Research Article Open Access
High Mobility Group Box 1 (HMGB1) is Associated with Progression and Poor Prognosis in Pancreatic Cancer
Xu Yang Liang1, Qian Qian Li1, Zong Li Zhang2, Chong Qi Jia3, Li Li4, Chun Tao Liu1, Yong Mei Yang5 and Bao Quan Cheng1*1Department of Gastroenterology, Shandong University, China
2Department of Surgery, Shandong University, China
3Department of Epidemiology and Health Statistics, Shandong University, China
4Department of Pathology, Medicine College, Shandong University, China
5Institute of Basic Medical Sciences, Qilu Hospital, Medicine College, Shandong University, China
- *Corresponding Author:
- Bao Quan Cheng, MD, PhD
Department of Gastroenterology
Qilu Hospital, Medicine College
Shandong University, Jinan 250012, China
Tel: +8613853198938
E-mail: drcbq@163.com
Received date: April 30, 2014; Accepted date: June 17, 2014; Published date: June 24, 2014
Citation: Liang XY, Li QQ, Zhang ZL, Jia CQ, Li LI, et al. (2014) High Mobility Group Box 1 (HMGB1) is Associated with Progression and Poor Prognosis in Pancreatic Cancer. J Gastroint Dig Syst 4:190. doi:10.4172/2161-069X.1000190
Copyright: © 2014 Liang XY, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Gastrointestinal & Digestive System
Abstract
Aim: Increased expression of High Mobility Group Protein Box 1(HMGB1) has been reported with several different tumor types, but the role of HMGB1 in pancreatic cancer is not fully understood. The aim of this study is to detect the expression of HMGB1 and evaluate the role of HMGB1 in pancreatic cancer. Methods: The expression level of HMGB1 in pancreatic cancer cell line PANC-1 was detected by western blot analysis and reverse transcription PCR. Furthermore, primary tumors tissues of 35 patients with resected pancreatic cancer(T2-4 N0-2 M0-1) were stained immune histochemically using the polyclonal anti-HMGB1 antibody. The prognostic relevance of HMGB1 expression was evaluated by univariate Kaplan-Meier among 35 patients. Results: HMGB1 is over expressed in pancreatic cancer.HMGB1 overexpression was significantly associated with clinical stage (p<0.05). Univariate Cox regression showed that the survival time was significantly different between groups with high and low expression of HMGB1, indicating that high level of HMGB1correlated with a shorter survival time (P<0.05; HR, 2.53; 95%CI, 1.14-5.64). Conclusions: HMGB1 might promote pancreatic cancer progression and HMGB1 expression was an independent prognostic indicator for patients’ survival
Keywords
High mobility group protein box1; Pancreatic cancer; Survival
Abbreviations
HMGB: High Mobility Group Box; PC: Pancreatic Cancer; RAGE: Advanced Glycation End Products; TNF: Tumor Necrosis Factor; IL: Interleukin
Introduction
Pancreatic cancer (PC) has the poorest prognosis among human tumors due to its highly aggressive and early metastasizing nature. Current diagnostic tools have difficulty in detecting the disease at early stage, and therapeutic applications often remain ineffective at advanced stage. Therefore, understandingthe mechanisms leading to progression of this cancer at molecular levelsis of great importance for early disease detection and new targeted treatment options [1].
High mobility group B 1(HMGB1) is a member of the high mobility group protein superfamily that consists of two other nuclear factors, HMGB2 and HMGB3. It has a molecular mass of approximately 27 kD, and it is a highly conserved protein with more than 95% similarity between rodents and humans [2]. While expression of HMGB2 and HMGB3 is more restricted, HMGB1 has been considered as ubiquitously expressed in all vertebrate nuclei [3]. In the nucleus, HMGB1 binds to the minor groove of linear DNA and bends it into a helical structure [4]. It has been implicated in a variety of biologically important processes, including transcription, DNA repair, recombination, differentiation, development, and extracellular signaling [5].
