Review Article Open Access
Hexachlorocyclohexane Contamination and Solutions: Brief History and Beyond. Emerging Model to Study Evolution of Catabolic Genes and Pathways
| Nayyar N and Lal R* | |
| Department of Zoology, University of Delhi, Delhi-110 007, India | |
| *Corresponding Author : | Lal R
Department of Zoology University of Delhi, Delhi-110 007, India Tel: +91112766625560 E-mail: ruplal@gmail.com |
| Received: January 27, 2016; Accepted: February 29, 2016; Published: March 05, 2016 | |
| Citation: Nayyar N, Lal R (2016) Hexachlorocyclohexane Contamination and Solutions: Brief History and Beyond. Emerging Model to Study Evolution of Catabolic Genes and Pathways. J Bioremed Biodeg 7:338. doi:10.4172/2155-6199.1000338 | |
| Copyright: © 2016 Nayyar N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | |
Visit for more related articles at Journal of Bioremediation & Biodegradation
Abstract
Recent revelation of the evolution of Hexachlorocyclohexane (HCH) degrading sphingomonads and their acquisition of lin genes for the degradation of HCH isomers at the HCH dumpsites and HCH contaminated sites has lead us to consider that bacteria employ science and chemistry beyond scientific imagination. The HCH contamination of the environment portrays one of the best examples to highlight the evolution of catabolic genes and pathways leading to survival of these bacteria at HCH concentrations as high as 450 mg/g soil. While contamination of the environment with HCH has created several health related issues, this compound has not only enabled us to study the marvel of HCH degradation that sphingomonads employ but has also emerged as a good model to study the evolution of catabolic genes, especially the lin genes. The potential of HCH degradation by these sphingomonads can be tapped in order to create a phenomenal and large scale bioremediation technology. This review describes briefly the massive contamination of our environment by HCH isomers, along with the spontaneous evolution of the versatile HCH degradation pathways in sphingomonads and lin genes in response to HCH.
| Keywords |
| Hexachlorocyclohexane; Bioremediation; Sphingomonads; Catabolic genes; Evolution |
| Abbreviations |
| HCH: hexachlorocyclohexane; PCCH: pentachlorocyclohexene; 1,4-TCDN: 1,3,4,6-tetrachloro-1,4-cyclohexadiene; 1,2,4-TCB: 1,2,4 trichlorobenzene; 2,4,5-DNOL: 2,4,5-trichloro-2,5- cyclohexadiene-1-ol; 2,5-DCP: 2,5-dichlorophenol; 2,5-DDOL: 2,5-dichloro- 2,5-cyclohexadiene-1,4-diol; 2,5-DCHQ: 2,5-dichlorohydroquinone; CHQ: Chlorohydroquinone; HQ: hydroquinone; γ-HMSA: gamma-hydroxymuconicsemialdehyde (Adapted from Ref. [11]). |
| Introduction |
| Hexachlorocyclohexane (HCH), a cyclic, saturated, chlorinated hydrocarbon insecticide had been extensively used and produced between early 1950’s to late 1980’s. The extensive use of this pesticide during the past 60 years or so had brought enormous benefits to mankind including the protection of crops from pests and by eradicating vector borne diseases [1,2]. However, the unusual and unregulated process of production and indiscriminate and extensive use of HCH over a large number of years has created a serious issue of contamination of the environment. Of grave concern is the open air stockpiling of HCH muck, improper management and illegal disposal coupled with continuous shifting of HCH muck from the HCH dumpsites. All this has resulted in wide spread and high level contamination of HCH. Due to the high toxicity, magnitude of contamination and persistence of these HCH isomers, they have been listed as the Persistent Organic Pollutants (POPs) by the Stockholm convention in 2009 [3,4]. Sadly, neither the ban of HCH production nor its restricted use was successful in reducing the levels of HCH in the environment. The problem is aggravated due to biotransformation and biomagnification though the food web and food chain of HCH residues that appear even in human milk, blood and fatty tissues [5,6]. |
| Apart from the worst effects of HCH, no other compound to our knowledge has produced such an interesting scientific data, depicting the appearance of sphingomonads at the HCH dumpsite and acquisition and evolution of HCH degrading catabolic genes in a very short time in these bacteria. While the use of HCH has been hardly for 60 years, but the rapid evolution of the catabolic genes and development of isomeric specific degradation pathways in Sphingomonads have portrayed an unexpected picture of bacterial evolution beyond scientific imagination. |
| The progress on research in this area was slow between 1900’s to 2000 [7-9] but moved with an extraordinary speed during the past 15 years or so. Although there is an excellent compilation of research describing the genetics and biochemistry of the degradation of HCH isomers, origins and evolution of lin catabolic genes [4,10,11], the past five years have provided vast information on the metagenomics analysis of microbial diversity present at the HCH dumpsites, isolation of several sphingomonads and their genome analysis, broad substrate range of lin genes and several functions these bacteria perform at the HCH dumpsites. Thus, the goal of this article is not to collect again such a wealth of information and to reinforce the points already made, but to review the situation as we see it now (most particularly with respect to the origins and evolution of lin genes and catabolic pathways) and to provide critical analysis of the status of research on this aspect to give directions for future research. However to make it cohesive and useful for even first time readers we have also tried to cover the good, bad, weird and wonderful aspects of HCH and Sphingomonads and brought in the forefront the impending bottlenecks that need to be resolved to develop an effective bioremediation technology. |
| HCH: an unusual pesticide: environmental risk remained unknown for many years |
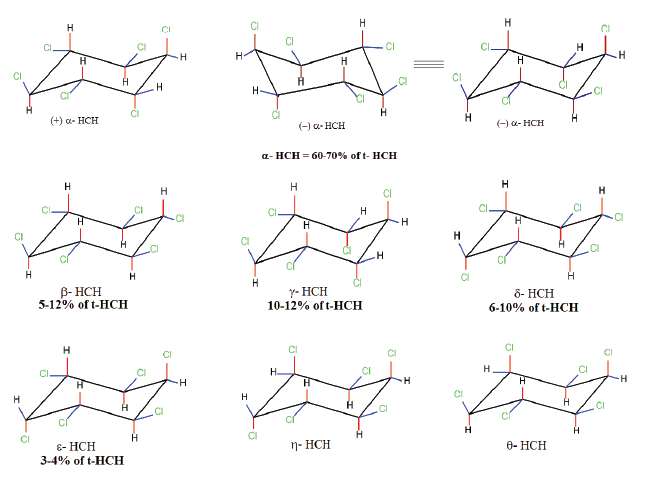
