Review Article Open Access
Global Situation and Trends of HIV, Influenza and Marburg Viruses: An Epidemiological Perspective
Wambani JR1*, Ogola PE2, Arika WM2, Rachuonyo HO3 and Burugu MW21Department of Medical Laboratory Sciences, Kenyatta University, School of Medicine, Nairobi, Kenya
2Department of Biochemistry and Biotechnology, Kenyatta University, School of Pure and Applied Sciences, Nairobi, Kenya
3Department of Microbiology, Kenyatta University School of Pure and Applied Sciences, Nairobi, Kenya
- *Corresponding Author:
- Japheth Rapando Wambani
Department of Medical Laboratory Sciences
Kenyatta University
School of Medicine, Nairobi, Kenya
Tel: +254 20 8710901
E-mail: wambanijapheth@gmail.com
Received date: December 15, 2015 Accepted date: January 08, 2016 Published date: January 18, 2016
Citation: Wambani JR, Ogola PE, Arika WM, Rachuonyo HO Burugu MW (2016) Global Situation and Trends of HIV, Influenza and Marburg Viruses: An Epidemiological Perspective. J Emerg Infect Dis 1:105. doi:10.4172/jeid.1000105
Copyright: © 2016 Wambani JR, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Infectious Disease and Pathology
Abstract
Viruses whose genome is RNA are capable of quickly adapting to different conditions because their polymerases have a higher error rate during replication. Most emerging and reemerging viruses such as HIV, Influenza and Marburg belong to this group of viruses. Different factors contribute to emergence of disease: The factors, coupled with the rapidly increasing human population have led to the emergence and reemergence of viral diseases in human populations over the years. This review discusses the global trends and epidemiology of some RNA viruses over the years to try and trace the epidemics of these diseases down the years and if there can be predictive capabilities.
Keywords
Emerging, Reemerging, HIV, Influenza, Marburg, Global trends
Introduction
HIV, Influenza and Marburg diseases are classified under emerging infectious diseases. Emerging Infectious Diseases are diseases that have recently increased in incidence or in geographic or host range such as tuberculosis, cholera, dengue fever, malaria, japanese encephalitis, yellow fever and west nile fever, OR are diseases caused by new variants due to evolution but are assigned to known pathogens such as the Human Immunodeficiency virus, new strains of Influenza virus, Severe Acute Respiratory Syndrome and Ebola [1].
Molecular mechanisms and cellular processes involved in disease pathogenesis, including transmission dynamics and epidemiology, of pathogens that cause reemerging infectious diseases like Marburg, Ebola, and Cholera have yet to be understood. Emerging diseases caused by novel or unrecognized pathogens such as Hantavirus, or those whose transmission modes are under study, such as Ebola and Nipah present yet another challenge [2].
The mechanisms of disease emergence involve a multiplicity of factors in addition to those at molecular and cellular levels. In addition to virus genetic variation (mutation, reassortment, and recombination), they also include environmental, ocean and air circulation patterns, extreme weather events as well as the ecological and demographic factors that increase the vulnerability of the people to previously unfamiliar microbes[3,4].
Demographic, social changes, environmental alterations and efforts to control diseases have added to the increasing problem of emerging infectious diseases like Influenza and Marburg [5]. The use of antibiotics, pesticides, and biological pathogen controls are effecting variations in pathogens, ecological systems, hosts while facilitating disease emergence and/or reemergence [6-8]. Prime examples are the antibiotic resistant Strepococcus A, Mycobacterium, and E. coli 0:157. Pathogenic microbes and their hosts also grow and reproduce while adapting in different environmental contexts. This is typified by the Avian influenza that has different hosts (chickens, pigs, ducks and humans in close confines).
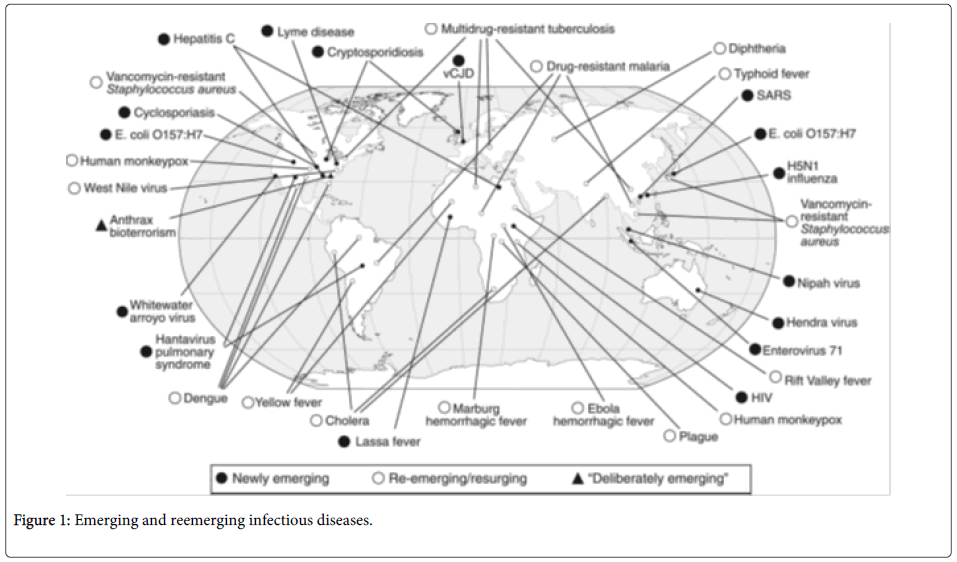
Majority of the emerging pathogens (75%) are zoonotic, that is, they can be passed on from animals to humans. Figure 1 shows the different emerging and reemerging diseases globally.
