Research Article Open Access
HPLC-DAD-ESI-MS/MS Identification and Characterization of Major Constituents of Iris crocea, Iris germanica and Iris spuria Growing in Kashmir Himalayas, India
Gulzar Bhat1, Abdul S Shawl1*, Zeeshan Shah1 and Mudasir Tantry21Natural Products Chemistry Division, CSIR-Indian Institute of Integrative Medicine, Srinagar, Kashmir, India
2Phytochemistry Laboratories, Centre of Research for Development, University of Kashmir, Srinagar, Kashmir, India
- *Corresponding Author:
- Abdul S Shawl
Natural Products Chemistry Division
CSIR-Indian Institute of Integrative Medicine Srinagar
Kashmir 190005, India
Tel: + 91 0194 2431509
Fax: + 91 0194 2441331
Email: asshawl@gmail.com
Received date: October 01, 2014; Accepted date: November 13, 2014; Published date: November 17, 2014
Citation: Bhat G, Shawl AS, Shah Z, Tantry M (2014) HPLC-DAD-ESI-MS/ MS Identification and Characterization of Major Constituents of Iris crocea, Iris germanica and Iris spuria Growing in Kashmir Himalayas, India. J Anal Bioanal Tech 5:223. doi: 10.4172/2155-9872.1000223
Copyright: © 2014 Bhat G, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Analytical & Bioanalytical Techniques
Abstract
Genus Iris has long history of use in various indigenous systems of medicine as alternative aperient, stimulant, cathartic, diuretic, gall bladder diseases, liver complaints, dropsy, purification of blood, venereal infections, fever, ringworms, bilious infections and for a variety of heart diseases. Rhizomes of genus Iris are rich source of secondary metabolites and most of these metabolites are reported to possess anticancer, antiplasmodial, anticholinesterase, enzyme inhibitor and immunomodulatory properties. In our study, high performance liquid chromatography with diode array detector coupled with electrospray ionization mass spectrometry (HPLC-DAD-ESI-MS/MS) method was established for simultaneous identification of flavonoids and other constituents in the rhizomes of three Iris species (Iris crocea, Iris germanica and Iris spuria) growing in Kashmir Himalayas, India. About twenty seven compounds were identified in these Iris species based on chromatographic retention time (tR), UV and MS/MS spectra and compared with those of isolated authentic compounds and literature data. About seven constituents (iridalglycoside 5b, iridalglycoside 7, irisquin B, irilin A, hesperetin, irisolone and irisolodone) were reported for the first time from Iris crocea. Our results demonstrate the developed method could be employed as a rapid and versatile analytical technique for identification of chemical constituents and quality control of Iris samples.
Keywords
HPLC-DAD-ESI-MS/MS; Iris; Rhizomes; Chemical constituents; Isoflavones; Flavones
Introduction
Genus Iris is well known rhizomatous herb belonging to family Iridaceae and is represented by 260 species [1]. Iris species have immense medical importance in indigenous systems of medicine and are used in the treatment of liver dysfunction, inflammation, bacterial and viral infections [2,3]. Externally root in powder or poultice is used as an application to sores and pimples [2]. Rhizomes of Iris nepalensis are used in Indian Indigenous System of medicine under the name of ‘Sosan’ for variety of heart diseases [4]. They are used as laxative, antidote, antispasmodic, emetic and haemoststic agents [5,6]. In traditional Chinese medicine (TCM) rhizomes of Iris dichotoma Pall. (called in Chinese ‘Bai She Gan’ or ‘Bai Hua She Gan’) have been used to treat inflammations and respiratory system disorders [7], as well as to clear heat and detoxify the organism [8]. The compounds isolated from various species are reported to possess antimicrobial, antioxidant, hepatoprotective, immunomodulatory, piscicidal, anticancer, antiplasmodial, antituberculosis, molluscicidal and antihelmintic properties [9-15].
Rhizomes of various Iris plants are rich source of bioactive molecules, which belong to flavonoids, xanthones, stilbenes, simple phenolics, iridal type triterpenoids, quinines and irones [16]. Isoflavonoids are the major constituents in almost all Iris species. About 50 different isoflavonoids in the form of diglucosides, triglucosides or aglycones, are reported in Iris [14,17-21]. Consumption of isoflavonoids are reported to reduce the risk of cardiovascular disease and cancer and have diverse biological activities like antimicrobial, oestrogenic and insecticidal [22,23].
Since genus Iris is rich in bioactive phenolics and other constituents, analysis of these components is significant for quality control of the plants samples belonging to genus Iris. Therefore, the aim of this study was to identify the flavonoids and other constituents in the rhizomes of three Iris species (Iris crocea , Iris germanica and Iris spuria ) growing in the Kashmir Himalayas. Electrospray ionization mass spectrometry (ESI-MS) combined with high performance liquid chromatography with diode array detector (HPLC-DAD) provides a simple and versatile approach to identify the constituents in crude plant extracts. HPLC-DAD-ESI-MS/MS simultaneously provide UV, retention time (tR), mass spectra which are important for analysis and structural characterization of known compounds by comparing chromatograms and mass spectra with authentic markers. However there are no reports on identification of chemical constituents by HPLC-DAD-ESI-MS/ MS in Iris crocea , Iris germanica and Iris spuria growing in Kashmir. Therefore, a rapid, sensitive and easy HPLC-DAD-ESI-MS/MS method was developed for identification of flavonoids and other constituents in the rhizomes of these species.
Materials and Methods
Plant material
Rhizomes of Iris crocea were collected from Yarikah (Tangmarg) region of Kashmir valley while the rhizomes of Iris spuria and Iris germanica were collected from Field station of Indian Institute of Integrative Medicine (CSIR) Srinagar of J & K. The plant species identified from Center for Biodiversity and Taxonomy (CBT), Department of Botany, University of Kashmir and voucher specimens were deposited.
200 g rhizomes of Iris crocea , Iris germanica and Iris spuria were cleaned, cut to small pieces, air dried and powdered. The powdered materials were extracted with methanol (3 × 0.5l) in an ultrasonic bath for 30 min each time. Combined extracts were filtered and concentrated in vacuum to obtain a crude extract (20 g) each of Iris crocea, Iris germanica and Iris spuria .
Eleven reference compounds such as apocynin, picied, tectoridin, tectorigenin-4`-glucoside, iridin, tectorigenin, irigenin, 5,2`,3`-trihydroxy-7-methoxyflavanone, irisflorentin, 5,7-dihydroxy- 6,2`-dimethoxyisoflavone and crocetenone were isolated from rhizomes of Iris species in our laboratory [17-19, 24]. The purity of each marker was found to be greater than 96% by HPLC.