| While the benefits accrued due to the use of technical HCH or lindanes are known, the secret of contamination of our environment by HCH isomers is not very well known. The problem of environmental contamination by HCH isomers has been mentioned very diligently in several articles from time to time [12-15]. Scientists, environmentalists and government agencies are only just beginning to understand the potential risks and consequences associated with contamination by HCH isomers especially at the HCH dumpsite. What was not known to the public was that the unusual process of HCH production and indiscriminate usage was leading to widespread environmental contamination. HCH is commercially synthesized by the chlorination of benzene in the presence of UV resulting in the formation of five major stable isomers namely α−, β−, γ−, δ− and ε− ΗCH (70: 14: 8: 6: 2) [16] (Figure 1). This mixture is also known as ‘technical HCH’ (t-HCH), of which the γ- isomer is purified and marketed as lindane, a well-known pesticide [17]. The purification of one ton of lindane from the HCH mix: α− ( 60−70%), β−( 5−12%), γ−( 10−12%), δ− ( 6−10%), ε− (3−4%) leads to the production of nearly 10 tons of HCH muck primarily containing α−, β−, δ−, ε−HCH. A rough estimate indicates that between 1950-2010 nearly 60,000 tons of γ−ΗCH was produced leaving behind nearly 4-6 million tons of HCH muck [4]. All the HCH isomers differ in their chemical and biological properties due to the different spatial arrangement of chlorine atoms around a central cyclohexane ring (Figure 1). This in turn has an impact on the environmental fate, and distribution of these isomers [16]. In case of the β- isomer, due to the presence of all equatorial chlorine atoms its susceptibility to bio-chemical transformation is reduced to a greater extent [16]. Additionally, the β- isomer has a very high melting point and lower water solubility due to its structural symmetry and greater crystal lattice energy, resulting in its greater stability and its persistence in the environment (Table 1, Figure 1) [18]. The major problem associated with HCH is the process of purification and concentration of the insecticidal γ - isomer from its technical mix that involves multiple steps thereby increasing its cost of production. Due to economic concerns the developing countries continued using technical HCH in place of the purified γ -HCH, while the developed nations could afford its heavy production cost. Purification and processing of lindane has thus led to production of large amount of HCH isomers with a predominance of α−, β−, δ− and ε− HCH (commonly referred as HCH muck) in contrast to lindane [19]. Consequently, the former i.e., the purification of lindane, has resulted in the release and spread of the HCH waste into the environment hence causing environmental pollution, while the latter has led to creation of stockpiles of HCH waste (Table 2). |
| HCH dumpsites and their shifting from one location to another |
| Approximately 10-12 tons of HCH muck (α-, β- , δ- and ε− ΗCH) is generated with the purification of one ton of γ-HCH [10]. For a very long period of time this muck has either been dumped in the open by the industries and in a few cases it has been pyrolysed to trichlorobenzene and hydrochloric acid [37]. It is estimated that between 4 to 7 million tons of bio-accumulative and toxic HCH muck should have been dumped at different locations around the globe. While several such HCH dumpsites are being discovered year after year but the sites with heavy HCH contamination (i.e. more than 50,000 tons) have been reported from The Brazil [20], Netherlands [21], US [28], India [29] Germany [38,39], France [40], Spain [41], Greece [42], Canada [28], Japan [43], and many other countries [11] are well documented (Table 2). |
| While the serious consequences of these dumpsites were not known at the time of their creation, the danger posed by these HCH isomers is being realized now. To our existing knowledge there seems to be no technology available to get rid of these chemicals. The first possible and feasible step for preventing the further addition of HCH to the environment was to halt its production and consumption. Consequently, realizing the danger posed by the HCH isomers, most of the nations banned the production of lindane and its usage was brought to a halt [4]. However, under the pretext of use of lindane for the control of malaria, India continued producing lindane until 2010. Prior to 1999, only technical HCH was used and its use peaked to 25000 million tons during 1990s. During the past 15 years, around 7000 – 10000 tons of lindane has been produced that should have led to stockpiles of HCH muck weighing nearly ~90000 million tons that still awaits clearance [29]. However, more accurate estimates of HCH residue and the number of HCH dumpsite are missing due to the lack of available data. Our frequent visits to this site revealed that HCH muck is continuously shifted from these dumpsites to other locations because of the use of the land that contained these dumpsites for other purposes. In most other countries especially in Bitterfeld in Germany the sites are completely sealed and there are very little human or animal activities there. The cause of concern today is the HCH muck, especially the notorious β-isomer, which is highly bioaccumulative due to its very low water solubility and reduced vapor pressure [8]. Moreover, HCH residues from the dump sites continuously enter the environment either through leaching into the nearby water bodies or too far off areas by wind. Until these HCH dump sites are cleared, the dirty play of HCH contamination will continue spreading. |
| Covert scientific aspects of HCH contamination |
| However, there is indeed a bright depiction of HCH contamination. Putting up HCH in the environment especially creating HCH dumpsites has led to the production of interesting scientific data. Research over the past decade has generated a vast pool of scientific information especially from the HCH dumpsites. It has been thus projected that a solution to the based on this vast scientific data may come in near future. Though there exist various alternatives for HCH decontamination like incineration, landfills, etc., the most effective, economical and greener alternative remains bioremediation– exploiting the catabolic abilities of microorganisms against this problematic pollutant this seems possible for HCH for number of reasons. While there are no good and plenty of microbes available that degrade persistent pesticides like DDT, endosulphan and many other persistent chemicals, there is now a vast pool of such bacteria isolated from the HCH dumsites has residues as high as 450 mg /g soil [29]. |
| Bioremediation involves three major approaches: biostimulation, bioaugmentation and enzymatic remediation. While bio stimulation involves the addition of oxygen and other inorganic nutrients to stimulate the existing bacterial activity against the contaminant, bio augmentation involves the introduction of other HCH degrading bacteria into the contaminated site which will act against the pollutant along with the resident bacteria. Enzymatic remediation, although not used extensively, seems to be feasible in this case because of the extensive characterization of two major enzymes that are the primary enzymes that initiate the process of degradation of HCH isomers. |