This review discusses the epidemiology of three RNA viral diseases (HIV, Influenza and Marburg) while tracing their global trends down the years.
HIV/AIDS
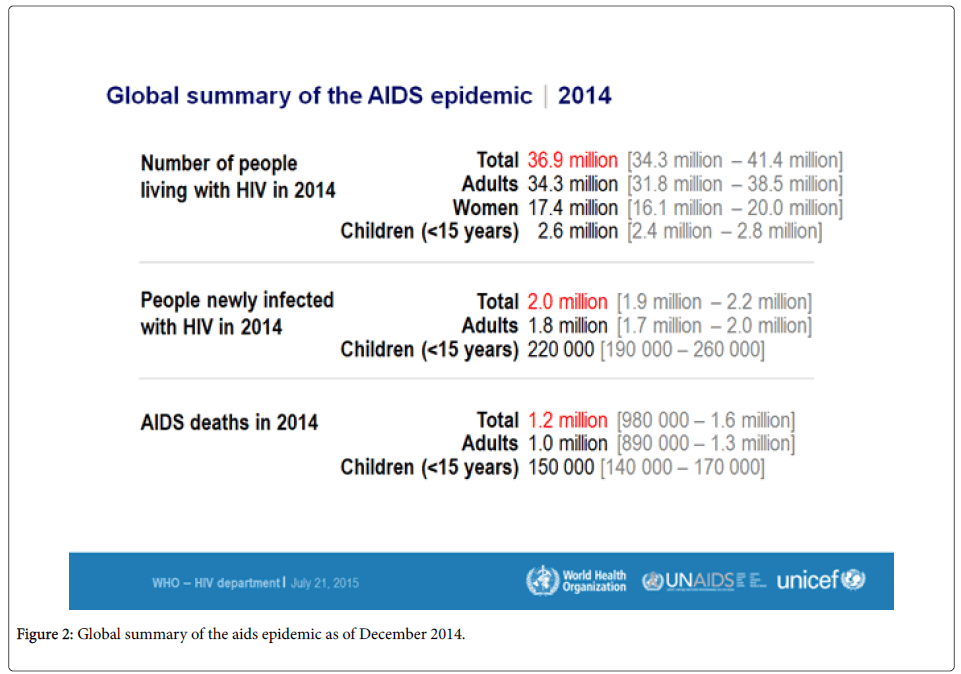
HIV was first described in June 1981. The pandemic is now 34 years old and it will soon be described as one of the matrix diseases. In 1981, it was an emerging disease and it has continued to reemerge since then because of the frequent viral mutations and evolution [9,10]. The epidemic has spread globally due to its subtle transmission routes such as sexual, blood-blood contact as well as through the parenteral route. Figure 2 shows the global summary of the epidemic by the year 2014 according to the world health organization.
HIV treatment has been strongly emphasized on to reduce the effects of the disease and in 2014, 1.9 million people enrolled for antiretroviral treatment. This number represents the largest increase ever and it points towards a disease burden that is on the increase. According to the world health organization, the HIV virus has infected close to 78 million people with about 39 million of them having died since the epidemic broke. Among adults aged 15-49 years, about 0.8% of them are living with the virus. However, the disease burden varies between countries and regions. Sub-Saharan Africa is the most affected, with about 1 in every 20 adults living with HIV. This accounts for about 71% of the people with HIV globally [11,12].