Sample Preparation and solvents
Analytical samples (methanol extracts and eleven reference compounds) at the concentration of 1 mg/ml were prepared by dissolving in methanol followed by filtration over 0.22 μm nylon HPLC filter. All samples were kept at 4°C before use. LCMS grade acetonitrile (Sigma Aldrich), LCMS grade water (Merck), analytical grade acetic acid (Sigma Aldrich) and LCMS grade methanol (Sigma Aldrich) were used in the analysis work.
HPLC procedures
Chromatographic separation was carried out using Enable RP C18 Column (250 mm × 4.5 mm, 5 μm) at 25°C. Elution was performed at a flow rate of 1 ml/min. Solvents used were acetonitrile (A) and 0.05% acetic acid in water (B). All solvents were filtered through a 0.45 μm nylon filter after ultrasonic degassing. Mobile Phase Gradient was as follows 5% B at 0 min, 10% B at 10 min, 25% B at 30 min, 40% B at 48 min and 50% B at 65 min. From 65 min to 75 min column was reequilibrated to initial condition. The chromatograms were detected at 270 nm and data was acquired using Lab solutions software.
Instrumentation
Liquid chromatography separation was performed using Nexera UHPLC composed of quaternary pump, prominence degassing unit, Autosampler, Column oven and Diode Array Detector (DAD).
ESI-MS parameter
Nexera UHPLC coupled with Shimadzu Co. ltd (Kyoto, Japan) LCMS-8030 was used in both positive and negative ion modes. The MS parameters were as follows: negative and positive ionization modes, scan range from m/z100 to 1000, DL temperature 250°C, Nebulizing gas flow 3 L/min, Heat block temperature 400°C and drying gas flow 15 L/min. For MS/MS positive ion fragmentation, collision energy of -35 V was used and for negative ion fragmentation, collision energy of +35 V was used.
Results and Discussion
Optimization of HPLC-DAD-ESI-MS/MS conditions
In order to achieve good resolution and separation of isoflavonoids and other constituents in the rhizomes of Iris crocea , Iris germanica and Iris spuria , different HPLC conditions such as column temperature, mobile phase and detection wavelength were examined. We started with gradient profile of acetonitrile-methanol at the column temperature of 20°C and detection wavelength of 269 nm but it does not show good separation of peaks. This was followed by gradient solvent system of acetonitrile (B) and water (A) at column temperature of 25°C and detection wavelength 270 nm. This solvent system showed good resolution of peaks within specific time and was finally selected as mobile phase system for analysis. Further, addition of 0.05 % acetic acid in waters (A), chromatographic separation of marker compounds and extracts was greatly improved with sharper peaks. There was small variation in retention times because of complicated gradient solvent system.
For MS analysis both positive and negative modes of ESI were examined with the scan range ofm/z 100-1000. All the marker compounds showed [M - H]¯ and/or [M + H]+ / [M + Na]+ ions of abundance and were subjected to MS/MS analysis. For positive ion fragmentation in MS/MS, collision energy in the range of -5 V to -35 V was used while for negative ion fragmentation collision energy was in the range of +5 V to +35 V. Fragmentation of negative ions was found best at +35 V and for positive ions at -35 V and the resulting product ions were used as fingerprints of each molecule.
HPLC-DAD-ESI-MS/MS of reference compounds
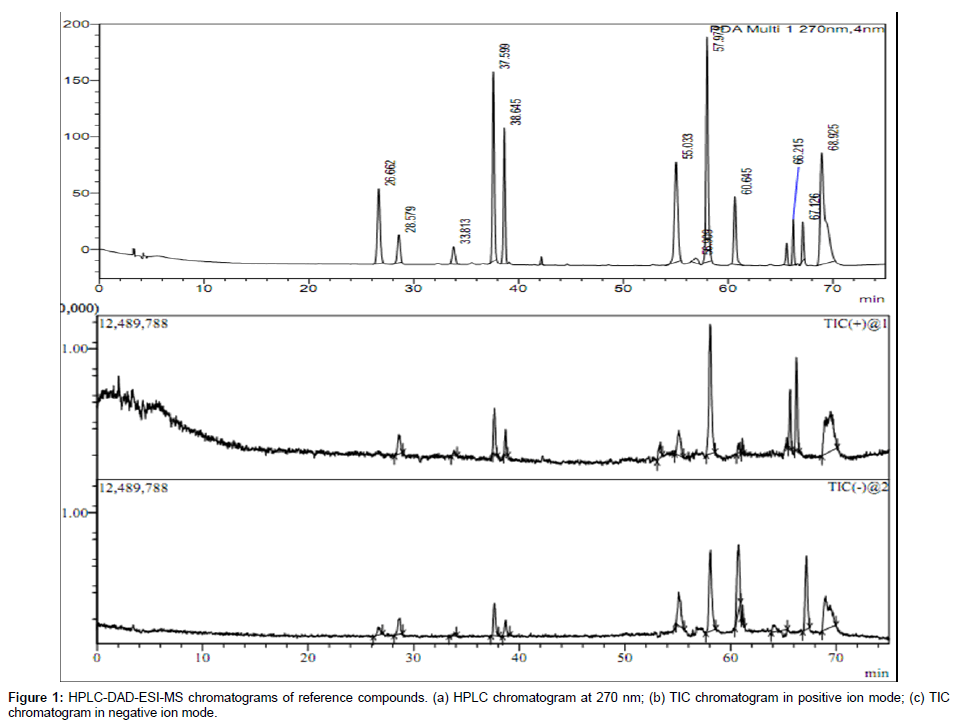
In this study, eleven compounds authentic markers were studied for their dominant fragmentation pathways. Most of the compounds in MS exhibited abundant [M + H]+ in the positive ion mode and [M - H]¯ in negative ion mode and were subjected to MS/MS analysis. UV, retention time (tR) and mass spectral characteristics of all markers compounds is given in Table 1. HPLC chromatogram and total ion currents (TIC) mass spectrum of marker compounds in positive and negative ion modes is shown in Figure 1.