| It will be pertinent to mention here that the problem of contamination is posed not only γ- but by α-, β-, δ- and ε-HCH that are discarded as HCH muck after the purification of lindane [10]. |
| Interestingly presence of these HCH isomers at the HCH dumpsites has resulted in the evolution and selection of various bacterial strains that are able to degrade these HCH isomers to varying degree. Several sphingomonads are now available from different HCH contaminated sites especially from HCH dumpsites that can now be exploited for their potential to degrade HCH isomers at the HCH dumpsite (Table 3). |
| Overview of the early research during 90s |
| After the concerns of persistent nature of HCH isomers, scientists began to look for bacterial strains that degraded HCH isomers and reports appeared on the anaerobic degradation of organic pesticides that were adequately described in two review articles [11,49]. After since the production of lindane until 1970, there was no concern of the toxicity, persistence and degradation of HCH isomers. The first bacterium known to be involved in the aerobic degradation of HCH was isolated from lindane contaminated soils in Japan [43] in 1989 and is now called Sphingobium japonicum UT26 [50]. Although UT26 was able to degrade the γ-isomer, it was then projected not to degrade β-HCH. The inability to discover microbes capable of degrading the β - isomer made the scientific community believe that there will not be any microbial system capable of degrading β - isomer due to its stability and equatorial position of chlorine atoms around the cyclohexne ring. However, immediately thereafter, the discovery of another bacterium now called Sphingobium indicum B90A from sugarcane rhizophere soils in India in 1990 [51] refuted this notion. Not only could this bacterium degrade α -, δ -, and γ -HCH but also the β - isomer. Although B90A became an important strain to study the biochemistry and genetic of degradation of HCH isomers, prior to 2000 majority of the work was done on UT26 and only with respect γ -HCH (lindane) degradation. But nothing much was known about the degradation of other four isomers i.e. α -, β -, δ - and ε -HCH that are really problematic and require immediate remediation at the HCH dumpsites. Until 1990 there were only two strains of sphingomonads that were available for the study, i.e., Sphingobium japonicum UT26 and Sphingobium indicum B90A, however there was a follow of bacterial strains especially sphingomonads that were discovered from the HCH dumpsites located in Germany [24], Spain [26], China [27], and India [52]. The discovery of several HCH degrading sphingomonads from HCH contaminated soils and identification of several HCH dumpsites, has opened up new avenues for a better understanding of the genetics and biochemistry of HCH degradation and develop bioremediation technologies. |
| A rich microbial diversity at the HCH dumpsite |
| Bacterial species for long have proven that they can play a significant role in putting to use their natural biochemical processes for the cycling of nutrients and detoxifying many harmful compounds. Although understanding the evolution of catabolic genes in bacteria is difficult, HCH pressure offers a very good example of bacteria acquiring catabolic genes in a very short span of time as the first dissemination of γ - HCH into the environment occurred around 65 years ago. Since then, under HCH pressure, bacteria not only adapted to the menace, but also acquired the ability to degrade it and found ways for the dissemination of HCH catabolic genes across themselves. As stated previously, after their isolation in 1990 both, S. japonicum UT26 and S. indicum B90A for at least 10 years remained the only two strains known which had the ability to degrade HCH. However, Boltner et al. [24] and Mohan et al. [26] reported a large number of HCH metabolizing bacteria from the dumpsites (to varying degrees) located in Germany and Spain. During the next five to six years or so, the literature was flooded with novel HCH degrading bacterial strains, mostly sphingomonads that were isolated from many such HCH dumpsites from India [30,31,44-46,53,54], Germany [24], Spain [26], Japan [43] and Czech Republic [36,47,48] and China [27]. These bacterial strains have helped in a better understanding of the biochemistry and genetics of degradation of HCH isomers [11] along with providing a way for remediating HCH by biological means. |
| The first basic requirement for developing a bioremediation technology is first to understand the gravity of the problem and then to look for suitable microorganisms with degradation potential. This seems to have been fulfilled to some extent. It is also essential to understand the basic biochemistry, genetics and physiology of degradation pathway. Suitable microbes can then be mass cultivated and used individually or in form of consortia followed by field studies. In this context, the above mentioned aspects to develop a bioremediation technology are nearly complete. |
| HCH degrading lin genes: A rapidly evolving system at the HCH dump sites |
| The catabolic genes involved in the degradation of HCH isomers, termed as lin genes, were first identified in Sphingobium japonicum UT26. Subsequently, other lin genes (lin A, B, C, DER, F, G, H, J, K, L, M, N and X) [55] and regulatory genes (linI and R) [56,57] were identified that were involved in either the uptake or the degradation of HCH isomers in this strain. LinA (HCH dehydrochlorinase, acting on γ-, α-, δ-, ε- HCH) and LinB (halidohydrolase, acting on β-, δ-, ε- HCH and also on α- HCH) are the key enzymes involved in the degradation. Interestingly, S. indicum B90A was found to contain two copies of linA genes (linA1 and linA2) [58] which enantioselectively transformed (+) α-HCH (by LinA1) and (-) α-HCH (by LinA2) [59]. Other genes involved in the pathway include linC (dehydrogenase), linD (reductive dechlorinase), linE/linEb (ring cleavage oxygenase), linF (maleylacetate reductase); linGH (acyl-CoA transferase), and linJ (thiolase). While linA to linC are constitutively expressed, linD to linJ are induced either by γ- and α-HCH or their metabolites. The constitutively expressed genes encode enzymes participating in the upper pathway while linD to linJ encode enzymes responsible for the lower pathway. In addition, various studies on S. japonicum UT26 [60] and S. indicum B90A [61,62] have confirmed that these catabolic genes are scattered throughout the genome (either on the chromosome or on the plasmids) instead of being organized in co-ordinately regulated compact operons. Studies [63] have also revealed the presence of four additional lin genes linKLMN which though not endowed with degradation potential are essential for utilization of γ-HCH in Sphingobium japonicum UT26. These genes encode a putative ABC transporter system consisting of four components – a permease, ATPase, periplasmic protein, and lipoprotein. The core of this ABC transporter system is formed by LinK homologue (a transmembrane protein) and the LinL homologue (a cytoplasmic ATPase). In addition, LinM and LinN homologues have been found to function in substrate-binding and serving as a lipobox respectively. Thus, apart from the catabolic genes and regulatory elements, this gene cluster acts as a cellular factor essential for uptake of γ-HCH or other HCH isomers by controlling the membrane hydrophobicity. Many members of proteobacteria possess orthologues of linKLMN which exist as a cluster and form a putative operon [63]. This also includes many species that have not been reported to have any association with xenobiotic degradation (Figure 2a and 2b). |
