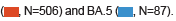Identification and Analysis of SARS-CoV-2 Mutations and Subtypes Using 2 × Tiled Primer Sets with Oxford Nanopore Technologies Sequencing
Received: 01-Jan-2024 / Manuscript No. JIDT-24-124187 / Editor assigned: 05-Jan-2024 / PreQC No. JIDT-24-124187 / Reviewed: 17-Jan-2024 / QC No. JIDT-24-124187 / Revised: 24-Jan-2024 / Manuscript No. JIDT-24-124187 / Published Date: 31-Jan-2024 DOI: 10.4172/2332-0877.1000580
Abstract
Introduction: Since its emergence in 2020, the SARS-CoV-2 virus, an RNA virus, has spread globally, causing a pandemic. It mutates frequently, and some mutations may weaken vaccine effectiveness. This research aims to enhance mutation detection using advanced sequencing techniques, crucial for developing strategies to control the spread and impact of COVID-19.
Methods: Here, RNA samples were collected from patients who tested positive for SARS-CoV-2 from April to July 2022, and validated; 613 samples were selected for sequencing.
Results: The results of the study showed that using a new method of next-generation sequencing with 2× tiled primer sets allowed for the accurate identification of new mutations within the SARS-CoV-2 genome. These mutations were analyzed in relation to the characteristics of patients who tested positive for the virus from April to July 2022. This approach improved understanding of how the virus’s genetic variations can influence its behavior and treatment, providing valuable insights for managing the pandemic.
Conclusion: The findings demonstrated the importance of long-read-based NGS analysis and 2× tiled primer sets for determining full SARS-CoV-2 genome sequence with new mutations and understanding the correlation between viral genotypes and patient characteristics for the effective management of SARS-CoV-2.
Keywords: Amplicon; Long-read sequencing; Mutations; Nextgeneration sequencing; SARS-CoV-2
Language Summary
This study explores better method to identify changes (mutations) and types (subtypes) of the COVID-19 virus. Using a technique called Oxford Nanopore Technologies sequencing, combined with a special approach called 2× tiled primer sets, researchers can more accurately read the complete genetic information of the virus. This is crucial for understanding how the virus changes and spreads. The findings help improve our understanding of the relationship between the virus’s genetic makeup and its effects on patients, aiding in the management of the COVID-19 pandemic. This summary aims to make the study’s findings understandable to everyone, including those not specialized in this field.
Key Summary Points
• To improve the detection and understanding of SARS-CoV-2 mutations and subtypes, addressing the need for better surveillance of the COVID-19 pandemic.
• Utilizing 2× tiled primer sets and Oxford Nanopore Technologies sequencing will yield more accurate and comprehensive SARSCoV- 2 genome sequencing.
• Demonstrated that this method allows for better identification of the full SARS-CoV-2 genome, including new mutations.
• This approach enhances the understanding of the correlation between viral genotypes and patient characteristics, informing better pandemic management strategies.
Introduction
Coronavirus disease, caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), was declared a global pandemic by the World Health Organization in March 2020. There were more than 460 million confirmed cases and more than 6.9 million associated deaths by March 2023 [1].
SARS-CoV-2 has RNA as its genetic material. RNA viruses are easily mutated during gene replication, and many mutations occur within a short period [2,3]. Usually, these mutations are ineffective; however, in rare cases, they create highly contagious and lethal variants, causing serious medical problems. In addition, spikes and surface proteins play important roles in infection and serve as target sites for vaccines [4,5]. Mutations in these proteins reduce the efficacy of existing vaccines [6-8].
The World Health Organization and U.S. Centers for Disease Control and Prevention closely monitor the occurrence of these mutant viruses and designate variants with high lethality and transmissibility. To date, variants ranging from alpha to Omicron have been classified as variants of concern and variants of interest [9].
Most current quantitative Polymerase Chain Reaction (qPCR) tests target the RdRp, N, and E genes [10,11]. These genomic regions are generally less mutated and can be used to confirm SARS-CoV-2 infection. However, the S genes with various mutations are difficult to utilize as qPCR targets, and if the mutation site overlaps with the PCR primer site, false negatives may occur, hindering the detection of new mutations. Therefore, it is impossible to accurately identify new mutations without Next-Generation Sequencing (NGS).
Current SARS-CoV-2 NGS analysis involves an amplicon-based enrichment method through multiplex PCR [12,13]. Typically, the primer set used for Illumina analysis (COVIDSeq Test) generates 98 amplicons, and the size of each amplicon is approximately 400 bp, whereas the primer set used for Oxford Nanopore Technologies (ONT) analysis (Midnight Amplicon panel) generates only 29 amplicons of 1,200 bp [14,15]. Amplicons cannot be generated if the primer site is mutated, and this is less likely to occur in ONT analysis because of the larger amplicon size and fewer amplicon counts than in Illumina analysis. While the base quality of ONT is inferior to that of Illumina, multiplex PCR amplicon-based analysis can generate sufficient reads to compensate for low base quality in both analysis methods [16,17]. In addition, the ONT analysis takes 8-24 h, whereas Illumina sequencing requires approximately 40 h or more [18].