| Peak No. | tR (min) | UV λmax (nm) | [M+H]+ (m/z) | Fragment ions (+) | [M-H]Ë? (m/z) | Fragment ions (-) | Compound |
|---|---|---|---|---|---|---|---|
| 1 | 26.66 | 229,275,305(sh) | NDa | NDa | 165 | 150,122,107 | Apocynin |
| 2 | 28.57 | 218,319(sh) | 391 | NDa | 389 | 227,182,143 | Picied |
| 3 | 33.81 | 265,332(sh) | 463 | 301,183,103,85 | 461 | 446,283,227,162,136 | Tectoridin |
| 4 | 37.59 | 213,263,339(sh) | 463 | 343,329,301,286,127,109,85 | 461 | 446,283,255,239,195,167 | Tectorigenin-4`-glucoside |
| 5 | 38.64 | 215,265,339(sh) | 523 | 361,346,310,282 | 521 | 491,359,343,313 | Iridin |
| 6 | 55.03 | 264,314 | 301 | 285,268,168,122 | 299 | 284,255,240,211,132,110 | Tectorigenin |
| 7 | 57.97 | 221,266 | 361 | 345,315,303,245,208,193,107 | 359 | 344,328,329,314,285,258,270,191 | Irigenin |
| 8 | 60.64 | 219,289,344(sh) | NDa | NDa | 301 | 286,253,166,155,140,120,110,87 | 5,2`,3`-trihydroxy-7-methoxyflavanone |
| 9 | 66.21 | 264,333(sh) | 387 | 357,342,326,311,279,206, 165,132,120 | NDa | NDa | Irisflorentin |
| 10 | 67.12 | 224,297,348(sh) | NDa | NDa | 313 | 298,224,213,192,166,138,123,110 | Crocetenone |
| 11 | 68.90 | 263,346(sh) | 315 | 299,285,269,254,169,133 | 313 | 298,283,267,255,211,183,167,141,117 | 5,7-dihydroxy-6,2`-dimethoxyisoflavone |
tR: Retention time.
Sh: shoulder peak.
aThe ion signals of this component investigated were not detected in this ion mode.
Table 1: Chromatorgraphic, UV and mass spectral characteristics of marker compounds.
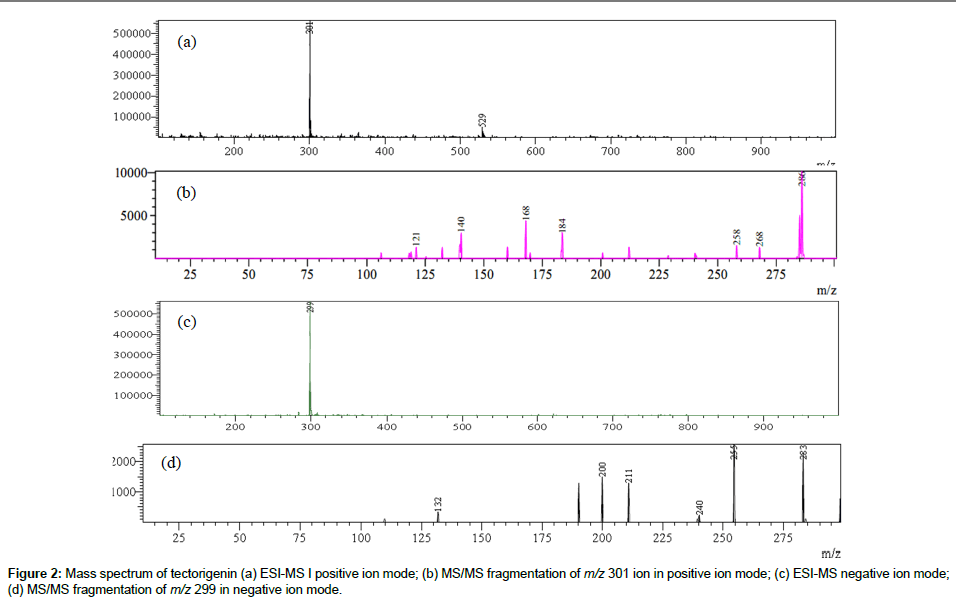
MS/MS data obtained was used for structural identification of each marker compound. Most of these markers were isoflavonoids and their MS/MS data revealed some common features such as neutral loss of CH3, OCH3, H2O and loss of glucose residue in case of isoflavone glycosides. Elimination of CH3 indicates the presence of methoxy group while loss of CO is due contraction of C-ring [25]. Besides Retro-Diels- Alder (RDA) fragment ions (pathway I) were also observed, which are important for determination of substitutions on -A and -B rings of flavonoid nucleus. The mass spectrum (both ESI-MS and MS/MS) of tectorigenin both in positive and negative ion modes is shown in Figure 2. The molecular ion peak at m/z 301 [M + H]+ at -35 V in the positive ion mode showed MS/MS fragment ions at m/z 286 [M+H-CH3]+ and 258 [M+H-CH3-CO]+, while as molecular ion peak at m/z 299 [M - H]¯ in negative ion mode showed MS/MS fragment ions at m/z 283 [M-2H-CH3;]- and m/z 240 [M-2H-CH3-CO]¯. In our study, loss of CH3 radical was dominant fragmentation in positive ion mode in case of isoflavones due to formation of stable radical structure [7,25,26]. Besides diagnostic Retro-Diels-Alder (RDA) ions of tectorigenin were observed in positive ion mode at m/z 184 followed by loss of CH3 (m/z 168), due to cleavage of two C-C bonds of C-ring. These RDA fragments indicate the presence of a methoxy group in A-ring of tectorigenin. The possible fragmentation pathway of tectorigenin is shown in Scheme 1.
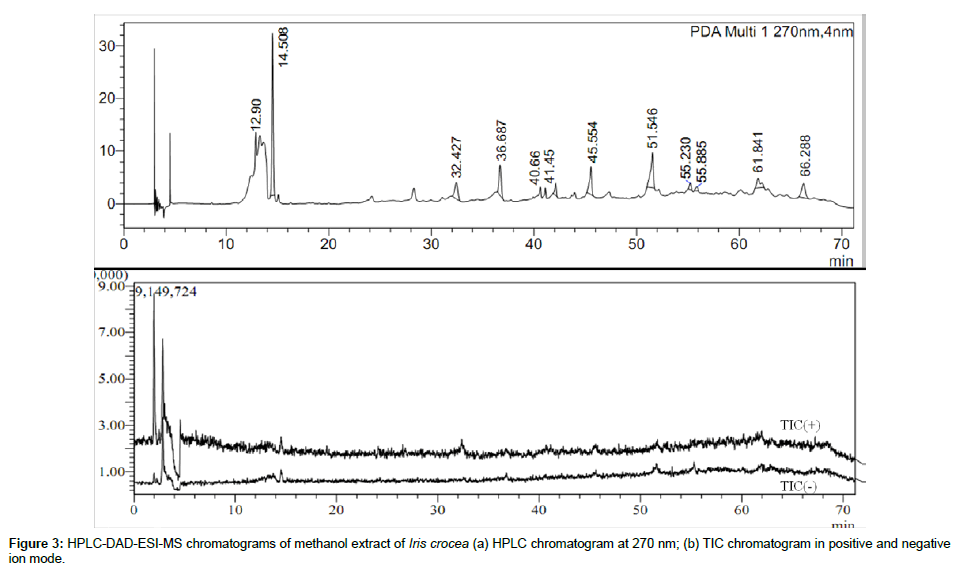
HPLC-DAD-ESI-MS/MS of Iris crocea
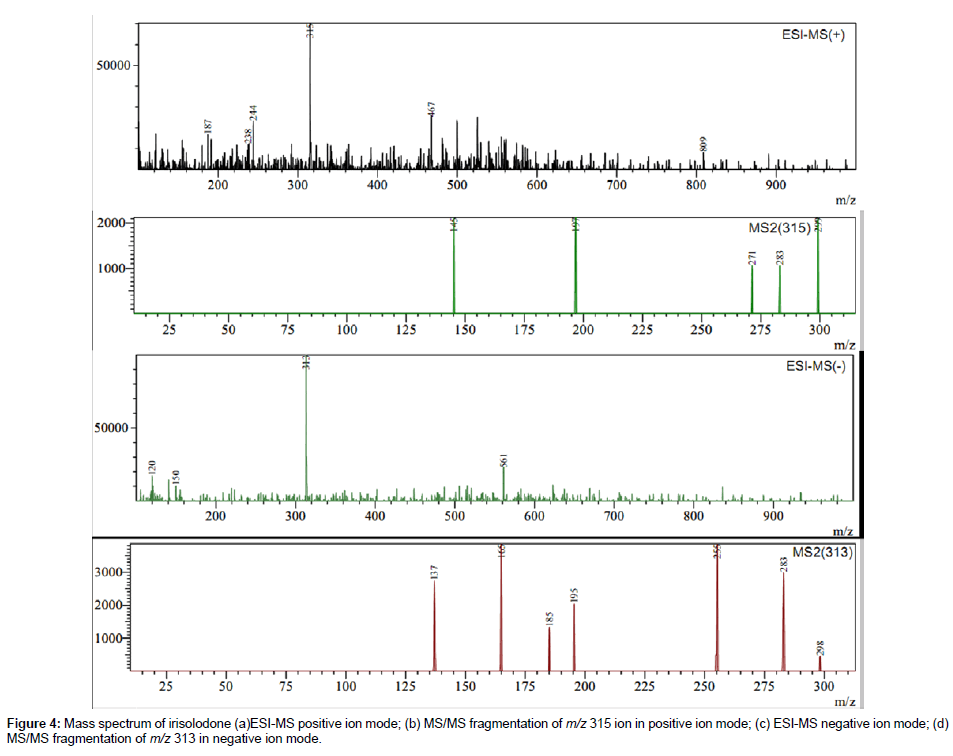