As a cytokine-like factor, HMGB1 is secreted by macrophages, mature dendritic cells, and natural killer cells stimulated by LPS, TNF-α, or IL-1 [6,7]. It was also recently identified as an inflammatory cytokine functioning as a late mediator of lethality in sepsis [7]. The study suggests that HMGB1 is a regulator of the inflammation seen following acute tissue damage, responses to ischemia, burn, infection, or sepsis and initiate tissue regeneration [8]. The protein is detected on the cell surface where it has been reported to contribute to cellular migration and invasion of many tumor types [9,10], such as breast cancer, hepatocellular carcinoma, melanoma, gastric cancer, and so on. HMGB1 is up-regulated in immature cells, migrating growth cones and edges of migrating malignant cells [11], but it is down-regulated as a consequence of cell-to-cell contact as a sensor for contact-dependent inhibition in migrating cells. The protein also binds tissue-type plasminogen activator and plasminogen through their lysine-binding kringle domains, resulting in plasmin production and enhancing penetration and invasion into tissues [12]. It has been officially grouped as a metastasis specific gene in PC [13]. Supported by these observations, here, we examined the HMGB1 protein expression in human PC in both clinical patient samples and tumour cell lines PANC-1 to evaluate the roles of HMGB1 in PC progression and clinical outcome of patients with PC.
Material and Methods
Cell culture and treatment
The human PC cell lines, PANC-1 were maintained in complete growth medium DMEM (High glucose, Keygenbio, Nanjing, China) supplemented with 10% fetal bovine serum, penicillin and streptomycin at 37°C in a 5% CO2 humidified incubator and 95% air.
Patient characteristics
Specimens of 35 patients with resected PC were collected after approval by the ethical committee of the Shandong University and after written informed consent. The patients were histologically and clinically diagnosed at the Qilu Hospital of Shandong University between 1999 and 2006. The tumors were classified according to the WHO TNM classification of tumors of the exocrine pancreas [14]. Clinical information about the samples is described in detail in Table 1. The postoperative staging of all patients had resulted in the diagnosis of resectable tumors (T2-4 N0-1 M0-1). Thirty patients whose primary tumors were classified as T2-4 N0-1 M0 received curative surgery, and the other five patients whose primary tumors were classified as T2-4 N0-1 M1 received palliative surgery. The 35 patients included 24 males and 11 females, ranging in age from 20 to 84 years (mean 59 years). The median postoperative survival was 16.5 months (ranged from 2 to 56.8 months) for all patients. Follow-up studies included physical examinations, abdominal X-ray, and blood tests in a 6-month interval and an additional abdominal computed tomography scan, abdominal ultrasound. Closing follow-up was documented by contacting family practitioners with questionnaires or telephones concerning local relapse, distant metastasis, and death (Table 1).
| Characteristics N (%) | |
| Gender | |
| Male | 24(68.57%) |
| Female | 11(31.43%) |
| Age | |
| ≥59 | 19(54.29%) |
| >59 | 16(45.71%) |
| Histopathology WHO Classification | |
| Well-differentiated | 6(17.14%) |
| Moderately-differentiated | 19(54.29%) |
| poorly-differentiated | 10(28.57%) |
| T classification | |
| T2 | 20(57.14%) |
| T3-T4 | 15(42.86%) |
| N classification | |
| N0 | 29(82.86%) |
| N1 | 6(17.14%) |
| M classification | |
| M0 | 30(85.71%) |
| M1 | 5(14.29%) |
| Clinical stage | |
| I-II | 24(68.57%) |
| III-IV | 11(31.43%) |
| Expression level of HMGB1 | |
| Low expression | 20(57.14%) |
| High expression | 15(42.86%) |
| Status at follow-up | |
| Alive and lost to follow-up | 7(20.00%) |
| Cancer relation death | 28(80.00%) |
Table 1: Clinicopathological characteristics of patient samples and expression of HMGB1 in PC, a Postoperative staging was classified according to the WHO TNM classification of tumours of the exocrine pancreas
Western blot analysis
Western blot analysis was performed according to standard protocols. Briefly, tumor cell line PANC-1 cells were washed twice with cold PBS and lysed on ice in RIPA buffer (RIPA buffer, Beyotime, Haimen, Jiangsu, China) with protease inhibitors. Protein lysate(40 ug) was separated in a sodiu dodecyl sulphate-12% polyacrylamide gel. Proteins were electrotransferred to nitrocellulose membrane (Beyotime, Haimen, Jiangsu, China) and blocked with 1% BSA (Beyotime, Haimen, Jiangsu, China) in Tris-buffered saline containing 0.1% Tween (TBST, pH7.6; 100 mM NaCl, 50 mM Tris, and 0.1% Tween-20). Membranes were immunoblotted with anti-HMGB1 polyclonal antibody (1:500) (Abcam, Cambridge, UK) and anti-β-actin antibody (1:1000) (Santa Cruz Biotechnology, Santa Cruz, CA, USA) overnight at 4°C, followed by their respective secondary antibodies conjugated to horseradish peroxidase (HRP) (1: 5000) (Zsgb-bio, Zsgb, Beijing, China). Lastly, the signals were detected by enhanced chemiluminescence (ECL; Pierce, Rockford, IL, USA), according to the manufacturer’s recommendations. Specific bands were visualized using a Kodak IMAGE STATION 2000 MM detection system (Rochester, USA).