| Rapid evolution of genes |
| The first HCH degrading bacterial strain discovered was known to degrade only the γ -isomer. Nothing was known about the degradation of the other isomers. However, continuous research for many years on sphingomonads, lead to the establishment of new pathways of HCH degradation. Additionally, the genes involved in the degradation pathways have undergone mutations and variations resulting in a wider scope to act on multiple isomers a well as further extending the pathway. The two enzymes that are known to be the key enzymes for HCH degradation are LinA (HCH dehydrochlorinase) and LinB (haloalkanedehalogenase) that are known to compete with each other for different substrates: HCH isomers or their intermediates for degradation. |
| It is known that these lin genes are associated with single or composite transposable element, IS6100 [61]. Another captivating aspect is the acquisition of lin genes specifically by Sphingomonads. Approximately, 90% of the HCH degrading bacteria known today are sphingomonads in which the lin genes also appear to be associated with IS6100 elements. These genes are absent from non-HCH degrading organisms indicating the horizontal movement of these genes in nature as well as the rapid evolution of these genes by genome rearrangements and gene duplications. |
| The HCH degradation system or potential in Sphingomonads can serve as a model to study the rapid evolution and adaptability of lin genes and its pathways. The existing data indicates that exploitation and studies on this system can lead to the development of bioremediation as a technology for decontamination of HCH muck from the environment. This would be the first system developed so far for large scale decontamination of hazardous pesticides. |
| LinA: HCH dehydrogenase |
| LinA, one of the first enzymes of HCH degradation pathway, is primarily a dehydrochlorinase coded by linA gene which belongs to the group of enzymes that remove HCl from the substrate molecule leading to the formation of double bond. Till date, three dehydrochlorinases have been reported that are involved in the degradation of recalcitrant toxic pollutants. These are DDT dehydrochlorinase, which catalyzes the conversion of 1,1,1-trichloro-2,2-bis (4-chlorophenyl) ethane (DDT) into 1,1-dichloro-2,2-bis (4-chlorophenyl) ethylene (DDE) [64]; 5-chloro-2-hydroxyhydroquinone (5-CHQ) dehydrochlorinase, which is involved in the biodegradation of 2,4,5-trichlorophenoxyacetate (2,4,5-T) and hexachlorocyclohexane dehydrochlorinase (LinA), an enzyme that gives pentachlorocyclohexenes (PCCH) from α-, γ-, δ-and ε-HCH (Figure 2) [57,59]. In S. indicum B90A two copies of linA viz., linA1 and linA2 [58] are found which preferentially degrade α -HCH: (+)-, and (-)- α -HCH to β -1,3(S),4(S),5(R),6(S)-PCCH and β -1,3(R),4(R),5(S),6(R)-PCCH respectively [59]. |
| linA gene was first cloned by Imai et al. from the organism Sphingobium japonicum UT26S and then expressed in Escherichia coli followed by its purification [65]. Further study has revealed its sub-cellular location in the periplasmic space [66]. The enzyme is 156 a.a. long, with GC content 53%. The activity of the enzyme depends on the stereochemistry of the enzyme and thus it was confined to only those isomers which possess 1,2-biaxial HCl group [67,68]. |
| linA gene from Sphingobium japonicum UT26S was analyzed for its stereochemistry to propose the absolute configuration, confirmation of the reactants as well as products and the reaction mechanism by Tratirek et al. [68]. The study brought to light few very interesting points like the most probable configuration of γ -HCH in the active site is its chair conformation and the HCl pair involved in the reaction is made to adapt the 1,2-biaxial configuration in the enzyme active site because the enzyme favors this spatial confirmation in order to prefer it as a substrate. The reaction mechanism of γ -HCH dechlorination is similar to E2-like dehydrochlorination mechanism. The role of amino acids is very crucial as His-73 is also assisted by Asp-25 which helps it in maintaining the proper orientation and also stabilizes the positive charge which develops on the histidine imidazole ring during this reaction. It has been also proposed that other residues like Lys-20 and Arg-129 also help in stabilizing the intermediates via non-bonding interactions with hydrogen and chlorine atoms. |
| Further, the linA protein shows catalytic mechanism similar to the enzyme scytalone dehydratase [69]. It is basically a homo-trimeric protein with a molecular weight of 16.5 kDa and it belongs to a group of folds called α+β rolls. It is proposed that the polypeptide chain of linA forms α + β barrel which forms a hydrophobic cavity that in turn forms a binding pocket. The putative catalytic residues H73-D25 and putative substrate binding residues K20 and R129-D115 are situated inside this cavity. K20 and R129 are supposed to interact with the chlorine atom in order to stabilize the leaving Cl- ion. Initially it was assumed that D115 assists R129 in its proper positioning, but after analyzing the clones with site directed mutagenesis, it was confirmed that D115 is not essential for linA activity. Substrate specificity of the enzyme was found to be narrow with the enzyme mediating dehydrochlorination reactions in α, γ, δ −HCH and β −, γ − PCCH. |
| LinB: A haloalkane dehalogenase |