In Iris crocea 12 peaks in HPLC-DAD and HPLC-MS (TIC) chromatograms were identified as shown in Figure 3, Table 2. Only three peaks were identified by comparing their retention time (tR), UV spectra and MS/MS data with those of reference compounds. The other nine peaks were tentatively identified as Tectorigenin 4`-gentiobioside- 7-glucoside, Tectoridin 4`-glucoside, Iridalglycoside 5b, Iridalglycoside 7, irisquin B, Irilin A, hesperetin, irisolone and irisolodone, by comparing molecular weight, UV spectra and characteristic MS/MS data with reference to literature [16,19,20,27-33]. Rhizome of Iris crocea was found rich in isoflavones and most of these isoflavones showed common fragmentation pathway as described previously. For example the ESI-MS of irisolidone in positive gave molecular ion peak atm/z 315 and in negative ion mode at m/z 313. MS/MS of both molecular ion peaks showed successive loss of two methyl residues which indicates the presence of two methoxy groups. In addition, there was neutral loss of CO which shows molecular ion peak at m/z255. RDA fragments were also observed in MS/MS of both positive and negative ion mode. The mass spectrum (MS & MS/MS) in both positive and negative ion mode is shown in Figure 4.
| Peak No. | tR (min) | UV λmax (nm) | [M+H]+ (m/z) | Fragment ions (+) | [M-H]Ë? (m/z) | Fragment ions (-) | Compound |
|---|---|---|---|---|---|---|---|
| 1 | 12.90 | 262 | 787 | 595,512,425,352,297, 266,234,151,174 |
785 | 481,469,452,344,329, 281,236,227,204,147 |
Tectorigenin 4`-gentiobioside-7-glucoside |
| 2 | 14.50 | 210,263,327 | 625 | 509,385,301,232,109,85 | 623 | 461,445,298,227 | Tectoridin 4`-glucoside |
| 3m | 32.42 | 263,329(sh) | 463 | 301,299,183,103,85, | 461 | 446,283,256,227,162,147, | Tectoridin |
| 4m | 36.60 | 262,339(sh) | 463 | 344,301,286,258 | 461 | 446,397,365,283,255,233,195,167 | Tectorigenin 4`- glucoside |
| 5 | 40.66 | 257 | 853b | 678,652,610,490,471,453 | 829 | 707,656,64 | Iridalglycoside 5b |
| 6 | 41.45 | 260 | 855b | 652,597,547,521,490,473, | NDa | NDa | Iridalglycoside 7 |
| 7 | 45.50 | 262 | 415b | 385,375,361,319,241,206 ,188,143,71 |
391 | 373,309,234,173,171,168 | Irisquin B |
| 8 | 51.58 | 262 | 301 | 280,217,168,151,142,56 | 299 | 269,120 | Irilin A |
| 9 | 55.01 | 242 | 303 | 238,182,148,141,53 | 301 | 154,152,140,138,124,117,114,111,87 | Hesperetin |
| 10 | 55.88 | 245 | 313 | 280,234,155,129 | NDa | NDa | Irisolone |
| 11 | 61.87 | 245 | 315 | 299,283,271,197,194,145 | 313 | 298,255,254,195,185,183,165,137,91 | Irisolodone |
| 12m | 66.01 | 224,297,348(sh) | NDa | NDa | 313 | 298,224,213,192,166,138,123 ,110,96,82,65 |
Crocetenone |
tR: Retention time.
m: Marker compounds.
sh: shoulder peak.
aThe ion signals of this component investigated were not detected in this ion mode.
bThis fragment ion was [M+Na]+
Table 2: Identified compounds in Iris crocea by HPLC-DAD-ESI-MS/MS.
Besides isoflavones, one alkylated p-benzoquinone was found and identified as irisquin B with ESI-MS molecule ion peak in positive ion mode at m/z 415 [M + Na]+ and in negative ion mode at m/z 391 [M - H]¯ . The molecule ion peak at m/z 391 showed MS/MS fragment ion at m/z 168 (100%), indicates the presence of quinoid moiety [32,34,35].