Reverse transcription PCR
Total RNA was extracted from PANC-1 cells using Trizol reagent(Invitrogen, Carlsbad, CA) followed by further purification with the RN easy RNA isolation system. Total RNA was converted to cDNA with the Easy Script First-Strand cDNA Synthesis Super Mix (Trans Gen Biotech, Trans Gen, Beijing, China). PCR were performed using 2×EasyTag PCR Super Mix (Applied Trans Gen Biotech, TransGen, Beijing, China) according to the manufacturer’s instructions. The human HMGB1 primers pairs used are listed as below: Sense: 5'-GGA CAA GGC CCG TTA TGA AAG AGA AAT GA-3'(residues 361-389) and Antisense: 5'-AGC AGA AGA GGA AGA AGG CCG AAG GAA-3'(residues 482-456). The human β-actin primers pairs are listed as below: Sense: 5'-AGC GAG CAT CCC CCA AAG TT-3'(residues 1379-1398) and antisense: 5'-GGG CAC GAA GGC TCA TCA TT-3'(residues 1663-1644). Amplification was performed for a predetermined optimal number of cycles. PCR products were separated by electrophoresis on 2% agarose gels, which were stained with ethidium bromide. To quantify the amount of DNA, each gel was analyzed on a Manutacl urea ay Panasonic System Soiutions (Suzhou, China).
Immunohistochemical (IHC) staining
Consecutive 4 µm sections were cut from each block. Immunohistochemical analysis of pancreatic cancer tissues was performed using standard protocols. Briefly speaking, all paraffin sections were baked for 1h at 65°C. Sections were deparafinized with xylene and rehydrated with graded ethanol to distilled water. Sections were submerged in Tris-EDTA antigenic retrieval buffer (10mMTris, 1mM EDTA solution, 0.05%Tween-20, pH 9.0) and subjected to microwave treatment. Then, Cool down room temperature naturally. After washing, the sections were treated with 5% normal goat serum confining liquid for 15 mins at 37°C to reduce non-specific binding and then rabbit polyclonal anti-HMGB1 antibody (1:300) (Abcam, Cambridge, UK) was incubated with the sections overnight at 4°C. After washing, the sections were treated with 0.3% H2O2 for 15 mins to block the endogenous peroxidase. After washing, the sections were incubated with biotinylated caprine antibody against rabbit immunoglobulin G (BOSTER, Wuhan, China) for 15 mins at 37°C. Washing in TBS, the sections incubated with enzyme labeling with streptavidin–horseradish peroxides (BOSTER, Wuhan, China) for 15 mins at 37°C. After washing, all sections were color-developed using diaminobenzidine (DAB) solution (BOSTER, Wuhan, China), then washed in water and counterstained with Meyer’s hematoxylin (Sigma Chemical, St. Louis, MO, USA) for 1min. After washed in water and hydrochloric acid-ethyl alcohol (1ml concentrated hydrochloric acid, 99 ml 75% ethyl alcohol) for 3s. All sections washed in water and dehydration with graded ethanol and transparence with xylene. Finally, all sections were mounted with neutrality GUM and coverslip. The negative control was consisted of TBS for the primary HMGB1 antibody. Controls were run with each batch of slides, at an average of approximately 10 slides per batch. Each entire slide was evaluated by inverted microscope (Olympus, Japan).
Evaluation of staining
The immunohistochemically stained tissue sections were scored separately by two pathologists blinded to the clinical parameters. The slides with discrepant evaluations were reevaluated and a consensus was reached (n=5). The slides were examined under light-microscopes using objectives with X10 and X40 magnification. For HMGB1 assessment, the entire tissue section was scanned to assign the scores. The ration of positive-staining tumor cells (0, none; 1,<1/100; 2, 1/100 to 1/10; 3, 1/10 to 1/3; 4, 1/3 to 2/3; and 5, >2/3). Next, an intensity score was assigned, which represented the average intensity of positive tumor cells (0, none; 1, weak, 2, intermediate; and 3, strong). The proportion and intensity scores were then added to obtain a total score, which ranged from 0 to 8. This relatively simple, reproducible scoring method gives highly concordant results between independent evaluators and has been used in previous studies [15]. For the purpose of statistical evaluation, tumors with a final staining score of 7 were considered to be low expression and a final staining score of ≥7 were considered to be high expression.
