous but the second is arboreal. In the past, herpetologists used the morphology of snakes for their classification. But currently, we are using more and more genetic codes of these reptiles. Chemists and biochemists, for their part, try to use appropriate analytical techniques for the identification and characterization of the constituents of venoms in order to discover the molecules of interest but also to facilitate the work of the systematicians
keywords
Poisonous; Miniaturization; Venom
Introduction
Modern analytical techniques have developed strongly in recent decades with respect to the growth of sensor miniaturization, data digitization and computing power to enable the use of high performance mathematical tools with equipment less and less bulky [5,6].
Biochemists currently have a wide variety of techniques for analyzing biomolecules such as snake venom proteins. The frenetic research of scientists for the high precision that must characterize the results of their work always pushes them to perfect their analysis techniques on the one hand and on the other hand, to combine them for a better performance [7]. Mass spectrometry has become a powerful analytical tool over the last 20 years. The discovery of new ionization modes (Electrospray ionization (ESI), matrix-assisted desorptionionization or MALDI), which earned their authors part of the Nobel Prize in Chemistry in 2002, made it possible to implement this family, and in almost all types of samples, and in particular to biological macromolecules [8]. In this study, we are interested in the development of an approach based on mass spectrometry to allow characterizing progressively the venoms of Congolese snakes. The results of this study will make it possible, on the one hand, to build a database of Congolese snake venom proteins and, on the other hand, to understand the differences between these two serpents in terms of the biochemical constitution of their respective venoms.
Biological material
The venoms of Puff Adder and African Bush Viper are taken manually from live specimens of the Lwiro serpentarium (South Kivu Province) in DR Congo. The venoms are then frozen and freeze-dried and stored at 4ºC.
Method
Our samples were analyzed using the Waters process with the assistance of MassLynx software (c: \ masslynx \ snake venom \ snake venom.pro \ acqudb \ 20151209_mse_60min) for data processing. The details of the different steps are as follows:
Run method parameters
Pump
Waters Acquity SDS
Run Time: 60.00 min
Comment:
Solvent Selection A: A1
Solvent Name B: 0.1% FA Acetonitrile
Switch 1: No Change
Switch 2: No Change
Switch 3: No Change
Seal Wash: 20.0 min
Chart Out 1: System Pressure
Chart Out 2: %B
System Pressure Data Channel: Yes
Flow Rate Data Channel: No
%A Data Channel: No
%B Data Channel: No
Primary A Pressure Data Channel: No
Accumulator A Pressure Data Channel: No Primary B Pressure Data Channel: No
Accumulator B Pressure Data Channel: No
Degasser Pressure Data Channel: No
Gradient Table
Time (min) Flow Rate %A %B Curve
1. Initial 0.300 98.0 2.0 Initial
2. 2.00 0.300 98.0 2.0 6
3. 40.00 0.300 50.0 50.0 6
4. 40.10 0.300 20.0 80.0 6
5. 50.00 0.300 20.0 80.0 6
6. 50.10 0.300 98.0 2.0 6
7. 60.00 0.300 98.0 2.0 6
Run Events: Yes
Gradient Start (Relative to Injection): 0 uL
Participate in pre-analysis: No
2D Repeat: No
Detector
Waters Acquity TUV
Run Time: 30.00 min
Wavelength Mode: Single Wavelength
Lamp On: On
Channel A..
Comment:
Wavelength: 205 nm
Sampling Rate: 20 points/sec
Data Mode: Absorbance
Time Constant: 0.1 sec
Auto Zero on Wavelength Change: Maintain Baseline
Auto Zero On Inject Start: Yes
Analog 1...
Sensitivity: 2.000 AUFS
Chart Polarity: Positive (+)
Voltage Offset: 0 mV
Enable Chart Mark: Yes
Run Events: Yes
Pulse Width: 1.0 sec
Rect Wave Period: 0.2 sec
Auto sampler
Waters ACQUITY FTN Auto Sampler
Run Time: 60.00 min
Comment:
Load Ahead: Disabled
Loop Offline: Automatic min
Wash Solvent Name: Water
Pre-Inject Wash Time: 0.0 sec
Post-Inject Wash Time: 6.0 sec
Purge Solvent Name: Water
Dilution: Disabled
Dilution Volume: 0 uL
Delay Time: 0 min
Dilution Needle Placement: 4 mm
Target Column Temperature: Off C
Target Sample Temperature: 8.0 C
Sample Temperature Alarm Band: Disabled
Syringe Draw Rate: Automatic
Needle Placement: Automatic
Pre-Aspirate Air Gap: Automatic
Post-Aspirate Air Gap: Automatic
Column Temperature Data Channel: No
Room Temperature Data Channel: No
Sample Temperature Data Channel: No
Sample Organizer Temperature Data Channel: No
Sample Pressure Data Channel: No
Preheater Temperature Data Channel: No
Seal Force Data Channel: No
End detector
Run Events: No
Sample Run Injection Parameter
Injection Volume (uL) - 4.00
End autosampler
End of experimental record..