HPLC-DAD-ESI-MS/MS of Iris germanica
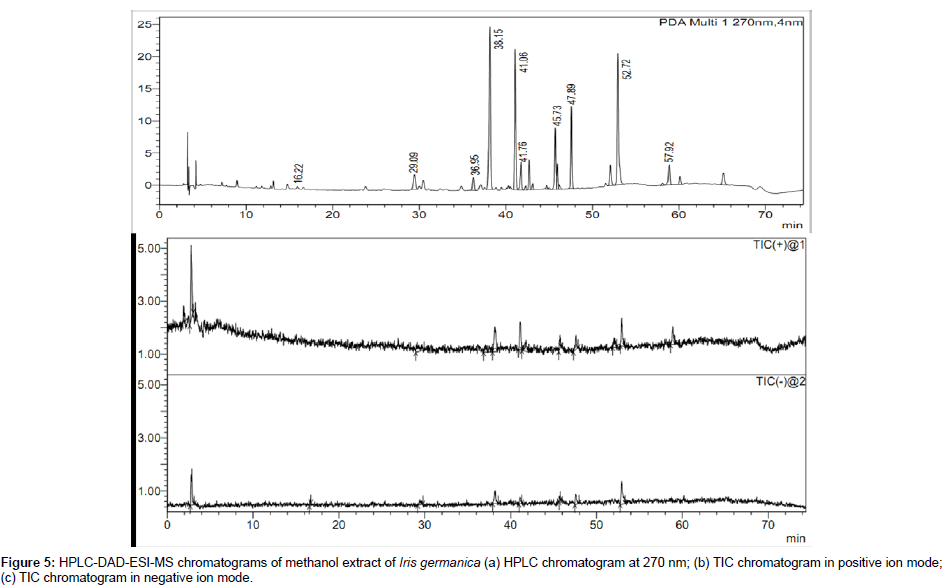
In Iris germanica about 10 constituents were tentatively identified, including 9 isoflavonoids, one xanthone and one stilbene. Only four constituents i.e., picied, tectorigenin 4`-glucoside, iridin and irigenin were identified by comparing their retention time, UV spectra, molecular weight and MSMS with that of reference compounds. The other six constituents i.e., mangiferin, germanaism B, germanaism A, irilone-4`-glucoside, irisolidone 7–glucoside and irisolone, were tentatively characterized on the bases of UV λmax, molecular weight and mass spectrum with reference to literature [18,19,36]. The HPLC-DAD chromatogram and TIC chromatogram in positive and negative ion modes of methanol extract of Iris germanica is shown in Figure 5. The identified compounds in rhizomes of Iris germanica are given in Table 3.
| Peak No. | tR (min) | UV λmax (nm) | [M+H]+ (m/z) | Fragment ions (+) | [M-H]Ë? (m/z) | Fragment ions (-) | Compound |
|---|---|---|---|---|---|---|---|
| 1 | 16.22 | 243,261,366 | NDa | NDa | 421 | 405,331,301,271,159,125,111 | Mangiferin |
| 2 | 29.09 | 235,293 | 391 | 307,267,248,43 | 389 | 227,182 | Picied |
| 3m | 36.95 | 217,263,339(sh) | 463 | 344,301,286,258 | 461 | 446,397,365,283,255,233,195,167 | Tectorigenin 4`- glucoside |
| 4m | 38.15 | 215,265,339(sh) | 523 | 361,346,310,282 | 521 | 491,359,343,313 | Iridin |
| 5 | 41.06 | 261,319 | 475 | 292,290,163,141 | NDa | NDa | Germananaism B |
| 6 | 41.76 | 246,262 | 505 | 313,248,175 | NDa | NDa | Germananaism A |
| 7 | 45.73 | 246,269 | 461 | 404,354,298,270,218,147 | NDa | NDa | Irilone 4`-glucoside |
| 8 | 47.59 | 265 | 477 | 399,217,183,157 | 475 | 299,297 | Irisolidone 7-glucoside |
| 9 | 52.02 | 246,259,315(sh) | 313 | 298,268,245,217,193,151,145 | NDa | NDa | Irisolone |
| 10m | 57.92 | 266 | 361 | 345,315,303,245,208,193,107 | 359 | 344,328,329,314,285,258,270,191 | Irigenin |
tR: Retention time.
m: Marker compounds.
sh: shoulder peak.
aThe ion signals of this component investigated were not detected in this ion mode.
bThis fragment ion was [M+Na]+.
Table 3: Identified compounds in Iris germanica by HPLC-DAD-ESI-MS/MS.
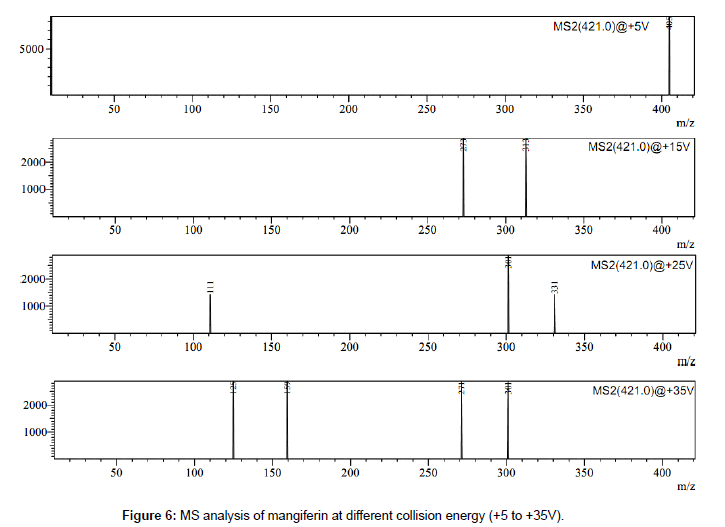
Mangiferin was only one C-glycosidic xanthone identified in Iris germanica by this method. The molecular ion of mangiferin at m/z 421 [M - H]¯ in negative ion mode showed fragment ions at m/z 301 [MFigure H-120]¯, 331[M-H-90]¯, 405 [M-2H-H20]¯, 273 [M-H-120-CO]¯, 313 [M-90-H2O]. Two fragment ions at m/z 331 [M-H-90]- and at m/z 301 [M-H-120]- are characteristic product ion in flavonoids C-glucosides, which are formed by cross-ring cleavages in glucose residue [25,37,38]. The MS analysis of mangiferin at different collision energy voltage (+5 V to +35 V) is shown in Figure 6.