We implemented 2× tiled primer sets for better coverage of SARSCoV- 2 genome sequences and adopted ONT to sequence SARS-CoV-2 genotypes using specimens from SARS-CoV-2-positive volunteers, living in the Seoul metropolitan area, South Korea. We also examined the association between clinical symptoms and SARS-CoV-2 subtypes, especially BA.2 and BA.5.
Materials and Methods
Compliance with ethics guidelines
All human samples used in this study were obtained from voluntarily participating patients after obtaining written informed consent. The research plan and processes were reviewed and approved by the Institutional Review Board of Wiltse Memorial Hospital (2022- W07).
Sample collection and validation
Specimens were collected from patients who tested positive for SARS-CoV-2 between April and July 2022 using nasopharyngeal swabs at Saint Peter’s Hospital, Seoul, Republic of Korea. The viral RNA was extracted using the Nextrator NX-48N and VN kit (Genolution, Seoul, Republic of Korea) and validated with qRT-PCR using the STANDARD M nCoV Real-Time Detection Kit (SD BioSensor, Suwon, Republic of Korea). Samples with a Ct value of 30 or less were selected for sequencing, resulting in a total of 613 samples.
cDNA synthesis and multiplex PCR for SARS-CoV-2-specific amplification
cDNA was synthesized from extracted viral RNA using the LunaScript® RT SuperMix Kit (NEB, Ipswich, MA) according to the manufacturer’s protocol. The synthesized cDNA was used as a template for SARS-CoV-2-specific multiplex PCR using the xGen™ SARS-CoV-2 Midnight Amplicon Panel (IDT, Coralville, IA) containing two primer pools designed for the emerging SARS-CoV-2 variants. Pools A and B included odd and even region primers, respectively, and were used to generate overlapping amplicons of approximately 1,200 bp in length. PCR using primer pools A and B was performed on separate plates to avoid the generation of overlapping amplicons. PCR was conducted using the Rapid Barcoding Kit 96 (ONT, Oxford, UK) with Q5® Hot Start High-Fidelity 2× Master Mix (NEB, Ipswich, MA) according to the manufacturer’s protocol designed for SARS-CoV-2.
Designing 2× tiled midnight primer sets for amplification failure prevention
An additionally designed primer C/D set was developed to improve the coverage of ONT sequencing using the Midnight primer (primer A/B) set. The primer C/D set comprised 15 and 14 pairs of primers for C and D, respectively, and produced an amplicon of approximately 1,200 bp. It avoids overlapping regions with the existing sets and considers factors such as primer redundancy, self-dimer formation, non-target (human genomic DNA) ratio, and amplicon size during primer design. We optimized the efficiency of the primer set by redesigning and adding primers to the set. Finally, a 2× tiled primer set (Midnight primer+primer C/D) was produced to mitigate unexpected drop-out region due to primer site mutation-based mis-amplification.
Library preparation and sequencing
The barcoding process involved pooling amplicons A and B from each sample. The pooled amplicons were then mixed with single set of 96 rapid barcodes from the Rapid Barcoding Kit 96 (ONT, Oxford, UK). The mixture was incubated at 30°C for 2 min and then at 80°C for 2 min. The barcoded samples were pooled and purified according to the manufacturer’s protocol. The library was prepared by combining the purified library with Rapid Adapter F from ONT native and incubating at room temperature (24°C) for 5 min. The library was loaded Onto a MinION sequencer using an R 9.4.1 flow cell (ONT).
SARS-CoV-2 genotyping and viral subtype classification
The sequenced reads were based on guppy2 and processed using the poreCov (v1.5.1) [19] and epi2me-labs/wf-artic (v0.3.11) tools. Using the two analysis tools, read quality, filtered FASTQ and BAM, consensus sequence, lineage (pangolin, scorpio), clade (NextClade), and genome coverage were analyzed. The more detailed and diverse poreCov results were used as the main results, and the epi2me-labs/ wf-artic analysis results were used for crosschecking.
The results from the poreCov and epi2me-labs/wf-artic tools were compared. Most of the mutations were detected by both tools, but some position-specific mutations were detected disconcordantly. For exclusive mutations, we inspected them with Integrative Genomics Viewer [20] to determine whether they were true or false.
To identify the subtype of complex recombinants, Lineage de Composition for SARS-CoV-2 was also performed within a poreCov pipeline [21].
Statistical analysis
Data on patient characteristics, including age, sex, and symptoms, were collected and combined with SARS-CoV-2 genotyping and subtype classification results. For statistical analysis, the features were analyzed using t-tests for age-related comparisons with inhouse Python code.