| Haloalkane dehalogenase LinB, belongs to α/β hydrolase family, initially identified as the enzyme which catalyzes the conversion of γ -1,3,4,6-tetrachloro-1,4-cyclohexadiene ( γ -TCDN) into 2,5-dichloro- 2,5-cyclohexadiene-1,4-diol (2,5-DDOL) in next subsequent steps of γ -HCH degradation in Sphingobium japonicum UT26 [70]. Later, the identification of its activity over β -HCH [71,72] which is considered as the most recalcitrant and persisting HCH isomer, makes it an important gene for genetic engineering, kinetic and bioremediation studies. Till today there are hundreds of linB variants have been sequenced and analyzed directly from the dumpsite and from the efficient β -HCH degraders [11]. Their activity over β -HCH with respect to the sequence variation has been studied in great detail [73]. Among these, Pandey et al. [74] recently have analyzed the residues critical for LinB activity. Their study has revealed one LinB variant i.e., LinBG2.2 with substitutions at V134L/T135L position was observed with ~80 fold increase in activity than LinBB90A. Sangwan et al. [75] has also shown that LinB is an ancestral gene in Sphingobium spp. Further, its association with insertion element i.e., IS6100 in Sphingobium spp. has been critically analyzed where the gene is found flanked with two copies of IS elements [76]. Also, the strains which are completely devoid of lin genes including linB were identified [36]. Hence, the evidence of linB being ancestral and presence of multiple copies of IS6100 in linB deficient strains proposes that during evolution they have lost the gene via intra-chromosomal single homologous recombination. |
| HCH degradation pathways |
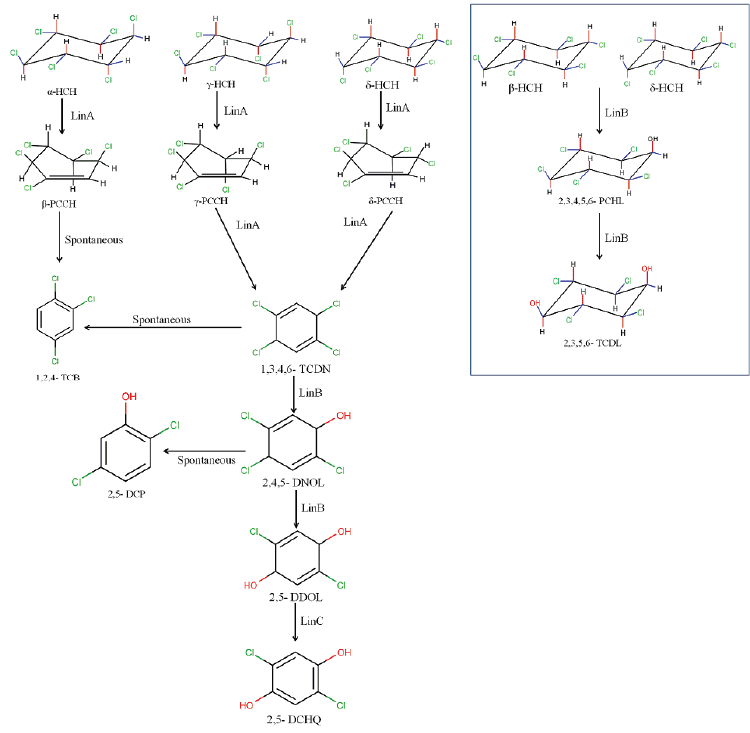
| Ever since the first report of a Pseudomonas paucimobilis strain capable of γ-HCH degradation [77] extensive research has been conducted by various groups to understand aerobic degradation pathways for major HCH isomers viz., α-, γ-, β-, δ - and ε -HCH [57,65,71,72,78-80] (Figure 2). Of all the lin genes that have been reported, it was found that linA and linB that encode HCH dehydrochlorinase and haloalkane dehalogenase respectively, are very important and play a major role in degradation of all the five HCH isomers.In addition to this, the relative proportion of LinA to LinB determines the metabolic routes to be followed during degradation of each of the HCH isomers [81]. |
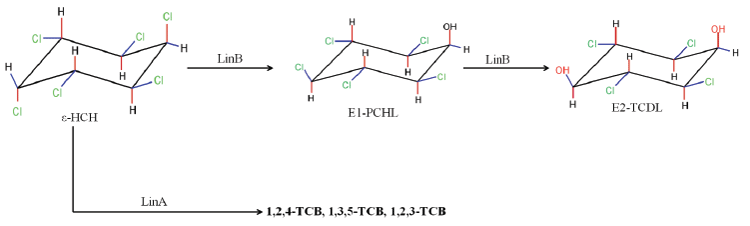
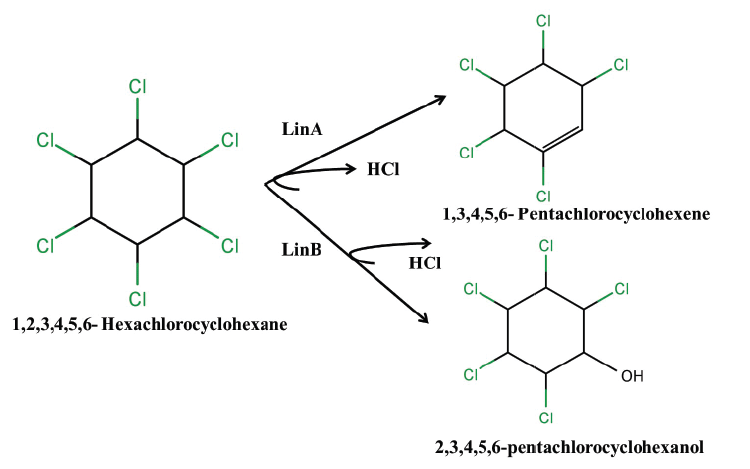
| The degradation of HCH isomers can be schemed into two types of metabolites: PCCHs produced by dehyrochlorination by LinA & hydroxylated derivatives produced by LinB. Although complete metabolism for γ-HCH has been deciphered in UT26 & B90A, the pathways for environmentally relevant isomers: α-, δ- & β -HCH are more important. Presently our understanding of the pathway for these isomers halts at the hydroxylated derivatives: A4 (α-trichloro-2-cyclohexene-1,4-diol), D4 (δ -trichloro-2- cyclohexene-1,4-diol), D2 (δ-tetrachlorocyclohexane-1,4-diol), and B2 (β -tetrachlorocyclohexane-1,4-diol). However metabolism of β- & δ-PCCH by LinA is also studied in detail as these can be substrate for LinA. Not only HCH, biotransformation of hexabromocyclododecanes (HBCD’s) was also observed with LinB [82], thus depicting its broad substrate range. For long the degradation pathway of ɛ -HCH could not be worked out for the simple reason that ɛ -HCH hardly contributes 2-3 % of the total technical mix (even this forms a huge amount at dumpsite) and its purification was difficult. Its degradation by LinA and LinB was not known. We now know that ɛ -HCH is hydroxylated by LinB to E1( ε -pentachlorocyclohexanol) and E2 ( ε -tetrachlorocyclohexane- 1,4-diol) and LinA dehydrochlorinates ɛ -HCH to TCBs (Figure 3). In conclusion LinA and LinB seem to be very important and form a network and intermediates at times compete with other for one substrate. |
| HBCD’s: A new substrate for LinA and LinB |
| Hexabromocyclododecans (HBCD’s) are the third most used brominated flame retardants. Generally these compounds are used as flame retardants for plastic and textiles. These compounds have been shown to exhibit properties of POP’s. HBCD’s and other flame retardants are considered as potential emerging POP’s. Concentration of brominated flame retardants along with HBCD’s have been found in various components of the environment. [83,84]. They are not only found near point sources, i.e. industrial sources but are also found in other remote areas indicating the diffuse sources of HBCD’s. [85]. HBCD’s are highly lipophilic and have been known to diffuse into biota. HBCD’s have been found in sediments of rivers, lakes and sea – indicating that HBCD’s are present in aquatic environment [85,86]. These compounds have been reported to have neurotoxic effects, suggesting that HBCD’s have an inhibiting effect on the uptake of neurotransmitters into synaptosomes and synaptic vesicles [87]. HBCD’s have also been reported to interfere with the Thyroid hormone system [88]. |
| Interestingly, the stereochemistry of 1,2,5,6,9,10 Hexabromocyclododecan seems to be similar to 1,2,3,4,5,6 Hexachlorocyclohexane [89]. In both the cases the six stereogenic centers are formed by complete halogenation of corresponding precursor molecule. Streogenic centers are formed at positions 1,2,5,6,9 and 10 (total six in number) during the bromination of 1,5,9- cyclododecatrienes. Cyclododecatrienes (CDT) are also present in four isomers, trans, trans, trans-; trans, trans; cis-, trans, cis, cis- and cis, cis, cis-. Technical grade CDT used in the synthesis of HBCD are a mixture of these four isomers. Depending upon the purity of the substrate different mixtures of HBCD’s are formed constituting various isomers. |
| HBCD’s are constituted by a total of 16 stereoisomers which include six diastereomeric pairs of enantiomers and four mesoforms. The stereochemistry of HBCD’s is far more complex than that of HCH. However, based on the striking structural and physiological similarities of HCH’s and HBCD’s, it was postulated and hypothesized that LinA and LinB might also accept HBCD’s as substrates. Heeb et al. [82] reported the biotransformation of α-, β-, and γ-HBCDs by LinB from Sphingobium indicum B90A. In 2014, the study was extended to LinA and even LinA was found to metabolize HBCD’s [90]. |
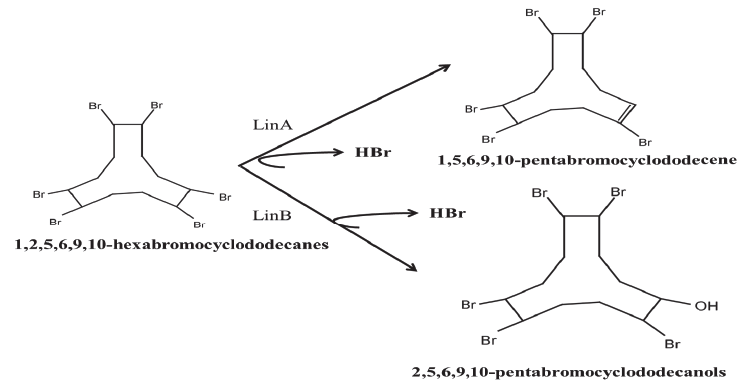
| As mentioned earlier, LinB being a haloalkane dehalogenase enzyme with broad substrate activity acts on a wide range of substrates. This includes β - and δ - HCH, heptachlorocyclohexanes, pentachlorocyclohexenes and tetrachlorocyclohexadienes to mono-hydroxymetabolites [91]. Hebb et al. [82] has introduced new class of its substrates i.e. hexabromocyclododecanes (HBCDs). In the study, they used LinB from Sphingobium indicum B90A, which is already known for its two step transformation of HCH isomers. Enzymatic transformation of HBCD’s by the activity of LinB was found comparable to HCH due to their similar structural and physiological properties [92,93]. Like HCH, in the first step, HBCD’s are hydrolyzed to monohydroxylated pentabromocyclododecanols (PBCDOHs) and were further transformed to di-hydroxylated tetrabromocyclododecadiols (TBCDDOHs) (Figure 4). Moreover, the study also highlighted that PBCDOHs is not naturally occurring compound however identified as by-product in HBCD’s mixtures. Hence, LinB haloalkane dehalogenase, which evolved at HCH dumpsites, might be useful in bioremediation of HBCD’s at HBCD’s contaminated sites. |