Generic Instrument Postrun Report
Software Version: 1.60.2782
Firmware Version: 1.60.2274 (Aug 10 2013)
Checksum: 0xd71e0eda
Serial Number: E10TUV015A
Lamp On/Off Event: No
Lamp Life: 1744.00
Lamp Serial Number: 1F8ABD2
Flow Cell Type: Analytical LG
Flow Cell Path Length: 10.000 mm
Flow Cell Volume: 0.50 micro liters
Flow Cell Serial Number: 10346
Flow Cell Part Number: 205015016
Optics Temperature Stabilization Setting: Normal Temperature
Waters Acquity SDS Postrun Report
IcsVersion: 1.60.1872
Firmware Version: 1.60.267 (Aug 19 2013)
Checksum: 0x36f220a9
Serial Number: C14BUR100G
Minimum System Pressure: 2890.5
Maximum System Pressure: 4731.5
Average System Pressure: 4248.0
Minimum Degasser Pressure: 0.5
Maximum Degasser Pressure: 0.5
Average Degasser Pressure: 0.5
Software Version: 1.60.1774
Firmware Version: 1.60.364 (Sep 20 2013)
Checksum: 0x35d5392b
Serial Number: B14USM471G
Needle Size: 15.0
Minimum Sample Temperature: 8.0
Maximum Sample Temperature: 8.3
Average Sample Temperature: 8.2
Minimum Column Temperature: -0.2
Maximum Column Temperature: 0.0
Average Column Temperature: -0.2
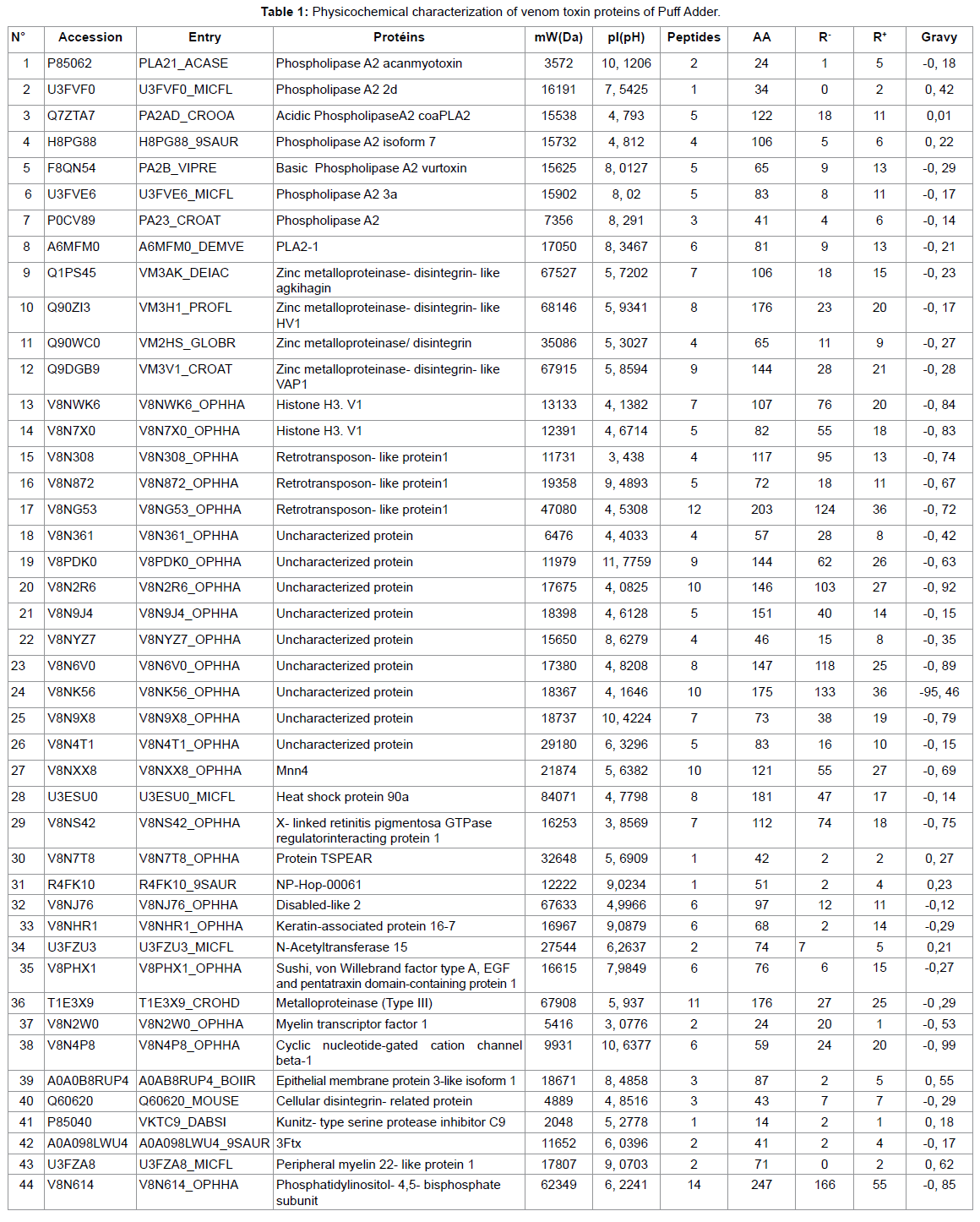
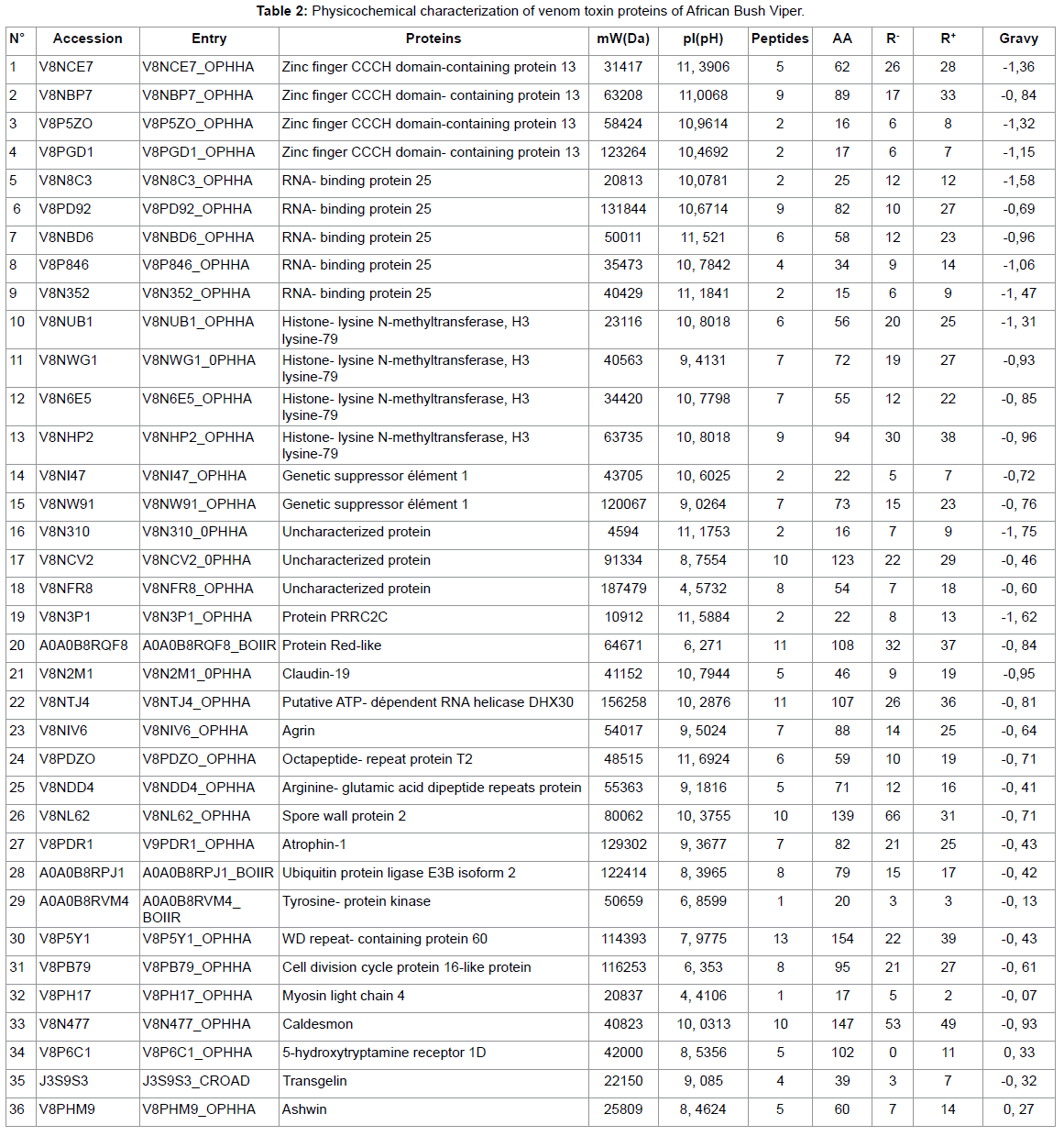
Results
HRMS Analysis of samples from snake venom