Results
Improved detection of variants with 2× tiled primer sets
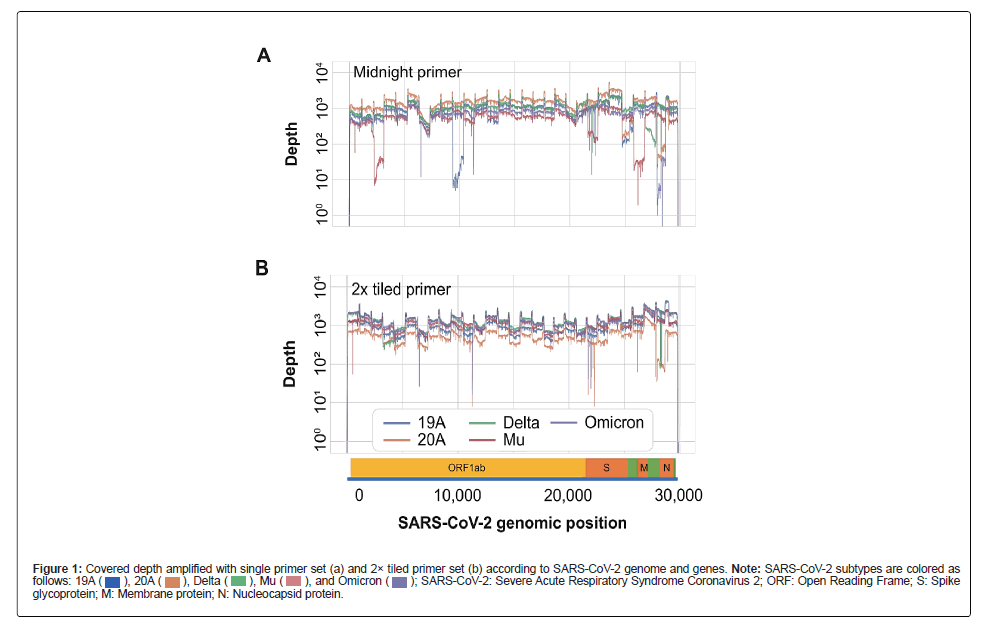
Compared to the conventional primer set (primer set A/B of Midnight primer), the 2× tiled primer set (primer set A+C/B+D) yielded better results for a range of mutants, including Omicron, Mu, and earlier strains such as 19A and 20A (Figure 1).
Figure 1: Covered depth amplified with single primer set (a) and 2× tiled primer set (b) according to SARS-CoV-2 genome and genes. Note: SARS-CoV-2 subtypes are colored as follows: SARS-CoV-2: Severe Acute Respiratory Syndrome Coronavirus 2; ORF: Open Reading Frame; S: Spike glycoprotein; M: Membrane protein; N: Nucleocapsid protein
SARS-CoV-2: Severe Acute Respiratory Syndrome Coronavirus 2; ORF: Open Reading Frame; S: Spike glycoprotein; M: Membrane protein; N: Nucleocapsid protein
These findings highlight the superiority of our approach over the existing methods and provide important insights into the detection of viral mutations. The 2× tiled primer set may enhance the detection and monitoring of the spread of SARS-CoV-2 and its variants.
We also identified large deletions in SARS-CoV-2 by modifying the composition of the primer set. By designing a set capable of amplifying fragments of approximately 3,600 bp, in addition to the standard 1,200 bp (Figure S1), we were able to successfully amplify previously failed regions.
Lineage and single-nucleotide variation analysis of omicron mutations
All analyzed SARS-CoV-2 sequences were reported to GISAID and all samples were classified as the Omicron subtype: The BA.2 Omicron subtype (n=506) and BA.5 Omicron subtype (n=87). In addition, there were 6 cases of the BA.1-like subtype, 2 cases of the BA.4-like subtype, and 12 cases of other variant subtypes. BA.2 was detected throughout the period of sample collection, whereas BA.5 was first found in May and had more confirmed cases than the BA.2 subtype in July. This pattern was also similar to that of the SARS-CoV-2 lineage type in Korea registered with the GISAID during the same period (April to July 2022) (Figure S2).