| Recently, Heeb et al. [90] has shown that the linA2 enzyme acts on hexabromocyclododecanes (HBCDs). This compound has some basic similarity with HCH. Like HCH, it is lipophilic, polyhalogenated and is having similar stereochemistry. This enzyme uses (-) β -isomer of HBCDs as a substrate and catalyzes the dehydrobromination of 1,2,5,6,9,10-hexabromocyclododecanes into 1,5,6,9,10-pentabromocyclododecanes (PBCDEs). The observed enzymatic conversion indicate that the enzyme although is very specific to HCH, but its activity depends on the stereochemistry of the substrate. HBCDs are highly produced chemicals in order to use as flame-retardants of polystyrenes [84] and has been shown to be wide spread in the environment [83,94-96]. HBCDs are strikingly similar to the HCH in its properties and thus preferred by the same enzyme. In future, the scope will be open for a wide variety of substrates that may act as a substrate for the same enzyme. |
| Bioremediation: The enzymatic aspect |
| In today’s date, the three approaches that are being worked out for bioremediation of HCH include (a) bio augmentation, (b) bio stimulation and (c) enzymatic bioremediation. Large number of sphingomonads can be used for bio augmentation either alone or in combination with each other, making bio augmentation a feasible option. Hence Sphingomonads are a suitable starting resource. Bio stimulation also seems to be quite a promising approach [52]. Sphingomonads can tolerate, survive and degrade HCH isomers at the HCH dumpsite where HCH pressure is approximately 450mg/g of soil [29]. For better results, these two approaches can be combined together to develop a suitable bioremediation technology. The third approach of enzymatic remediation although at a very initial stage of development, also seems to be feasible. Enzymes are a source of significant biotechnological potential and form the basis of several bioremediation strategies intended to reduce the environmental impacts of pesticide residues [97]. Enzymatic bioremediation has a high potential for application at locations of highly contaminated point sources. It involves direct application of detoxifying enzyme formulations to clean up residues in contaminated environments. Some of the prerequisites of using enzymatic bioremediation include cofactor independency, high catalytic efficiency, and stability of the enzyme and economical mass production of enzyme. |
| The use of free enzymes for decontamination of pesticides has been till date the most promising approach. These enzymes are selected based on addressing its stability requirements. These enzymes in majority cases are heterologously expressed in expression vectors and surpass the requirement for microbial growth for activity making them effective in a short period of time. The use of enzymes depends on the availability of a moist environment and appropriate conditions to aid in the action of the enzyme. LinA and LinB are two enzymes which can be considered as potential candidates for a starting resource for a bioremediation technology. Both these enzymes are stable and easy to work with and do not require cofactors for their activity. The activity of these enzymes at every step leads to the removal of a chlorine atom thus reducing the toxicity of the metabolite produced by several folds. Another factor to be considered while using these enzymes as a resource for bioremediation is the fact that these enzymes have many variants. The kinetics and activity of all these variants can be tested and put to use to select the best variant for bioremediation. Various applications have tested optimized organophosphate-degrading enzymes [97]. The key to a successful enzymatic bioremediation program is the availability of highly active and specialised enzymes. Protein engineering and modelling techniques can also be employed to improve the activity of existing enzymes to ensure its applicability as well as to improve its substrate range and isomer specificity. |
| Carbon Stable Isotope Analysis (CSIA): Implications in HCH bioremediation |
| Biodegradation of HCH remains to be a major source of intrinsic remediation in addition to the physical processes of volatilization, dispersion and sorption [98]. Although a lot of serious efforts have been conducted towards bioremediation of HCH with an endeavor to pace up the degradation processes [52] yet they suffer a common major drawback in lacking a methodology to quantify the approximate amount of degradation. CSIA is an analytical technique developed over 30 years with the potential to measure the isotopic markers of substrates and their intermediate degradation products. This conventional method promises to evaluate the success of biodegradation by monitoring the concentration of pollutant especially at the dumpsites. CSIA is based upon the preferential transformation of molecules consisting of light isotopes during the biodegradation reaction and thus leading to the enrichment of molecules with heavier isotopes in the residual (non-degraded) phase during the course of biodegradation. US-EPA [99] in its reports has also considered CSIA as a potential technique for assessing biodegradation at contaminated field sites not only qualitatively but also quantitatively. |
| Stable carbon isotope analysis based studies with reference to HCH have reported reductive dechlorination to be the major degradation process under anaerobic conditions in contrast to the aerobic ones being dominated by biodegradation and photochemical degradation [100,101]. In addition to these studies, CSIA is being increasingly considered for in–situ degradation [102] with an aim to measure 13C/12C fractionation [103] occurring during microbial degradation. Recent revelation of differential fractionation being measured even for the two enantiomers of chiral α - HCH isomer [104,105] has further widened the precinct for future investigation and applications of this technique for assessing HCH bioremediation. |
| Genomes of HCH degrading Sphingomonads: What do they reflect? |