Statistical methods
All statistical analyses were carried out with STATA version 9.2 (Stata Corporation, College Station, TX, USA). Intergroup comparisons of the clinical and pathologic variables were analyzed using two-tailed P which was calculated by Pearson’s χ2 test (Fisher exact probability) for discrete variables. The follow-up of patients was completed at death or at the cut-off date, January1, 2009. At the cut-off date, 28 patients (80.00%) was death, 5 patients (14.29%) lost to follow-up, 1 patient (2.86%) was operative death and 1 patient (2.86%) was still alive. The median follow-up interval, for all patients at the cut-off date, was 16.5 months (range 2-56.8 months). The cases lost to follow-up and operative deaths were treated as censored data for the survival analysis. For analysis of follow-up data, life table curves were calculated using the Kaplan-Meier method, and survival distributions were compared by log-rank statistics. The primary end point was cancer-related survival, as measured from the date of surgery to the time of the last follow-up (January1, 2009) or cancer-related death. The threshold for statistical significance was chosen at P<0.05.
Results
Analysis of the expression of HMGB1 by western Blot and reverse transcription PCR in PC cells
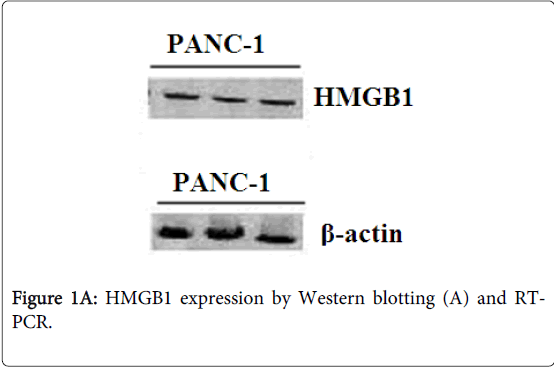
Identification of HMGB1 was confirmed by Western blot and reverse transcription PCR. The protein product of the cell line PANC-1 reacting with polyclonal HMGB1 antibodies was blotted to a nitrocellulose membrane for Edman degradation. The cellines PANC-1 showed a strong expression of HMGB1 protein (Figure 1A) and mRNA in PC cells (Figure 1B). β-actin expression was used as a loading control.
HMGB1 is overexpressed in PC tissues
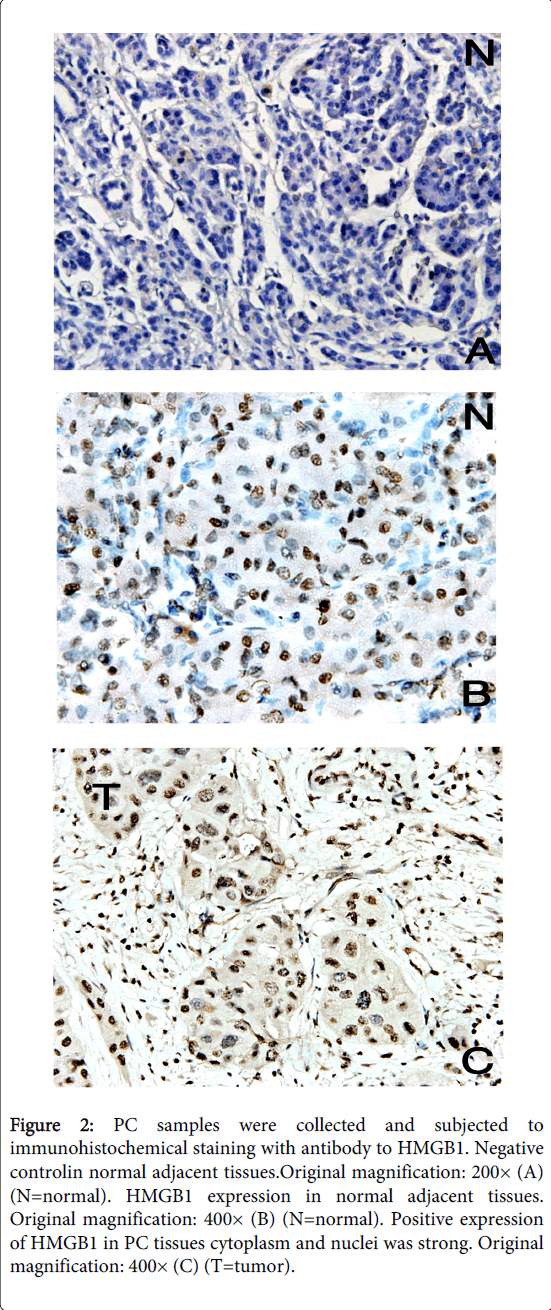
As an alternative method of detecting HMGB1 protein expression in tumor tissue, we performed immunohistochemical staining of sections derived from 35 different pancreatic cancer tissues and normal adjacent tissues. HMGB1 staining was mostly observed in the nuclei and cytoplasm of carcinoma cells (Figure 2). Figure 2 shows two representative examples of the thirty-five carcinomas analyzed by microscope, all showed profound HMGB1 protein upregulation in tumor cells compared with normal adjacent tissues of the same pancreatic cancer patient. These studies confirm the correlation between elevated HMGB1 protein expression and neoplastic transformation in human pancreatic cancer.