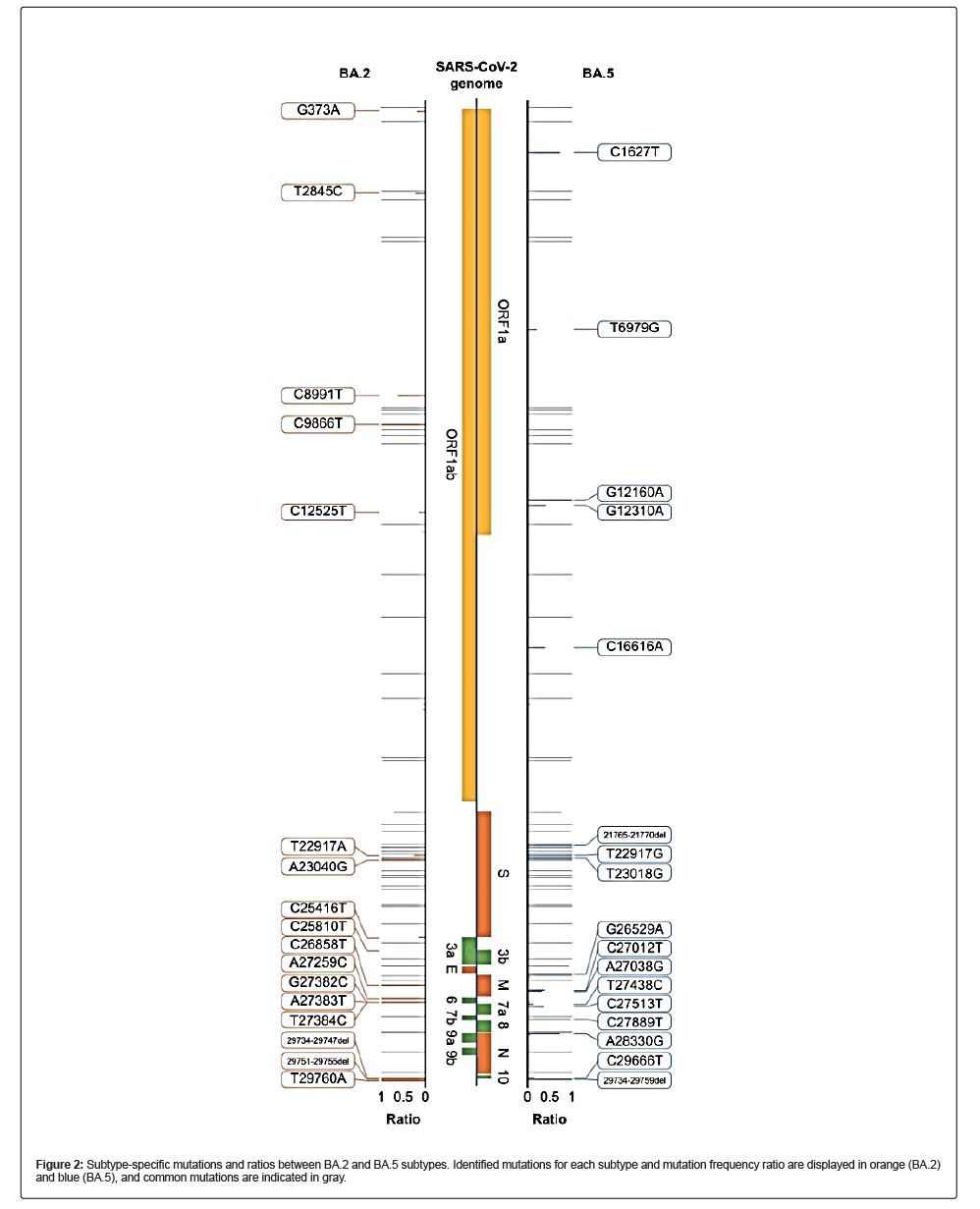
We covered the full SARS-CoV-2 genome and identified a set of Single-Nucleotide Variations (SNVs) unique to the BA.2 and BA.5 lineages. The locations of these BA.2- and BA.5-specific SNVs were consistent with publicly available data, including the Cov lineage and NextClade, implying that although ONT sequencing has low base quality, its results can be used to detect mutations and classify viral subtypes (Figure 2).
During the analysis, the subtypes of some samples were not properly identified. In one sample identified as a recombinant sample, although it had an Omicron mutation, a clear constellation and sublineage could not be identified. Lineage deComposition for SARS-CoV-2 analysis revealed that the sample was a mutant in which BA.1 and BA.2 were recombined (Figure S3), and it was later named as XE variant (2022- 01-19, recombinant lineage of BA.1 and BA.2) [22]. Furthermore, no amplicons were generated in six samples, even though the 2× tiled primer sets were used. In these cases, we found that 2–3 amplicons were not generated among the 29 amplicon pools. We modified the primer composition for detecting the large deletion at that location and sequenced again by separately applying a primer set covering the unamplified region. Separate sequencing results were concatenated and reanalyzed (Figure S4). We sequenced and identified 936-, 929-, 851-, and 425-bp deletions in the SARS-CoV-2 genome. There were three large deletions, which comprised ORF7a, ORF7b, and ORF8 genic regions, and the 425-bp deletion overlapped with ORF8. These SARS-CoV2 variants have also been reported in GISAID (EPI_ISL_14435402, EPI_ISL_14467955, EPI_ISL_15269593, and EPI_ISL_17463497).