| With increase in number of HCH degraders, there is a need to explore their inbuilt capability to participate in HCH bioremediation. Among them, strains belonging to Sphingobium spp. are prevalent and well known. Hence, to analyze the ongoing phenomenon between them and study their ancestory, Verma et al. [106] performed comparative genome analysis of nine Sphingobium strains. In their study, they examined the evolution and diversification of the lin pathway under HCH pressure, by comparing genomes of six newly sequenced Sphingobium spp. (Strains LL03, DS20, IP26, HDIPO4, P25 and RL3) with were already available in the database as well as the sequence of three reference genomes (Strains UT26S, B90A and SYK6). Among these six newly sequenced strains, five (except DS20) were identified as HCH degraders. The whole of the lin pathway study revolves around UT26S and B90A genomes, as majority of the work has been performed by using them as model organisms. The analysis revealed the mosaic distribution of lin genes and IS6100 elements within Sphingobium spp. The foreign origin of lin genes have been already reported [55,61] based on codon usage and GC content. This was further extended by the two proposed mechanism of recruitment i.e., via IS6100 as well as plasmid mediated. Here, the association of linA with IS6100 was found consistent in all HCH degrading sphingomonads except the two copies of IS6100 that were flanking on both sides of linA in strain RL3. Till now the exact mechanism of action of IS6100 is unknown but the evidences suggest its role is very crucial in the establishment as well as spread of lin pathway [60,61,106]. As far as linB gene is concerned, it is found flanked by IS6100 on both sides in strain IP26 including the resolvase gene which is flanking both the ends of these IS6100. Furthermore, this gene is missing in two newly sequenced Sphingobium strains i.e., strain LL03 and P25. This type of scenario clearly indicates the loss of these genes occur via intra-chromosomal homologous recombination events, although further investigations are needed. Another sphingomonad known as Sphingomonas sp MM1 [76] is very interesting in context of its genomic content. The organism harbours one chromosome and five plasmids (named from pISP0 to pISP4) and the complete set of lin genes are arranged in four of its plasmids. In the comparative analysis of Sphingobium spp., plasmids of strain MM1 were used as reference to delineate the intra genus plasmid pool between genus Sphingobium and Sphingomonas. They observed that the arrangement of lin genes i.e., linB, linC and a truncated linF in pISP4 is also present in newly sequenced Sphingobium sp. HDIPO4. Hence, the two having similar arrangement profile of lin genes may represent the presence of same type of plasmid suggesting the evidence of plasmid mediated recruitment of lin pathway within sphingomonads. |
| While major research in context to microbial decontamination of HCH has been diverted in a direction of developing a deeper understanding of the genetics, physiology and biochemistry, the genomic science has emerged as a strong support. Presently, the two completely sequenced genomes (Sphingobium japonicum UT26 [60] and Sphingomonas sp. MM1 [76] and several partially sequenced, HCH degrading, sphingomonad genomes available in the database (Table 4). Although the sequence of these HCH degrading bacteria opened up a lot of avenues for propagating this research, the sequence of several other sphingomonads was done in order to have a better understanding of the mechanisms involved and genes acquired by these strains [107-113]. The availability of the genome inventory of these microorganisms has opened the window for comparative analysis of the genome content and architecture.For instance, the presence of genes for phenol and pentachlorophenol degradation in S. japonicum UT26 and S. indicum B90A was not reported until their genome was sequenced needs special mention. Though the two organisms are phylogenetically closely related but are different both in terms of their potential to degrade HCH (B90A being efficient than UT26) and presence of degradation pathway for anthranilate and homogentisate (present in UT26 but absent in B90A). Though the genomes of these organisms need to be studied and compared with scrutiny, there is also a need to expand this resource which would lend a scope to study the differences at the genomic level. |
| Recent metagenomic analyses of HCH dumpsite has further brought into some of the interesting changes in the microbial community at the HCH dumpsite as a result of HCH selection pressure. Apart from sphingomonads that were previously thought to be predominant at the dumpsite, there appear to be several other bacteria including Marinobacter, Chromohalobacter [114] that work in association with sphingomonads to degrade HCH isomers. This further reflects that instead of bioaugmentation, biostimulation could be the better choice for HCH remediation at the HCH dumpsite.In an extension to this study, the combination of genomics and metagenomics has brought together very insightful results [75]. The combination of the metagenomic data from the HCH dumpsite as well as the genomic data of two closely related HCH degraders, B90A and UT26 has enabled the reconstruction of the Last Common Ancestor genotype. This analysis revealed that almost 20% of the genes present in B90A and UT26 were absent in the LCA. These include the genes involved in the upper pathway of HCH metabolism, i.e, linA and linC. The absences of these key genes in the LCA suggest the horizontal acquisition of these genes through transposon mediated lateral gene transfer. |
| Metagenomic information: A point to rethink designing bioremediation strategy |
| Metagenomics of stressed environments holds great promise in resolving the unexplored indigenous composition and structure of microbial communities, given that the extreme conditions limit the microbial community composition thus leading to assembling of near complete genomes [115,116]. Recent comparative metagenomic survey of heavily contaminated HCH dumpsite (450 mg/g of soil), 1 km away (0.7 mg/g) and 5 Km away (0.04 mg/g) soil samples performed by Sangwan et al. [114], revealed interesting results in terms of microbial diversity as well as community potential of HCH degradation. Bacterial diversity at the three sites revealed notable differences at genera level, with Pseudomonas, Sphingomonas, Novosphingobium, Sphingopyxis, Marinobacter, Chromohalobacter among the most abundant at HCH dumpsite. Out of these, Sphingomonas, Novosphingobium, Sphingopyxis are already well established HCH degraders. Marinimicrobium, Idiomarina and Salinisphaera were among the unique genera present at the dumpsite, which although lacks any link with HCH degradation/ tolerance potential but have been reported earlier from hyper saline conditions. This suggests that high saline conditions at the dumpsite along with HCH contamination together act in shaping up the microbial community. This also substantiates that besides sphingomonads, other indigenous bacterial community should also be biostimulated for the development of an effective bioremediation technology [114]. |