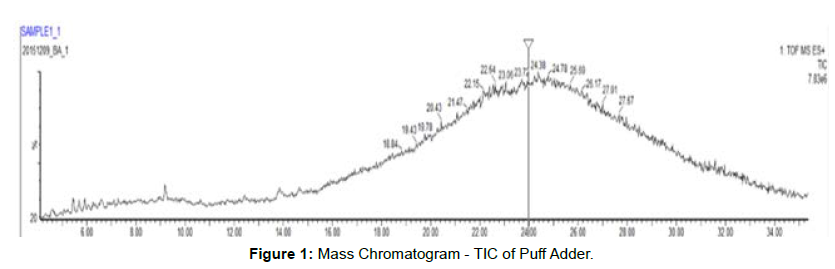
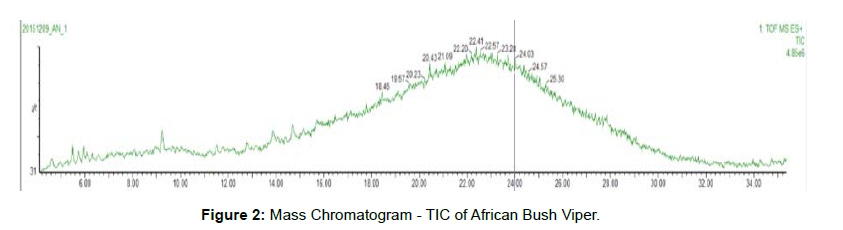
This data contains an overlay of total ion chromatogram of all two samples [Figures 1 and 2].
• Peptides have eluted from ~ 6 min to ~ 35 min.
• Due to presence of high number of peptides, the TIC gives a bellshaped curve.
•Every scan of the TIC gives mass spectral information of eluted/ co-eluted peptides.
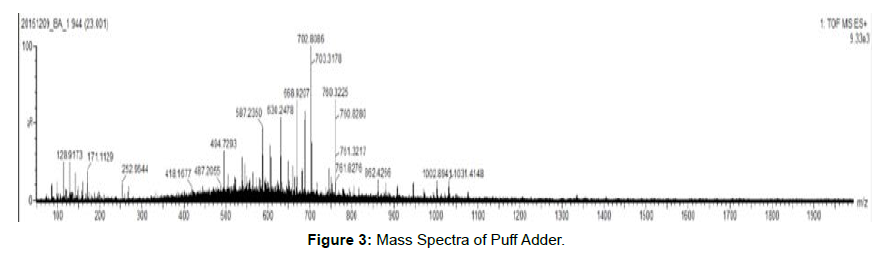
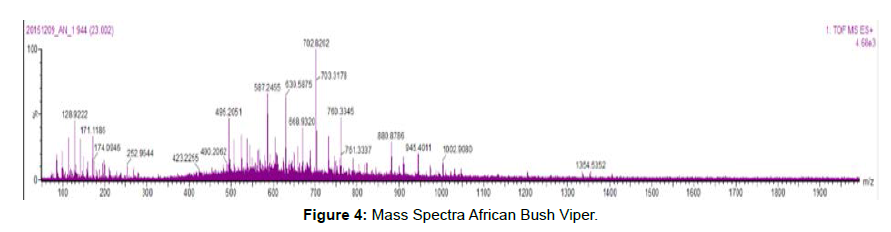
• This data contains an overlay of mass spectra of combined scan [Figures 3 and 4]

At ~24 min of the mass chromatogram, of all two samples.