Figure 2: PC samples were collected and subjected to immunohistochemical staining with antibody to HMGB1. Negative controlin normal adjacent tissues.Original magnification: 200× (A) (N=normal). HMGB1 expression in normal adjacent tissues. Original magnification: 400× (B) (N=normal). Positive expression of HMGB1 in PC tissues cytoplasm and nuclei was strong. Original magnification: 400× (C) (T=tumor).
Increased expression of HMGB1 correlates with progression of PC
The association between HMGB1 expression and clinicopathological characteristics of tumors was examined. As summarized in Table 2, there was no significant correlation between HMGB1 expression and gender, age or N classification, However, HMGB1 expression was positively correlated with T classification (p<0.05), M classification (p<0.01), and clinical stage (WHO Pancreatic Cancer stage) (p<0.01).
| HMGB1(%) | ||||
| characteristics | n | Low expression | High exprssion | P |
| gender | ||||
| Male | 24 | 15(62.50) | 9(37.50) | |
| Female | 11 | 5(45.45) | 6(54.55) | 0.467 |
| Age(years) | ||||
| ≥59 | 19 | 11(57.89) | 8(42.11) | |
| <59 | 16 | 9(56.25) | 7(43.75) | 1 |
| T classification | ||||
| T2 | 20 | 15(75.00) | 5(25.00) | |
| T3-4 | 15 | 5(33.33) | 10(66.67) | 0.019* |
| N classification | ||||
| N0 | 29 | 19(65.52) | 10(34.48) | |
| N1 | 6 | 1(16.67) | 5(83.33) | 0.064 |
| M classification | ||||
| M0 | 30 | 20(66.67) | 10(33.33) | |
| M1 | 5 | 0(0.00) | 5(100.00) | 0.009* |
| Clinical stage | ||||
| I-II | 24 | 18(75.00) | 6(25.00) | |
| III-IV | 11 | 2(18.18) | 9(81.82) | 0.003* |
Table 2: Correlation between the clinicopathological features and expression of HMGB1 protein *Statistically significant difference.
a HMGB1 expression was categorized according to the ration (0, none; 1, 1/100; 2, 1/100 to 1/10; 3, 1/10 to 1/3; 4, 1/3 to 2/3; and 5, 2/3) and the average intensity (0, none; 1, weak, 2, intermediate; and 3, strong) of positive-staining tumor cells into low expression (i.e., a final staining score of 7), high expression (i.e., a final staining score of ≥ 7).
B Two-sided P were calculated by Pearson’s χ2 test (Fisher exact propability) to determine the significance of the correlation between clinicopathological parameters and expression level of HMGB1. Bold printing indicates statistical significance.
C Post-operative staging was classified according to the WHO TNM classification of tumours of the exocrine pancreas.