| Functional assignment of the metagenomic sequence data, revealed enrichment of 12 genes at the HCH dumpsite and 1Km away samples, already reported to be involved in HCH degradation i.e. linA, linB, linC, dehydrochlorinase, chlorocatechol 1,2-dioxygenase, 2,4,6-trichlorophenol monooxygenase, 2,6-dichloro-p-hydroquinone 1,2-dioxygenase, 2,5-dichloro-2,5-cyclohexadiene-1,4-diol, (chloro) muconate-cycloisomerase, LysR family transcriptional regulator (LinR), TRAP-type mannitol/chloroaromatic compound transport system and periplasmic component (ttg2 gene) [114]. This indicates higher metabolic potential of the dumpsite and 1 Km away soil samples to degrade HCH isomers, compared to 5 Km away soil samples where these genes were almost negligible. Also, the enrichment of benzoate, toluene, naphthalene and aromatic ring opening genes at HCH dumpsite hints at further degradation of end products of HCH degradation pathway. This further supplements the on-going efforts to develop the bioremediation technology for HCH contamination by suggesting the use of both known HCH degraders (Sphingomonads) and non-degraders (indigenous community) to improve remediation efforts. |
| Furthermore, metagenome data at the dumpsite was used along with sequence data of two HCH degrading Sphingobium species (Sphingobium japonicum UT26 and Sphingobium indicum B90A) to reconstruct last common ancestor (LCA) genotype [75]. Comparison of LCA genotype with these two subspecies in terms of genes repertoire revealed that >20% of the genes of the subspecies were absent in LCA. This included two genes i.e. linA and linB encoding for enzymes involved in the upper pathway of HCH degradation, which indicates that the descendants acquired these genes under the stress via transposon mediated lateral transfers [75]. This further supports that the lower pathway (linD, linE and linR) genes makes for an evolutionary “long lived” event [75]. This study hence demonstrates the potential of metagenome data to provide insights into development of ancestral phenotypes. |
| Future Prospects |
| HCH waste continues to thrive in the environment with even the addition of new waste till date despite the ban on its production and use. To stop further addition of HCH in the environment, urgent measures are required to stop more release of HCH into the environment as well as to treat and remediate the HCH dumpsites which serve as the major reservoir of pollution. As already discussed, among the remediation methods available, enzymatic bioremediation seems to be the best choice, hence, it becomes essential to get rid of the problems that are being faced to use enzymes as an option. Thus, the best variants that have been identified so far need to be stabilized and mass produced before being used in the form of formulations for applying to the fields directly that are contaminated with tons of HCH waste. This will be just a step ahead in the long journey of HCH decontamination and targets like an effective and cheap technology to be used on a mass-scale still needs to be achieved. But, nevertheless, with all the information and the research that has already been done so far in such a short span of time, the targets are quite very much achievable. |
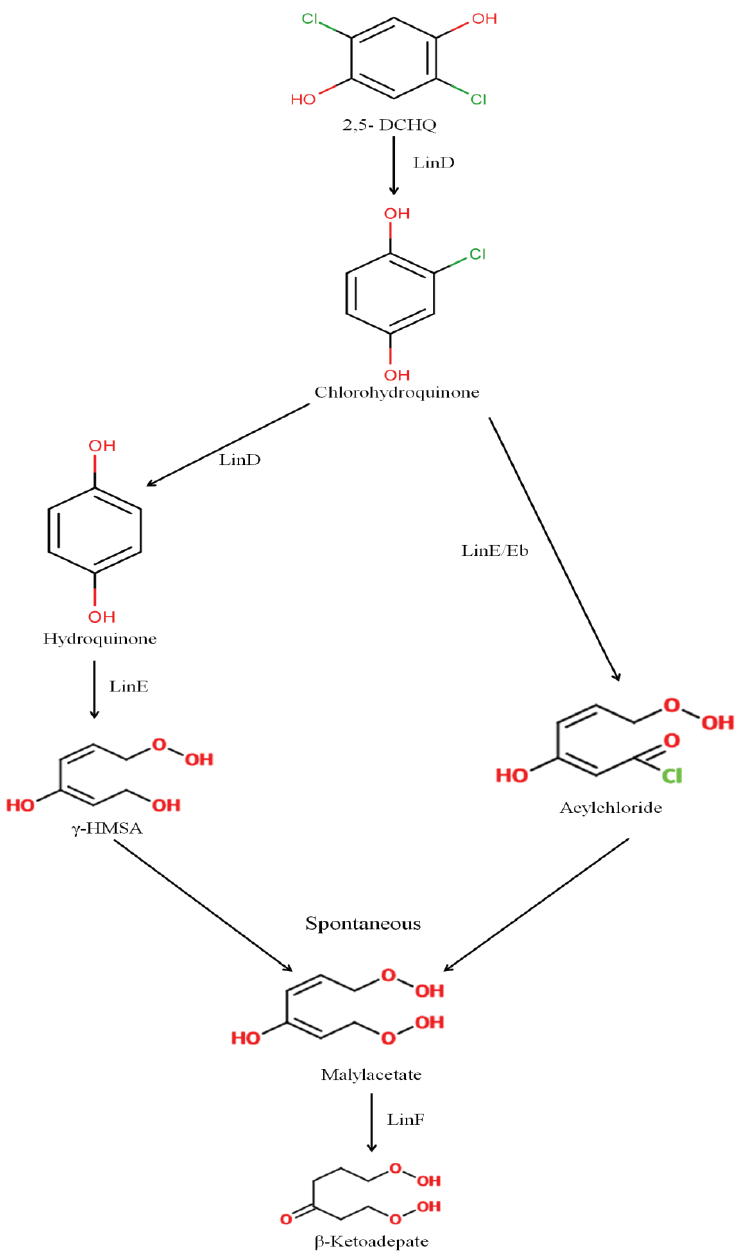
| While we need to devise strategies to resolve this problem that is of pandemic existence, we also need to address the issue of decontamination of HCH muck that has been dumped at open places. Developing fields like Carbon Stable Isotope analysis also can be tapped to devise strategies for decontamination of HCH. This technique can be exploited for assessment of HCH degradation and concentration of contaminant present at the sites. Also, metagenomics and genomic approaches can also be undertaken further to understand the community dynamics active at the HCH dumpsite. The data generated on the genetics, biochemistry and physiology of degradation provide sufficient indication that development of bioremediation for decontamination of HCH isomers is close to happen. During the degradation process two types of metabolites are produced: PCCHs by action of LinA (dehydrochlorination) & hydroxylated derivatives produced by LinB. Although complete metabolism for γ -HCH has been deciphered in S. japonicum UT26 & S. indicum B90A, the pathways for environmentally relevant isomers: α-,δ-, β- HCH are more important. Figure 2a: α – HCH pathway: LinA1 preferentially turns over the (+)-enantiomer, whereas LinA2 prefers the (-)-enantiomer of α -HCH. The metabolite β -PCCH thus produced is spontaneously degraded to TCBs, γ -HCH pathway: Three different dechlorination reactions are sequentially involved in the γ- HCH degradation. These are dehydrochlorination (γ-HCH to 1,4-TCDN), hydrolytic dechlorination (1,4-TCDN to 2,5- DDOL) and reductive dechlorination (2,5-DCHQ to HQ) resulting in chlorohydroquinone (CHQ) as the central intermediate which then undergoes ring cleavage to produce further intermediates that are degraded to β -Ketoadipate (Figure 2b). 1,2, 4- TCB; 2,5-DCP and γ -HMSA are predicted to be a dead-end product [57]. β - and δ -HCH pathway: LinB acts on both the isomers to produce β/δ - PCHL and β/δ - TCDL by hydroxylation reactions in S. indicum B90A. δ - HCH is also acted upon by linA producing δ - PCCH (similar to γ-HCH) which is further metabolized to 1,2,4,5- TCDN by linB by reductive dechlorination. Subsequent action of linB leads to 2,4,5- DNOL and 2,4,5-DDOLs. At times, LinA and LinB seem to compete with each other for this single substrate. |
References
|
Tables and Figures at a glance
| Table 1 | Table 2 | Table 3 | Table 4 |
Figures at a glance
 |
 |
 |
| Figure 1 | Figure 2a | Figure 2b |
 |
 |
 |
| Figure 3 | Figure 4a | Figure 4b |
Relevant Topics
- Anaerobic Biodegradation
- Biodegradable Balloons
- Biodegradable Confetti
- Biodegradable Diapers
- Biodegradable Plastics
- Biodegradable Sunscreen
- Biodegradation
- Bioremediation Bacteria
- Bioremediation Oil Spills
- Bioremediation Plants
- Bioremediation Products
- Ex Situ Bioremediation
- Heavy Metal Bioremediation
- In Situ Bioremediation
- Mycoremediation
- Non Biodegradable
- Phytoremediation
- Sewage Water Treatment
- Soil Bioremediation
- Types of Upwelling
- Waste Degredation
- Xenobiotics
Recommended Journals
Article Tools
Article Usage
- Total views: 14591
- [From(publication date):
March-2016 - Jul 15, 2025] - Breakdown by view type
- HTML page views : 13538
- PDF downloads : 1053