HPLC-DAD-ESI-MS/MS of Iris spuria
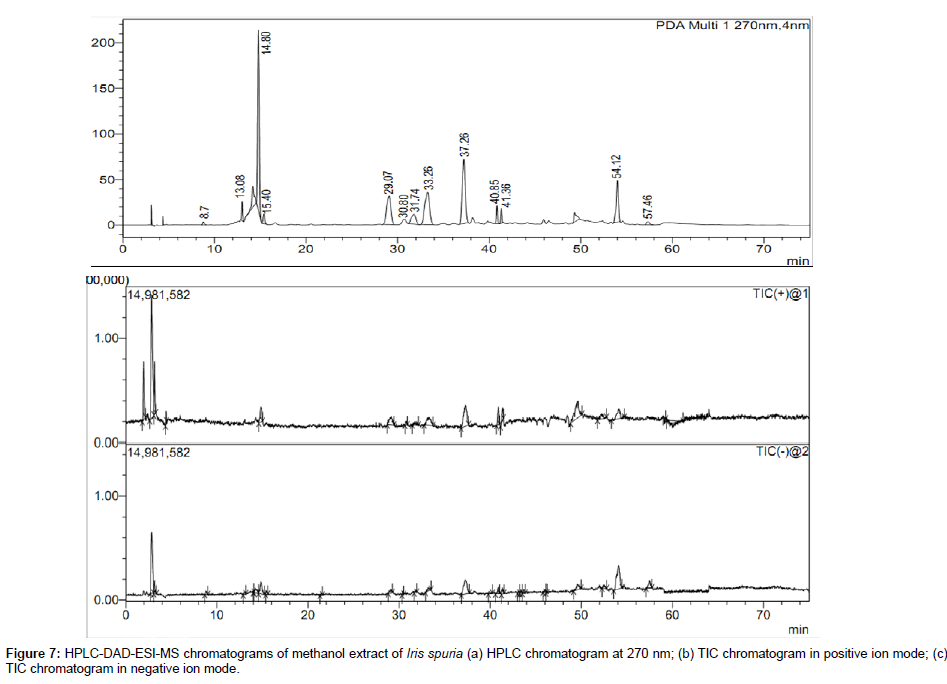
The HPLC-DAD chromatogram and total ion chromatogram of methanol extract of rhizomes of Iris spuria in both positive and negative ion modes are shown in Figure 7. A total of 13 constituents were identified including 10 flavonoids, 2 iridalglycosides and one rotenoid. Four compounds were unambiguously identified by comparing retention time (tR), UV and MS data with those of reference compounds, while other eleven peaks were tentatively identified and characterized on the basis of MS/MS fragmentation and UV spectra obtained by HPLC-DAD-ESI-MS/MS and by comparing with literature data [16,19,25-29,39]. The UV λmax, retention time (tR) and MS/MS fragments in both positive and negative ion modes of all identified compounds in methanol extract of Iris spuria is given in Table 4. Previously we have reported the hepatoprotective activity of LC-MS standardized Iris spuria extract and the sample for that was collected in the month of November, while in our study all the samples were collected in the month of March. It is worth to mention that the time of collection and processing plays an important role as is evident from our previous work on Iris spuria [10].
| Peak No. | tR (min) | UV λmax (nm) | [M+H]+ (m/z) | Fragment ions (+) | [M-H]Ë? (m/z) | Fragment ions (-) | Compound |
|---|---|---|---|---|---|---|---|
| 1 | 8.7 | 206,253,288 | NDa | NDa | 329 | 295,255,204,186,155,108,85 | Irispurinol |
| 2 | 13.08 | 262 | NDa | NDa | 785 | 469,452,344,236,204,147 | Tectorigenin 4`-gentiobioside-7-glucoside |
| 3 | 14.80 | 210,263,329(sh) | 625 | 509,232,109 | 623 | 461,446,359,298,90 | Tectoridin 4`-glucoside |
| 4 | 15.40 | 231,264 | NDa | NDa | 653 | 613,532,513,490,442,387 ,363,310,254,231,182 |
Iristectoregenin B 4`-diglucoside |
| 5m | 29.07 | 263 | 625 | 621,463,356 ,331,301,273 |
623 | 477,419,257,213,155 | Tectorigenin 7-diglucoside |
| 6 | 30.80 | 229,266,340(sh) | 493 | 331,324,316 ,256,215,180,168 |
NDa | NDa | Iristectorin B |
| 7 | 31.74 | 260 | 433 | 392,352,348,304,280, 271,243,225,153,130 |
NDa | NDa | Gentenstein 7-glucoside |
| 8m | 33.26 | 264,329(sh) | 463 | 301,183,103,85 | 461 | 446,283,227,162 | Tectoridin |
| 9m | 37.26 | 217,263,339(sh) | 463 | 343,329,301 ,286,127,109,85 |
461 | 446,283,255,239,195,167 | Tectorigenin 4`-glucoside |
| 10 | 40.85 | 257 | 853b | 678,652,610 ,490,471,453 |
829 | 707,656,227,64 | Iridalglycoside 5b/6c |
| 11 | 41.36 | 260 | 855b | 652,597,547,521,490 ,454,393,309,262 |
830 | 668,601,476,315,258 | Iridalglycoside 7/8 |
| 12m | 54.12 | 265,314 | 301 | 285,281,268, 189,168,133,122 |
299 | 269,55 | Tectorigenin |
| 13 | 57.46 | 243,264,334(sh) | NDa | NDa | 301 | 252,207,80 | 5,8,2`-trihydroxy-7-methoxyflavnone |
tR: Retention time.
m: Marker compounds.
sh: shoulder peak.
aThe ion signals of this component investigated were not detected in this ion mode.
bThis fragment ion was [M+Na]+.
Table 4: Identified compounds in Iris spuria by HPLC-DAD-ESI-MS/MS.
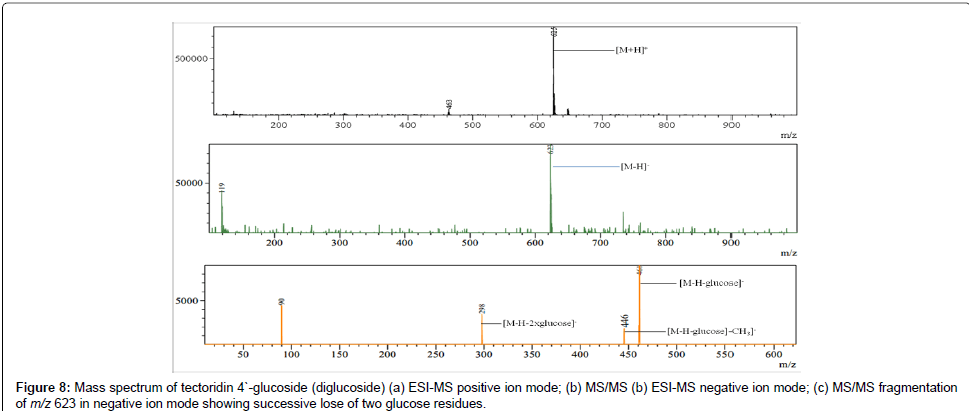
Based on the above data it was found that isoflavones and their glycosides are main constituents in the rhizomes of Iris spuria . In MS/MS data of isoflavone glycosides there was successive loss of one or more hexose residues (162 Da) which indicates the presence of one or more hexose residues. This successive loss of hexose units is characteristic of O-glycosides [40]. For example the molecular ion peak of tectoridin 4`-glucoside (a diglucoside of tectorigenin) at m/z 623 in negative ion mode showed the MS/MS fragments at m/z 461 and m/z 298, due to successive loss of two glucose units as shown in Figure 8. Further some neutral losses of CH3, CO and H2O in both positive and negative ion modes was observed in almost all compounds in Iris spuria . Most of the fragmentation pathways were identical to those observed by Shawl et al. (1985), Plazonic et al. (2009), Zhang et al. (2011), Shu et al. (2010) and Chen et al. (2009) [33,37,40-42]. Two iridalglycosides (iridalglycoside 5b and 7) were also identified in Iris spuria. Both have identical UV λmax=257 nm, which is characteristic for iridal ring system with α, β-unsaturated aldehyde group [29].