HMGB1 expression is inversely correlated with prognosis in PC patients
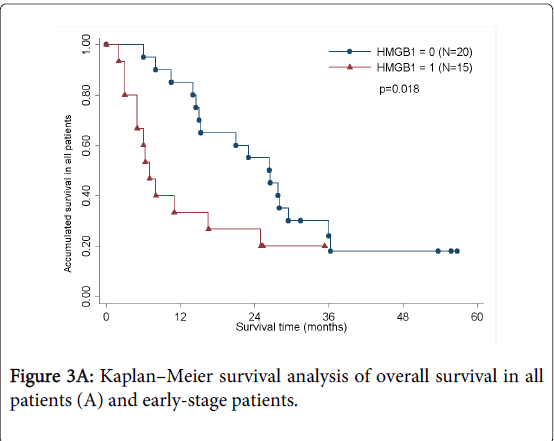
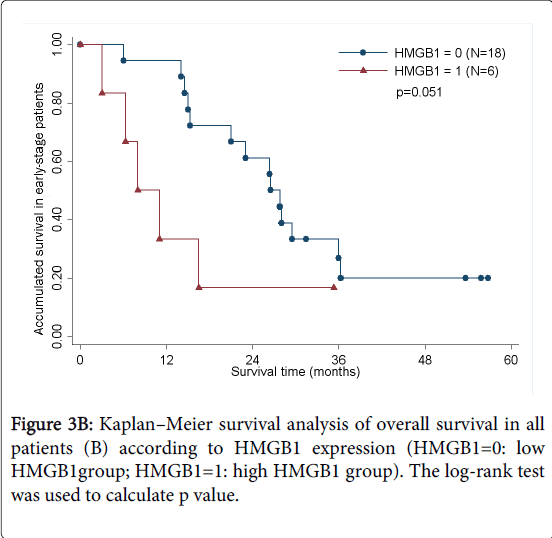
As determined by the Kaplan–Meier method, HMGB1 expression in pancreatic cancer was significantly correlated with overall survival (p<0.05; Figure 3A). The results of univariate analysis showed that the survival time was significantly different between groups with high and low expression of HMGB1, with high level of HMGB1 correlating with a shorter survival time (Table 3). No statistically significant difference(HR=1.460,p=0.535) was observed by multivariate analyses(Table 4). We also analyzed the prognostic value of HMGB1 expression in selective patient subgroups stratified according to the tumor clinical stage (WHO Pancreatic Cancer stage). In early-stage pancreatic cancer (i.e. stages I and II), patients with a high level expression of HMGB1 had a significantly shorter overall survival compared with patients with a low level of expression of HMGB1 (n=24; log-rank, p=0.051; Figure 3B).
| Univariate Analysis Pb | ||||
| Risk factor | HR | 95%CI | ||
| Age(years) | ||||
| ≥59vs<59 | 1.26 | 0.593 | 2.675 | 0.546 |
| Gender | ||||
| Males vs female | 0.788 | 0.354 | 1.751 | 0.564 |
| T classification | ||||
| T2 vs T3-T4 | 1.745 | 0.81 | 3.758 | 0.162 |
| N classification | ||||
| N0 vs N1 | 2.149 | 0.783 | 5.9 | 0.166 |
| M classification | ||||
| M0 vs M1 | 3.257 | 1.079 | 9.828 | 0.064 |
| Clinical stage | ||||
| I-II vs III-IV | 2.929 | 1.22 | 7.035 | 0.021* |
| HMGB1 expression | ||||
| Low vs high | 2.533 | 1.138 | 5.639 | 0.026* |
Table 3: Univariate analysis of cancer-related survival in the total populations* Statistically significant difference.a. P of univariate analyses were determined by Cox regression analysis. b. HMGB1 expression was categorized according to the ration and the average intensity of positive-staining tumor cells into low expression (i.e., a final staining score of 7), high expression (i.e., a final staining score of ≥ 7).
| Multivariate Analysis Pb | ||||
| Risk factor | HR | 95%CI | ||
| Age(years) | ||||
| ≥59vs<59 | 1.338 | 0.546 | 3.281 | 0.524 |
| Gender | ||||
| Males vs female | 0.798 | 0.326 | 1.954 | 0.622 |
| T classification | ||||
| T2vs T3-T4 | 1.414 | 0.539 | 3.71 | 0.482 |
| N classification | ||||
| N0vs N1 | 1.025 | 0.174 | 6.028 | 0.978 |
| M classification | ||||
| M0vs M1 | 1.445 | 0.217 | 9.619 | 0.704 |
| Clinical stage | ||||
| I-II vs III-IV | 1.964 | 0.287 | 13.448 | 0.492 |
| HMGB1 expression | ||||
| Low vs high | 1.46 | 0.442 | 4.819 | 0.535 |
Table 4: Multivariate analysis of cancer-related survival in the total populationsa. P of multivariate analyses were determined by Cox regression analysis. b. HMGB1 expression was categorized according to the ration and the average intensity of positive-staining tumor cells into low expression (i.e., a final staining score of 7), high expression (i.e., a final staining score of ≥ 7).