SARS-CoV-2 subtype information and clinical characteristics of patients
Table 1 summarizes the viral variants and clinical characteristics of 613 individuals infected with SARS-CoV-2. Most cases had BA.2 subtype (n=506, 82.6%), followed by BA.5 (n=87, 14.2%). The mean age of the participants was 40.3 (standard deviation=20.4) years, with a slightly higher number of female individuals (n=321) than male individuals (n=292); however, there was no significant difference in the severity of infection or symptom manifestation between the sexes. The most reported symptoms were sore throat (n=362; 59.1%), persistent cough/sneezing (n=262; 42.7%), and sputum production (n=171; 27.9%). Fever (n=164, 26.8%), headache, muscle aches, chills, and runny nose were also reported by a significant proportion of the patients. Asymptomatic cases were relatively rare (4.4%; n=27). Owing to a small number of patients, data for some variants, such as BA.1 and BA.4, were excluded from the subsequent analysis (Table 1).
| Characteristic | Total lineage | BA.1 | BA.2 | BA.4 | BA.5 | Others |
|---|---|---|---|---|---|---|
| Total no. | 613 | 6 | 506 | 2 | 87 | 12 |
| Sex | - | - | - | - | - | - |
| M | 292 | 4 | 242 | 1 | 39 | 6 |
| F | 321 | 2 | 264 | 1 | 48 | 6 |
| Mean age, years (SD) | 40.3 (20.4) | 55.2 (15.5) | 41.3 (20.2) | 17.5 (14.8) | 35.0 (20.0) | 31.7 (20.4) |
| symtom | - | - | - | - | - | - |
| Asymptomatic | 27 | 0 | 23 | 0 | 2 | 2 |
| Sore throat | 362 | 2 | 305 | 2 | 48 | 5 |
| Persistent cough/sneezing | 262 | 4 | 219 | 1 | 33 | 5 |
| Sputum | 171 | 1 | 136 | 1 | 30 | 3 |
| Fever | 164 | 0 | 136 | 0 | 26 | 2 |
| Headache | 110 | 0 | 85 | 1 | 22 | 2 |
| Muscle aches | 99 | 1 | 80 | 0 | 17 | 1 |
| Chills | 80 | 1 | 61 | 0 | 15 | 3 |
| Runny nose | 73 | 1 | 63 | 0 | 9 | 0 |
| Hoarseness | 9 | 0 | 9 | 0 | 0 | 0 |
| Sickness | 8 | 0 | 7 | 0 | 1 | 0 |
| Low fever | 8 | 0 | 7 | 0 | 1 | 0 |
| Diarrhea | 7 | 0 | 7 | 0 | 0 | 0 |
| Dizziness | 6 | 0 | 6 | 0 | 0 | 0 |
| Chest pain | 4 | 0 | 4 | 0 | 0 | 0 |
| Nausea | 3 | 0 | 3 | 0 | 0 | 0 |
| Foreign object sensation in throat | 2 | 0 | 2 | 0 | 0 | 0 |
| Hoarse voice | 3 | 0 | 3 | 0 | 0 | 0 |
| Tiredness severe fatigue | 3 | 0 | 2 | 0 | 1 | 0 |
| Vomiting | 2 | 0 | 2 | 0 | 0 | 0 |
| Shortness of breath | 2 | 0 | 2 | 0 | 0 | 0 |
| Hypofunction | 2 | 0 | 2 | 0 | 0 | 0 |
| Cold sweating | 2 | 0 | 1 | 0 | 1 | 0 |
| Sore eyes | 1 | 0 | 1 | 0 | 0 | 0 |
| Loss or change of sense of taste | 1 | 0 | 1 | 0 | 0 | 0 |
| Abdominal pain | 1 | 0 | 1 | 0 | 0 | 0 |
| Flank pain | 1 | 0 | 1 | 0 | 0 | 0 |
| Lethargy | 1 | 0 | 1 | 0 | 0 | 0 |
| Rhinalgia | 1 | 0 | 1 | 0 | 0 | 0 |
| Lumbago andlLow back pain | 1 | 0 | 1 | 0 | 0 | 0 |
SD, standard deviation
Table 1: Characteristics and counts of patients by subtype, sex, age, and symptom.