• Mass spectral information is obtained for peptide present
Discussion
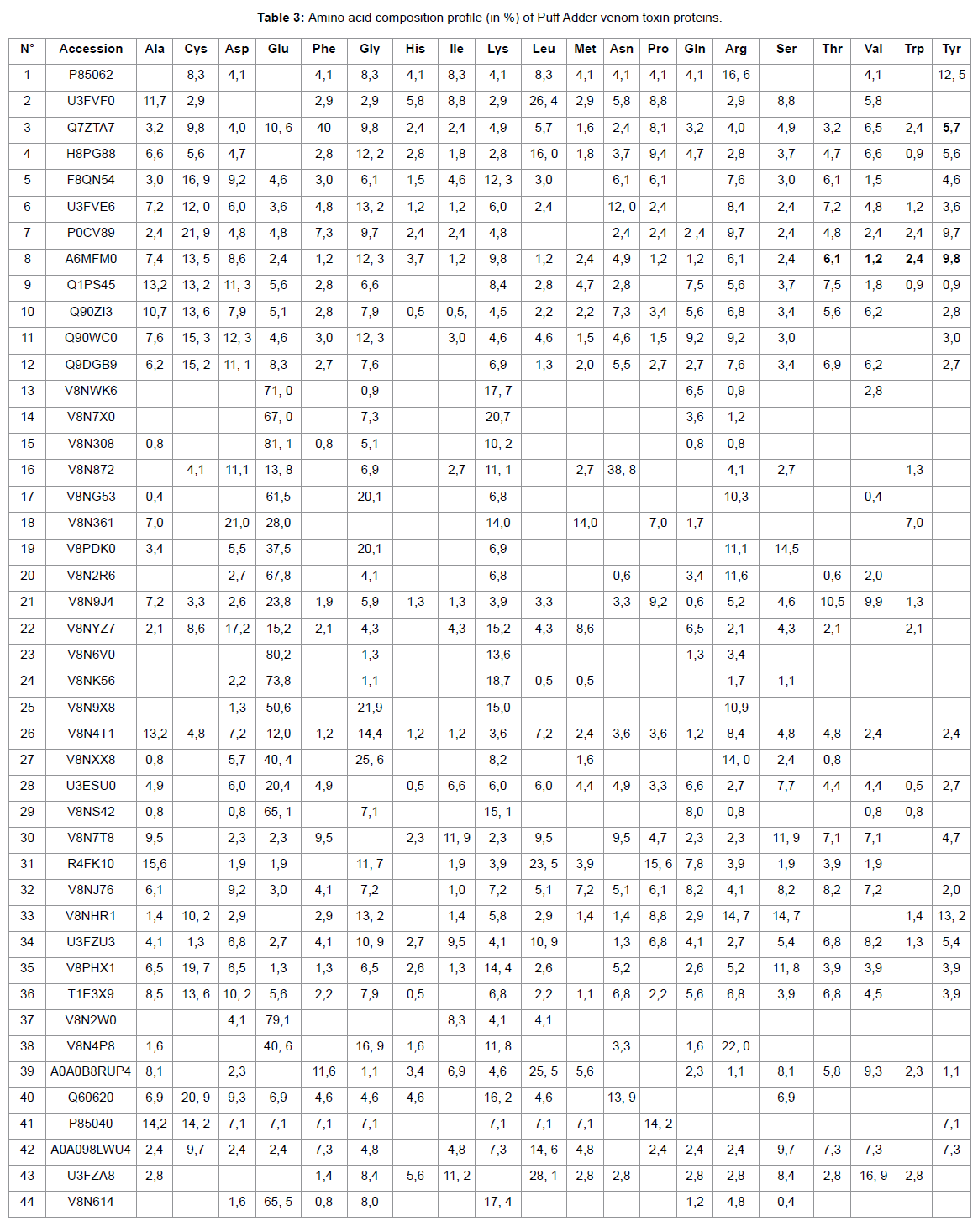
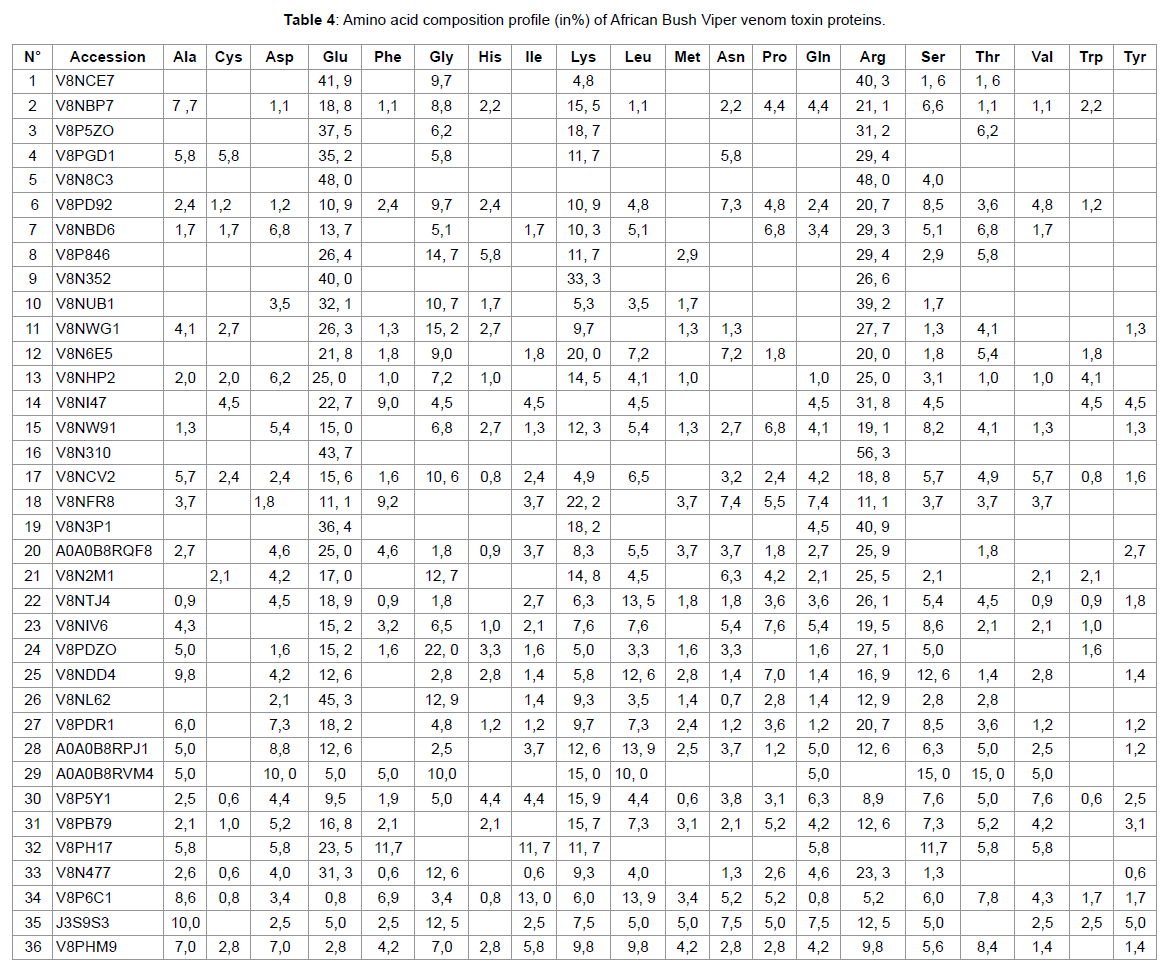
In the present study, snake venom toxin proteins of Puff Adder and African Bush Viper were analyzed with the help of Mass Spectrometric. The venom of Puff Adder contains 44 % of proteins against 36 % for African Bush Viper. Generally, snake venoms are considered to contain mainly proteins, ranging from 70 to 90 % (10). In their study by using the two-dimensional gel electrophoresis, Vijayan J. et al were reported the protein content of eight Malaysian snake venom was between 30 and 80% (11). According the molecular weight (Mw) of proteins, the Puff Adder venom contains 15, 9% of Mw (1000-9000 Da), 68, 2% of Mw (10000-49000 Da), 15, 9% of Mw (50000-99000 Da) and 0, 0% of Mw (100000-190000 Da). However, African Bush Viper venom comprise 2,8% of Mw (1000-9000Da), 44,4% of Mw (10000-49000 Da), 27,8% of Mw (50000-99000 Da) and 25,0% of Mw (100000-190000 Da). Both venoms under study belong to the Viperidae family. Several studies show that enzymes are high molecular weight proteins [9] this is what we find in this work. It should be noted that the African Bush Viper venom contains many enzymes of very high molecular weight compared to that of Puff Adder. The hydropathy index is a measure that allows knowing the hydrophilic or hydrophobic character of a region of a protein through the amino acid sequence. A positive value of the index corresponds to a hydrophobic behavior and a negative value corresponds to a hydrophilic behavior.