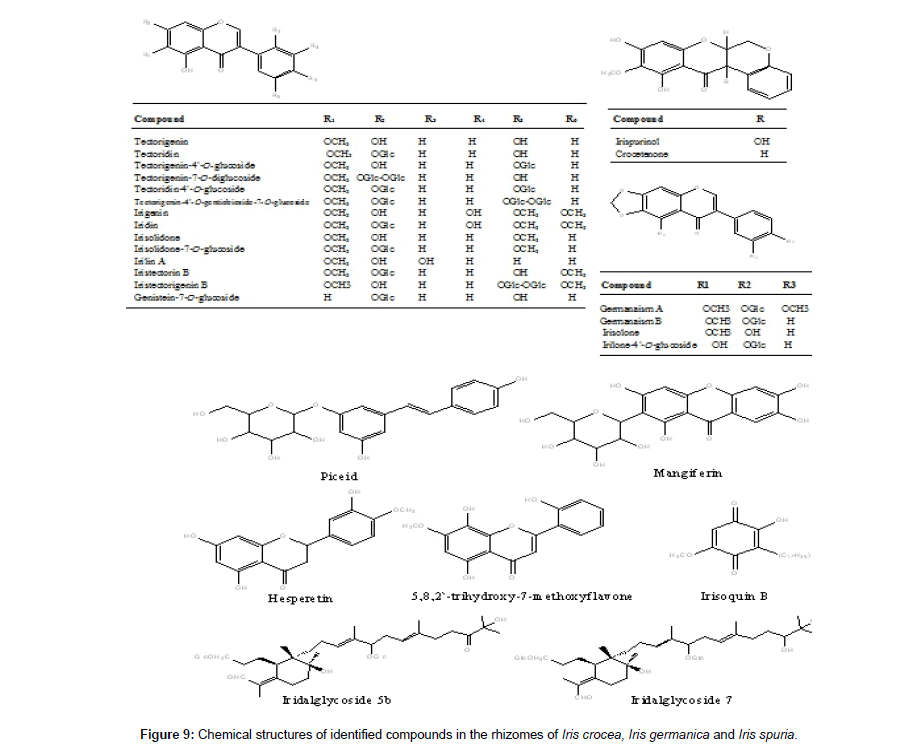
Tectorigenin is reported to possess promising analgesic, anti- inflammatory, hepatoprotective and aldose reductase inhibitory activities [43-45]. In our study tectorigenin was found to be major isoflavone and its 4`-glucoside was identified in all the three Iris species. Chemical structures of all the identified compounds in Iris crocea , Iris germanica and Iris spuria is given in Figure 9.
Conclusion
A simple and versatile analytical method was developed for qualitative identification of major constituents in the rhizomes of Iris crocea, Iris germanica and Iris spuria , by using HPLC-DAD-ESI-MS/ MS in both positive and negative ion modes. About 27 constituent including isoflavones, isoflavone glycosides, xathones, flavones, rotenoids, iridalglycosides, alkylated benzoquinones and stilbenes, were identified based on retention time (tR), UV and MS spectra compared with those of authentic compounds and literature data. In our study isoflavone glycosides were found major constituents. These isoflavone glycosides could be considered as chemotaxonomic markers of these Iris species. This method was successfully applied to identify 8 constituents in rhizomes of Iris crocea, which were not previously reported from this species. Due to high sensitivity of this method, some constituents in minor amount were also identified. Furthermore, the results demonstrate that this method could provide full qualitative information of genus Iris. Further use of innovation studies like MALDI-TOFD-MS and HPLC-DAD-ESI-MS/MS-NMR are necessary for detection and characterization of minor/unidentified compounds.
Acknowledgements
Abdul S. Shawl and Gulzar Bhat thank Council of Scientific and Industrial Research (CSIR), India for Emeritus Scientistship and Junior Research fellowships, respectively. The authors are thankful to Director IIIM-Jammu for providing necessary facilities.
The authors have declared no conflict of interest.
References
- Kassak P (2012) Secondary metabolites of the choosen genus Iris species. Acta cto universilatis agriculturae et silviculturae mendelianae hrunensis 32: 269-280.
- Nadkarni AK (1954) Indian Materia Medica. 3rd edition, Volume 1, Popular Book Depot, Bombay, 695.
- Evans WC, Evans D (2002) Trease & Evans Pharmacognosy. Volume 15, 15th Edition, Saunders Elsvier, 417.
- Vaya J, Tamir S (2004) The relation between the chemical structure of flavonoids and their estrogen-like activities. Curr Med Chem 11: 1333-1343.
- Seki K, Haga K, Kaneko R (1995) Phenols and a dioxotetrahydrodibenzofuran from seeds of Iris pallasii. Phytochem 38: 965-973.
- Seki K, Tomihari T, Haga K, Kaneko R (1994) Iristectorene B, a monocyclic triterpene ester from Iris tectorum. Phytochem 36: 433-438.
- Wei Y, Shu P, Hong J, Qin M (2012) Qualitative and quantitative evaluation of phenolic compounds in Iris dichotoma Pall. Phytochem Anal 23: 197-207.
- Huang L, Ma WH, Liu YZ, Yang JS, Peng Y, et al. (2011) Irisdichotins A-C, three new flavonoid glycosides from the rhizomes of Iris dichotoma Pall. J Asian Nat Prod Res 13: 744-748.
- Rahman AU, Nasim S, Baig I, Jalil S, Orhan I, et al. (2003) Anti-inflammatory isoflavonoids from the rhizomes of Iris germanica. J Ethnopharmacol 86: 177-180.
- Akther N, Andrabi K, Nissar A, Ganaie S, Chandan BK, et al. (2014) Hepatoprotective activity of LC-ESI-MS standardized Iris spuria rhizome extract on its main bioactive constituents. Phytomedicine 21: 1202-1207.
- Tariq KA, Chishti MZ, Ahmad F, Shawl AS, Tantray MA (2008) Evaluation of anthelmintic activity of Iris hookeriana against gastrointestinal nematodes of sheep. J Helminthol 82: 135-141.
- Hanawa F, Tahara S, Mizutani J (1991) Isoflavonoids produced by Iris pseudacorus leaves treated with cupric chloride. Phytochemistry 30: 157-163.
- Miyake Y, Ito H, Yoshida T (1997) Identification of iridals as piscicidal components of iridaceous plants and their conformations associated with CD spectra. Can J Chem. 75:734-741.