Comparison of patient information
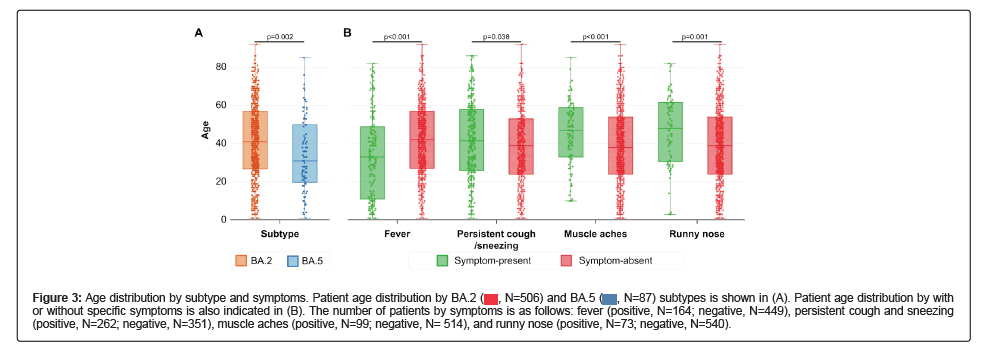
Based on a comparison of patient characteristics and SARS-CoV-2 subtypes, we found several correlated features. First, the age distribution of patients with BA.2 and those with BA5 was significantly different. The age distribution of patients with BA.5 was younger than that of patients with BA.2 (Figure 3A). Each mutation exhibited a similar pattern of specific mutations (Figure S5). Second, significant differences were found in some symptoms according to patient age. Patients with fever were younger than those without fever, whereas patients with persistent cough, sneezing, muscle aches, and runny nose tended to be older than those without symptoms (Figures 3A and 3B).
Figure 3: Age distribution by subtype and symptoms. Patient age distribution by BA.2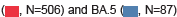 subtypes is shown in (A). Patient age distribution by with or without specific symptoms is also indicated in (B). The number of patients by symptoms is as follows: fever (positive, N=164; negative, N=449), persistent cough and sneezing (positive, N=262; negative, N=351), muscle aches (positive, N=99; negative, N= 514), and runny nose (positive, N=73; negative, N=540).
subtypes is shown in (A). Patient age distribution by with or without specific symptoms is also indicated in (B). The number of patients by symptoms is as follows: fever (positive, N=164; negative, N=449), persistent cough and sneezing (positive, N=262; negative, N=351), muscle aches (positive, N=99; negative, N= 514), and runny nose (positive, N=73; negative, N=540).
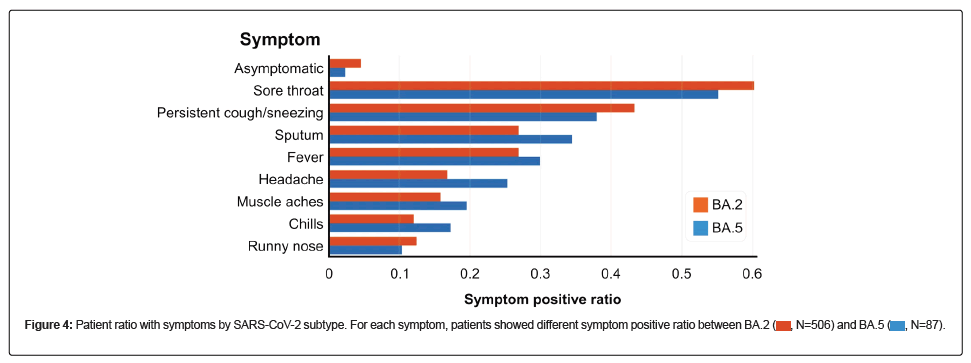
Finally, differences in the symptoms of each variant were noted. For symptoms of respiratory infection, such as sore throat, persistent cough, sneezing, and runny nose, the ratio of symptoms was higher in patients with BA.2, whereas systemic symptoms such as fever, headache, muscle aches, and chills were more frequent in patients with BA.5 (Figure 4).
However, the relationship between symptom presence and age was observed independent of the virus subtype and age, and the relationship with symptoms was also observed independent of the virus subtype and age.
Discussion
Here, the use of a 2× tiled primer set enabled more accurate detection of SARS-CoV-2 mutations than using a single primer set, suggesting that this method may be a promising approach for detecting and monitoring the spread of SARS-CoV-2 and classifying its subtypes. In addition, we applied a modified primer composition set similar to one previously implemented to help amplify large deletions in the SARS-CoV-2 genome [23,24]. Furthermore, we highlighted the benefits of employing ONT sequencing for this type of analysis. With longer reads, we obtained complete sequences of the amplified products without assembly and time- or resource-intensive processes. Moreover, ONT sequencing makes the primer set design simpler than other sequencing platforms with shorter reads. In addition, while ONT sequencing is known to have a high error rate, amplicon sequencing allowed us to identify the mutations with high accuracy, making it possible to find specific locations and frequencies of SNVs within the SARS-CoV-2 genome [25].
The observed differences in symptom profiles and age distribution between BA.2 and BA.5 subtypes in this study indicate distinct pathogenic mechanisms underlying their transmission and clinical presentation [26,27]. These findings suggest that different variants may require different diagnostic and treatment approaches to manage symptoms and the associated complications. For example, targeted interventions may be necessary to address the specific symptom profiles associated with each variant, which could help improve patient outcomes and reduce the burden on the healthcare system.