Discussion
HMGB1 has for a long time been known to enhance the activity of transcriptional activators and repressors by binding to transcription factors, and by simultaneously interacting with and distorting chromosomal DNA [16]. HMGB1 plays dual roles as both a chromatin structural protein and a cytokine. Furthermore, several publications have reported up-regulated HMGB1 protein in different tumor types [10]. A significant number of studies indicate that HMGB1 promotes the development of metastasis and also induces a poor prognosis to cancer [17,18], although the precise mechanism of action remains unknown.
In present study, we show that HMGB1 protein was expressed in human PANC-1 cells and overexpressed in human pancreatic cancer tissues. Furthermore, HMGB1 overexpression was significantly associated with T classification, M classification, and clinical stage, but not with patient gender, age, N classification. There were several receptors identified for HMGB1 including advanced glycation end products (RAGE), toll-like receptors (TLR) 2 and 4, syndecan, a specific receptor, tyrosine phosphatase, and thrombomodulin [18,19]. HMGB1 is a ligand for the receptor for RAGE [20]. HMGB1-RAGE signaling triggers activation of key cell signaling pathways, such as MAPK, NF-κB, and Rac/Cdc42 [21], thereby reprogramming cellular properties; Blockade of the HMGB1-RAGE interaction by soluble or mutated RAGE can suppress activation of p44/p42, p38, and SAP/JNK MAPK kinases and inhibition of migration of the glioma, forcing the tumor to undergo prolonged dormancy with decreased proliferation, invasion, and matrix metalloproteinase (MMP) activity [10]. Furthermore, Park et al. [22] studies indicate that both TLR2 and TLR4 are also involved in cellular activation by HMGB1. The participation of multiple receptors in HMGB1 signaling is somewhat surprising, similar results have been reported by other investigators, who also found that HMGB1 could activate cells through TLR-dependent and TLR-independent pathways [23]. In contrast, RAGE played only a minor role in the activation of macrophages by HMGB1 [22]. Consequently, HMGB1 clearly plays a role in cancer development and metastasis. However, the full extent of that role remains unknown along with the possibility of its interaction with other signaling systems. These findings represent a current challenge in modern medicine to exploit its capability to promote reparative responses while preventing its pathological potential.
In this report, we demonstrated the prognostic significance of HMGB1 protein in PC, and HMGB1 protein expression was inversely correlated with overall survival. In univariate analyses, high level expression of HMGB1 was a significant predictor of poor prognosis for patients with PC. This tendency suggested that HMGB1 might be an important prognostic marker for early-stage patients with PC. Prospective clinical studies are necessary to confirm that HMGB1 is one of the reliable clinical predictors of outcome for individual early-stage PC patients.
Although the results of univariate analyses showed that the survival time was significantly different between groups with high and low expression of HMGB1(HR=2.533,p=0.026), no statistically significant difference(HR=1.460,p=0.535) was observed by multivariate analyses, including the age, gender, T\N\M classification, clinical stage were used to analyze. The main reason of this phenomenon maybe was that our sample size was too small to show the significant difference.
In summary, this study certified that the expression of HMGB1 was highly increased in human PC. Furthermore, HMGB1 expression was inversely correlated with survival and directly correlated with the malignant status of PC. The discoveries in this study provide new insight into understanding the molecular mechanism involved in PC progression and prognosis, and may lead to the development of new approaches for available diagnosis and therapy.
Acknowledgements
We would like to thank Zhou Chengjun Professor, Department of Pathology, The Second Hospital of Shandong University, for his help with the pathology analysis.
References
- Sener SF, Fremgen A, Menck HR, Winchester DP (1999) Pancreatic cancer: a report of treatment and survival trends for 100,313 patients diagnosed from 1985-1995, using the National Cancer Database. J Am CollSurg 189: 1-7.
- Maher JF, Nathans D (1996) Multivalent DNA-binding properties of the HMG-1 proteins. ProcNatlAcadSci U S A 93: 6716-6720.
- Andersson U, Erlandsson-Harris H, Yang H, Tracey KJ (2002) HMGB1 as a DNA-binding cytokine. J LeukocBiol 72: 1084-1091.
- Bianchi ME (2004) Significant (re)location: how to use chromatin and/or abundant proteins as messages of life and death. Trends Cell Biol 14: 287-293.
- Czura CJ1, Wang H, Tracey KJ (2001) Dual roles for HMGB1: DNA binding and cytokine. J Endotoxin Res 7: 315-321.
- Andersson U, Wang H, Palmblad K, Aveberger AC, Bloom O, et al. (2000) High mobility group 1 protein (HMG-1) stimulates proinflammatory cytokine synthesis in human monocytes. J Exp Med 192: 565-570.
- Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, et al. (1999) HMG-1 as a late mediator of endotoxin lethality in mice. Science 285: 248-251.
- Scaffidi P, Misteli T, Bianchi ME (2002) Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 418: 191-195.
- Taguchi A, Blood DC, del Toro G, Canet A, Lee DC, et al. (2000) Blockade of RAGE-amphoterinsignalling suppresses tumour growth and metastases. Nature 405: 354-360.
- Brezniceanu ML, Völp K, Bösser S, Solbach C, Lichter P, et al. (2003) HMGB1 inhibits cell death in yeast and mammalian cells and is abundantly expressed in human breast carcinoma. FASEB J 17: 1295-1297.
- Rauvala H, Huttunen HJ, Fages C, Kaksonen M, Kinnunen T, et al. (2000) Heparin-binding proteins HB-GAM (pleiotrophin) and amphoterin in the regulation of cell motility. Matrix Biol 19: 377-387.
- Parkkinen J, Rauvala H (1991) Interactions of plasminogen and tissue plasminogen activator (t-PA) with amphoterin. Enhancement of t-PA-catalyzed plasminogen activation by amphoterin. J BiolChem 266: 16730-16735.
- Takada M, Hirata K, Ajiki T, Suzuki Y, Kuroda Y (2004) Expression of receptor for advanced glycation end products (RAGE) and MMP-9 in human pancreatic cancer cells. Hepatogastroenterology 51: 928-930.
- Paul K, Sobin LH (1999) World Health Organization Classification of Tumours. Chapter 10. France: IARC 220-230.
- Harvey JM, Clark GM, Osborne CK, Allred DC (1999) Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J ClinOncol 17: 1474-1481.
- Dintilhac A, Bernués J (2002) HMGB1 interacts with many apparently unrelated proteins by recognizing short amino acid sequences. J BiolChem 277: 7021-7028.
- Nestl A, Von Stein OD, Zatloukal K, Thies WG, Herrlich P, et al. (2001) Gene expression patterns associated with the metastatic phenotype in rodent and human tumors. Cancer Res 61:1569-1577.
- Milev P, Chiba A, Haring M, Rauvala H, Schachner M, et al. (1998) High affinity binding and overlapping localization of neurocan and phosphacan/protein-tyrosine phosphatase-zeta/beta with tenascin-R, amphoterin, and the heparin-binding growth-associated molecule. J BiolChem 273:6998-7005.
- Mitola S, Belleri M, Urbinati C, Coltrini D, Sparatore B, et al. (2006) Cutting edge: extracellular high mobility group box-1 protein is a proangiogenic cytokine. J Immunol 176: 12-15.
- Huttunen HJ, Fages C, Kuja-Panula J, Ridley AJ, Rauvala H (2002) Receptor for advanced glycation end products-binding COOH-terminal motif of amphoterin inhibits invasive migration and metastasis. Cancer Res 62: 4805-4811.
- Liu L, Zhao L, Zhang Y, Zhang Q, Ding Y (2007) Proteomic analysis of Tiam1-mediated metastasis in colorectal cancer. Cell BiolInt 31: 805-814.
- Park JS, Svetkauskaite D, He Q, Kim JY, Strassheim D, et al. (2004) Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J BiolChem 279: 7370-7377.
- Tsung A, Sahai R, Tanaka H, Nakao A, Fink MP, et al. (2005) The nuclear factor HMGB1 mediates hepatic injury after murine liver ischemia-reperfusion. J Exp Med 201: 1135-1143.
Relevant Topics
- Constipation
- Digestive Enzymes
- Endoscopy
- Epigastric Pain
- Gall Bladder
- Gastric Cancer
- Gastrointestinal Bleeding
- Gastrointestinal Hormones
- Gastrointestinal Infections
- Gastrointestinal Inflammation
- Gastrointestinal Pathology
- Gastrointestinal Pharmacology
- Gastrointestinal Radiology
- Gastrointestinal Surgery
- Gastrointestinal Tuberculosis
- GIST Sarcoma
- Intestinal Blockage
- Pancreas
- Salivary Glands
- Stomach Bloating
- Stomach Cramps
- Stomach Disorders
- Stomach Ulcer
Recommended Journals
Article Tools
Article Usage
- Total views: 15117
- [From(publication date):
June-2014 - Apr 02, 2025] - Breakdown by view type
- HTML page views : 10472
- PDF downloads : 4645