- Hideyuki I, Yoko M, Takshi Y (1995) New piscicidal triterpenes from Iris germanica. Chem Pharm Bull 43: 1260-1262.
- Bhat G, Ganai BA, Shawl AS (2014) New phenolics from the root of Scutellaria prostrata JACQ. ex BENTH. Nat Prod Res 28: 1685-1690.
- Wirginia KK, Sieniawska E, Widelski J, Urjin O, Glowniak P, et al. (2013) Major secondary metabolites of Iris spp. Phytochemistry Reviews.
- Shawl AS, Kumar T (1992) Isoflavonoids from Iris crocea. Phytochemistry 3: 1399-1401.
- Shawl AS, Vishwapaul, Zaman A, Kalla AK (1984) Isoflavones from Iris spuria. Phytochemistry 23: 2405-2406.
- Shawl AS, Mengi N, Kaul MK, Vishwapaul (1988a) Flavonoids of Iris spuria. Phytochemistry 27: 1559-1560.
- Farag SF, Backheet EY, El-Emary NA, Niwa M (1999) Isoflavonoids and flavone glycosides from rhizomes of Iris carthaliniae. Phytochemistry 50: 4114-4118.
- Atta-ur-Rahman, Nasim S, Baig I, Orhan I, Sener B, et al. (2003) Isoflavonoid glycosides from the rhizomes of Iris germanica. Helvetica Chimica Acta 86: 3354-3362.
- Bolad GM, Donnelly DMX (1998) Isoflavonoids and related compounds. Natural Product Reports 15: 241-260.
- Watanabe S, Uesugi S, Kikuchi Y (2002) Isoflavones for prevention of cancer, cardiovascular diseases, gynecological problems and possible immune potentiation. Biomed Pharmacother 56: 302-312.
- Shawl AS (1993) Piceid, a stilbene glucoside from the rhizomes of Iris hookeriana. Ind J Pharm Sci. 55: 197-198.
- Cuyckens F, Claeys M (2004) Mass spectrometry in the structural analysis of flavonoids. J Mass Spectrom 39: 1-15.
- Prasain JK, Jones K, Kirk M, Wilson L, Smith-Johnson M, et al. (2003) Profiling and Quantification of Isoflavonoids in Kudzu Dietary Supplements by High-Performance Liquid Chromatography and Electrospray Ionization Tandem Mass Spectrometry. J Agric Food Chem 51: 4213-4218.
- Shawl AS, Mengi N, Misra LN, Vishwapaul (1988b) Irispurinol A 12a-Hydroxyroteniod from Iris spuria. Phytochemistry 27: 3331-3332.
- Farag SF, Kimura Y, Ito H, Takayasu J, Tokuda H, et al. (2009) New isoflavone glycosides from Iris spuria L. (Calizona) cultivated in Egypt. J Nat Med 63: 91-95.
- Marner FJ, Singab AN, Al-Azizi MM, El-Emary NA, Schäfer M (2002) Iridal glycosides from Iris spuria (Zeal), cultivated in Egypt. Phytochemistry 60: 301-307.
- Asghar SF, Habib-ur-Rehman, Atta-ur-Rahman, Choudhary MI (2010) Phytochemical investigations on Iris germanica. Nat Prod Res 24: 131-139.
- Choudhary MI, Nur-e-Alam M, Baig I, Akhtar F, Khan AM, et al. (2001) Four new flavones and a new isoflavone from Iris bungei. J Nat Prod 64: 857-860.
- Singh N, Mahmood U, Kaul VK, Gupta AP, Jirovetz L (2006) A new alkylated benzoquinone from rhizomes of Iris kumaonensis. Nat Prod Res 20: 75-78.
- Shu P, Hong JL, Wu G, Yu B-Y, Qin M-J (2010) Analysis of Flavonoids and Phenolic acids in Iris tectorum by HPLC-DAD-ESI-MSn. Chinese Journal of Natural Medicines 8: 202-207.
- Kubo I, Kim M, Ganjan I, Kamikawa T, Yamagiwa Y (1987) Isolation, structure and synthesis of maesanin, a host defense stimulant from an african medicinal plant Maesa lanceolata. Tetrahedron 43: 2653-2660.
- Mahmood U, Kaul VK, Jirovetz L (2002) Alkylated benzoquinones from Iris kumaonensis. Phytochemistry 61: 923-926.
- Xie GY, Chen YJ, Wen R, Xu JY, Wu SS, et al. (2014) [Chemical constituents from rhizomes of Iris germanica]. Zhongguo Zhong Yao Za Zhi 39: 846-850.
- Zhang YY, Wang Q, Qi LW, Qin XY, Qin MJ (2011) Characterization and determination of the major constituents in Belamcandae Rhizoma by HPLC-DAD-ESI-MS(n). J Pharm Biomed Anal 56: 304-314.
- Zhou Y, Han QB, Song J-Z, Chun-Feng Q, Hong-Xi X, (2008) Characterization of polyprenylated xanthones in Garcinia xipshuanbannaensis using liquid chromatography coupled with electrospray ionization quadrupole time-of-flight tandem mass spectrometry. Journal of Chromatography A 1206: 131-139.
- Harborne J, Mabry TJ, Mabry H (1975) The Flavonoids. 1st edition, Academic press, NewYork, San Francisco, USA, Part I, Chapter 3.
- Plazonic A , Franz B, Zeljan M, Ana M, Biljana N, et al. (2009) Identification and Quantification of Flavonoids and Phenolic Acids in Burr Parsley (Caucalis platycarpos L.), Using High-Performance Liquid Chromatography with Diode Array Detection and Electrospray Ionization Mass Spectrometry. Molecules 14: 2466-2490.
- Shawl AS, Dar BA (1985) Isoflavones from Iris hookeriana. J Nat Prod 48: 849-850.
- Chen L, Qi J, Chang YX, Zhu D, Yu B (2009) Identification and determination of the major constituents in Traditional Chinese Medicinal formula Danggui-Shaoyao-San by HPLC-DAD-ESI-MS/MS. J Pharm Biomed Anal 50: 127-137.
- Ha le M, Que do TN, Huyen do TT, Long PQ, Dat NT (2013) Toxicity, analgesic and anti-inflammatory activities of tectorigenin. Immunopharmacol Immunotoxicol 35: 336-340.
- Moon HI, Jung JC, Lee J (2006) Aldose reductase inhibitory effect by tectorigenin derivatives from Viola hondoensis. Bioorg Med Chem 14: 7592-7594.
- Lee HU, Bae EA, Kim DH (2005) Hepatoprotective effect of tectoridin and tectorigenin on tert-butyl hyperoxide-induced liver injury. J Pharmacol Sci 97: 541-544.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 16990
- [From(publication date):
December-2014 - Apr 05, 2025] - Breakdown by view type
- HTML page views : 12179
- PDF downloads : 4811