In addition to the clinical implications, these findings highlight the importance of ongoing surveillance and monitoring of the SARS-CoV-2 subtypes. As new subtypes emerge and spread, it is critical to understand their pathogenic mechanisms, including differences in symptom profiles and age distributions, to inform public health measures and control the spread of the virus [28]. Further research is necessary to explore the underlying genetic, immune, and environmental factors that contribute to these differences and develop targeted interventions based on this knowledge.
Large-scale studies are necessary to confirm and expand these findings. These studies should include a diverse range of patients with different ages, comorbidities, and symptom profiles to ensure that the results are generalizable and applicable to a wider population. By conducting large-scale studies, researchers can gain a more comprehensive understanding of the pathogenic mechanisms of SARSCoV- 2 and its subtypes.
Conclusion
This study provides important insights into the characteristics of the SARS-CoV-2 subtypes and their effect on patients using long-read ONT sequencing with 2× tiled primer sets. The identification of distinct symptom profiles and age distribution associated with different viral lineages underscores the importance of continued surveillance and monitoring of variants and the need for targeted interventions based on specific symptom profiles. Further research is necessary to confirm and expand on these findings and develop effective strategies to control the spread of SARS-CoV-2 and its subtypes.
Funding
No funding or sponsorship was received for this study or publication of this article.
Other Assistance
We thank the staff at Saint Peter’s Hospital for their support.
Author Contributions
Conceptualization: Giyoun Han, Hyunsoo Kim, Kang-Jun Yoon, Minlee Kim; Methodology: Giyoun Han, Jaemyun Lyu, Hyunsoo Kim, Minlee Kim; Investigation: Giyoun Han, Sojung Lee, YaeEun Kwon; Data curation: Giyoun Han, Sojung Lee, YaEun Kwon, Kang-Jun Yoon; Writing – original draft preparation: Giyoun Han; Writing – review and editing: Jaemyun Lyu, Hyunsoo Kim, Kang-Jun Yoon, Minlee Kim; Supervision: Jaemyun Lyu, Hyunsoo Kim, Minlee Kim.
Conflicts of Interest
The authors declare no conflict of interest.
Ethics/Ethical Approval
human samples used in this study were obtained from voluntarily participating patients with written informed consent. The research plan and processes were reviewed and approved by the Institutional Review Board of Wiltse Memorial Hospital (2022-W07). Informed consent was obtained from all individuals involved in the study.
Data Availability
The sequencing data that support the findings of this study are openly available in BioProject (ID: PRJNA980725).
Thanking Patient Participants
We appreciate the patients for their participation
References
- World Health Organization (2023) WHO COVID-19 Dashboard.
- Duffy S (2018) Why are RNA virus mutation rates so damn high? PLOS Biol 16: e3000003.
- Sanjuán R, Domingo-Calap P (2016) Mechanisms of viral mutation. Cell Mol Life Sci 73:4433- 4448.
- Huang Y, Yang C, Xu XF, Xu W, Liu SW (2020) Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol Sin 41:1141-1149.
- Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R (2020) COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J Adv Res 24:91- 98.
- Candido KL, Eich CR, de Fariña LO, Kadowaki MK, da Conceição Silva JL, et al. (2022) Spike protein of SARS-CoV-2 variants: A brief review and practical implications. Braz J Microbiol 53:1133-1157.
- Mengist HM, Kombe Kombe AJ, Mekonnen D, Abebaw A, Getachew M, et al. (2021) Mutations of SARS-CoV-2 spike protein: Implications on immune evasion and vaccine-induced immunity. Semin Immunol 55:101533.
- Dai L, Gao GF (2021) Viral targets for vaccines against COVID-19. Nat Rev Immunol 21:73-82.
- World Health Organization (WHO) (2021) Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern.
- Abbasi H, Tabaraei A, Hosseini SM, Khosravi A, Nikoo HR (2022) Real-time PCR CT value in SARS-CoV-2 detection: RdRp or N gene? Infection 50:537-540.
- Valadan R, Golchin S, Alizadeh-Navaei R, Haghshenas M, Zargari M, et al. (2022) Differential gene expression analysis of common target genes for the detection of SARS-CoV-2 using real time-PCR. AMB Express 12:112.
- Carpenter RE, Tamrakar V, Chahar H, Sharma R (2022) Confirming multiplex RT-qPCR use in COVID-19 with next-generation sequencing: Strategies for epidemiological advantage. Glob Health Epidemiol Genom. 2270965.
[Crossref] [Google Scholar] [PubMed]
- John G, Sahajpal NS, Mondal AK, Ananth S, Williams C, et al (2021) Next-Generation Sequencing (NGS) in COVID-19: A tool for SARS-CoV-2 diagnosis, monitoring new strains and phylodynamic modeling in molecular epidemiology. Curr Issues Mol Biol 43:845-867.