The GRAVY (medium hydropathy) of these two venoms shows that 18.2% of the Puff Adder venom proteins are hydrophobic and 81.8% are hydrophobic. However, in the African Bush Viper venom, 5.6% is hydrophobic and 94.4% hydrophilic. The amino acid profiles of the venom proteins examined show that Adder Puff possesses a venom rich in negatively charged amino acids (Asp + Glu) while that of African Viper has many positively charged amino acids (Arg + Lys). Lysine and arginine and their adequate presence help to become effective in the bio-molecule [10]. The most striking element in this study is that both venoms are made up of proteins of different natures. This leads some authors to talk about the variability of venoms that can be observed even within the same family. This variability of the venoms would be observed on the symptomatology related to the local necrosis for example, presented during an envenomation by these two different vipers. We also find that the African Bush Viper venom is free of phospholipase A2. However one of the most important protein superfamilies present in snake venoms are the phospholipasesA2 (PLA2, E.C. 3.1.1.4), a class of heat-stable and highly homologous enzymes, which catalyze the hydrolysis of the 2-acyl bond of cell membrane phospholipids releasing arachidonic acid and lysophospholipids.
Conclusion
At the end of our analysis, we realize that although both snakes belong to the same family of vipers, they have fundamentally different venoms from the point of view of their biochemical compositions. This leads specialists to talk about the variability of venom. This variability is genetic, that is, specific to each individual. The variations concern both the concentration of the different fractions and their biochemical structures [11]. Mass spectrometry has been an undeniable aid to us. This allowed us to scrutinize the secrets of the venoms of these two snakes. As our samples came from the eastern part of the country, it perhaps would be useful to extend this study to all geozoological areas of the Democratic Republic of Congo but also to other venomous species that this vast territory has.
References
- Rosselot B (1978) Les Serpents dangereux du Burundi, Ministère Français de la Coopération.
- Laura Droctove (2018) First vasopressin type 2 receptor antagonist Kunitz toxins: pharmacodynamics study and structure-activity relationships. Biochemistry, Molecular Biology. Paris-Saclay University.
Indexed at, Google Scholar, Crossref
- Chippaux J-P (2002) Venin’s et envenomation’s, IRD Paris 288.
- Chippaux J-P (2001) the Serpents of West and Central Africa, IRD Editions Paris 292.
- Gilles Ohanessian La (2008) mass spectrometry for chemical and biological analysis, X-ENS-ESPCI-UPS days.
- Nkinin Sw, Chippaux Jp, Pietin D, Doljanski Y, Tremeau O et al. (1997) genetic origin of venom variability: impact on the preparation of antivenom serums. Bull Soc Path Exot 90:277-281.
- Rodrigo G Stabeli, Rodrigo Simoes-Silva, Anderson M Kayano, Gizeli S Gimenez, Andrea A Moura, et al. (2020) Purification of phospholipases A2 from American Snake Venoms.
- Subhamay Panda, Goutam Chandra (2012) physicochemical characterization and functional analysis of some snake venom toxin proteins and related non-toxin proteins of other chordates. Bio information 8:891-896.
Indexed at, Google Scholar, Crossref
- Vejayan J, Shine Yee L, Ponnudurai G, Ambu S, Ibrahim I et al. (2010) Protein profile of Malaysian snake venoms by two-dimensional gel electrophoresis. J Venom Animal Toxin incl Trop Dis.
Citation: Ngambongo J (2022) Identification ET Comparison of Venom Toxin Proteins of Puff Adder and African Bush Viper by Mass Spectrometrum. Epidemiol Sci, 12: 453.
Copyright: © 2022 Ngambongo J, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Usage
- Total views: 1646
- [From(publication date): 0-2022 - Apr 26, 2025]
- Breakdown by view type
- HTML page views: 1268
- PDF downloads: 378