[Crossref] [Google Scholar] [PubMed]
- Freed NE, Vlková M, Faisal MB, Silander OK (2020) Rapid and inexpensive whole-genome sequencing of SARS-CoV-2 using 1200 bp tiled amplicons and Oxford nanopore rapid barcoding. Biol Methods Protoc 5:bpaa014.
[Crossref] [Google Scholar] [PubMed]
- Tyson JR, James P, Stoddart D, Sparks N, Wickenhagen A, et al. (2020) Improvements to the Artic multiplex PCR method for SARS-CoV-2 genome sequencing using Nanopore. bioRxiv 283077.
[Crossref] [Google Scholar] [PubMed]
- Cheng C, Fei Z, Xiao P (2019) Methods to improve the accuracy of next-generation sequencing. Front Bioeng Biotechnol. 11:982111.
[Crossref] [Google Scholar] [PubMed]
- Grädel C, Terrazos Miani MA, Barbani MT, Leib SL, Suter-Riniker F, et al.(2019) Rapid and cost-efficient enterovirus genotyping from clinical samples using Flongle flow cells. Genes 10:659.
- Horiba K, Torii Y, Aizawa Y, Yamaguchi M, Haruta K, et al. (2022) Performance of Nanopore and Illumina metagenomic sequencing for pathogen detection and transcriptome analysis in infantile central nervous system infections. Open Forum Infect Dis. 9:ofac504. [Crossref]
[Google Scholar] [PubMed]
- Brandt C, Krautwurst S, Spott R, Lohde M, Jundzill M, et al. (2021) poreCov-an easy to use, fast, and robust workflow for SARS-CoV-2 genome reconstruction via Nanopore sequencing. Front Genet 12:711437.
[Crossref] [Google Scholar] [PubMed]
- Robinson JT, Thorvaldsdóttir H, Wenger AM, Zehir A, Mesirov JP (2017) Variant review with the Integrative Genomics Viewer. Cancer Res 77:e31-4.
[Crossref] [Google Scholar] [PubMed]
- Valieris R, Drummond RD, Defelicibus A, Dias-Neto E, Rosales RA, et al. (2022) A mixture model for determining SARS-Cov-2 variant composition in pooled samples. Bioinformatics 38:1809-1815.
[Crossref] [Google Scholar] [PubMed]
- Thakur P, Thakur V, Kumar P, Singh Patel SK. (2022) Emergence of novel Omicron hybrid variants: BA (x), XE, XD, XF more than just alphabets. Int J Surg 104:106727.
[Crossref] [Google Scholar] [PubMed]
- Mentes A, Papp K, Visontai, D, Stéger J, Csabai I, et al. (2022) Identification of mutations in SARS-CoV-2 PCR primer regions. Sci Rep 12:18651.
[Crossref] [Google Scholar] [PubMed]
- Mazur-Panasiuk N, Rabalski L, Gromowski T, Nowicki G, Kowalski M, et al (2021) Expansion of a SARS-CoV-2 Delta variant with an 872-nt deletion encompassing ORF7a, ORF7b and ORF8, Poland, July to August 2021. Euro Surveill 26:2100902.
[Crossref] [Google Scholar] [PubMed]
- Player R, Verratti K, Staab A, et al. (2020) Comparison of the performance of an amplicon sequencing assay based on Oxford Nanopore technology to real-time PCR assays for detecting bacterial biodefense pathogens. BMC Genom 21:166.
[Crossref] [Google Scholar] [PubMed]
- Player R, Verratti K, Staab A, Bradburne C, Grady S, et al. (2022) Differences in clinical presentations of Omicron infections with the lineages BA.2 and BA.5 in Mecklenburg-Western Pomerania, Germany, between April and July 2022. Viruses 14:2033.
[Crossref] [Google Scholar] [PubMed]
- Nakakubo S, Kishida N, Okuda K, Kamada K, Iwama M, et al. (2023) Associations of COVID-19 Symptoms with Omicron subvariants BA.2 and BA.5, host status, and clinical outcomes: A registry-s in Sapporo, Japan. medRxiv. 2023.
[Crossref] [Google Scholar] [PubMed]
- Poovorawan Y, Pyungporn S, Prachayangprecha S, Makkoch J (2013) Global Alert to avian influenza virus infection: From H5N1 to H7N9. Pathog Glob Health 107:217-223.
[Crossref] [Google Scholar] [PubMed]
Citation: Han G, Lee S, Kwon YE, Lyu J, Kim H, et al. (2024) Identification and Analysis of SARS-CoV-2 Mutations and Subtypes Using 2× Tiled Primer Sets with Oxford Nanopore Technologies Sequencing. J Infect Dis Ther 12: 580. DOI: 10.4172/2332-0877.1000580
Copyright: © 2024 Han G, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 687
- [From(publication date): 0-2024 - Nov 21, 2024]
- Breakdown by view type
- HTML page views: 632
- PDF downloads: 55