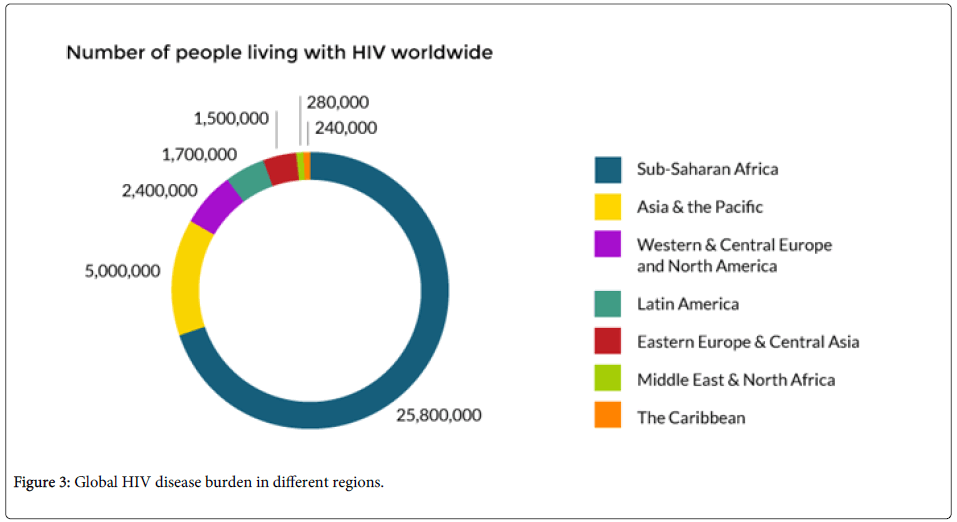
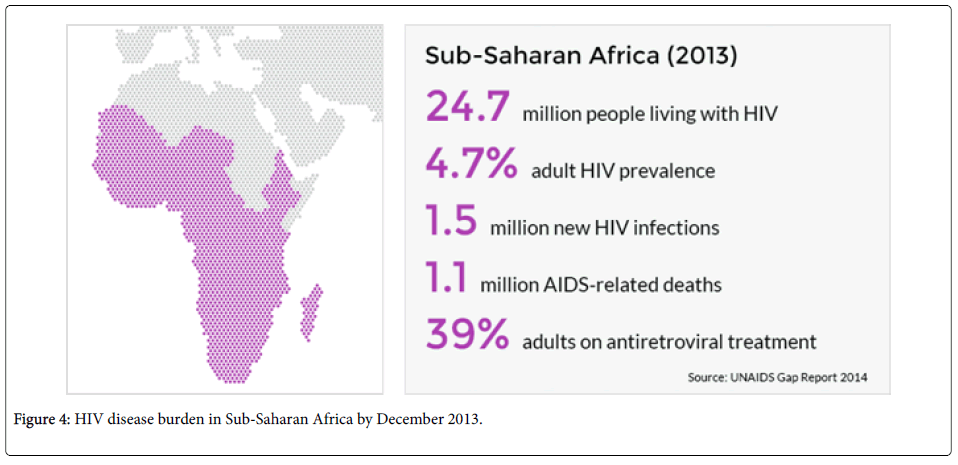
Sub Saharan Africa and Asia have the highest concentrations of HIV in the world. Sub-Saharan Africa has been the hardest hit by the HIV pandemic. About 25 million individuals with HIV reside in Sub Saharan Africa. Of every 10 children living with HIV/AIDS, nine have been reported to be from Sub-Saharan Africa. Political and financial commitments have been endeavored in the fight against the scourge. In 2013, 1.5 million people joined the ART treatment programed bringing the total number of those on treatment to over 9 million. This has reduced the deaths from HIV/AIDS in the region [13,14].
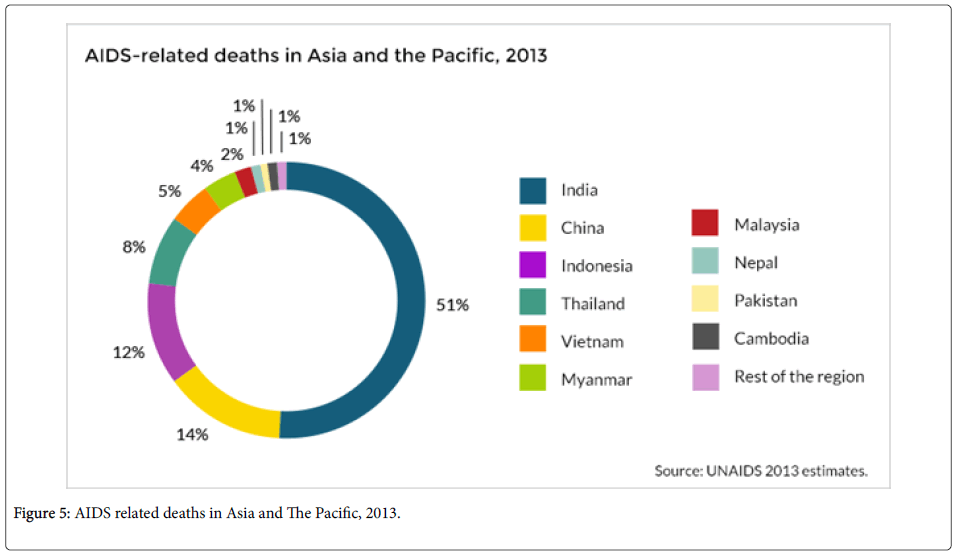
Despite HIV concentration in Sub-Saharan Africa, Asia is likely to be the next disease epicenter even though Asia and the Pacific have made great progress in tackling the epidemic. In Asia, many of the people living with the disease live in twelve nations; Cambodia, China, Indonesia, India, Myanmar, Malaysia, Pakistan, Nepal, Philippines, Papua new guinea, Vietnam and Thailand. India accounts for over 2.1 million people of those living with the disease [15]. New infections on the continent have reduced since 2001 by 26% [16].
Despite the decrease, the number of new HIV infections in the region has remained unchanged in the last five years. Those going for antiretroviral treatment in the area rose to 1.25 million people by end of 2012. Those going for ART treatment have slowed down since then. This slowdown may have a net effect in the rise of HIV epidemic in the Asia. A rise in the prevalence rate in Asia, with more than one billion people in population, would be catastrophic [16].
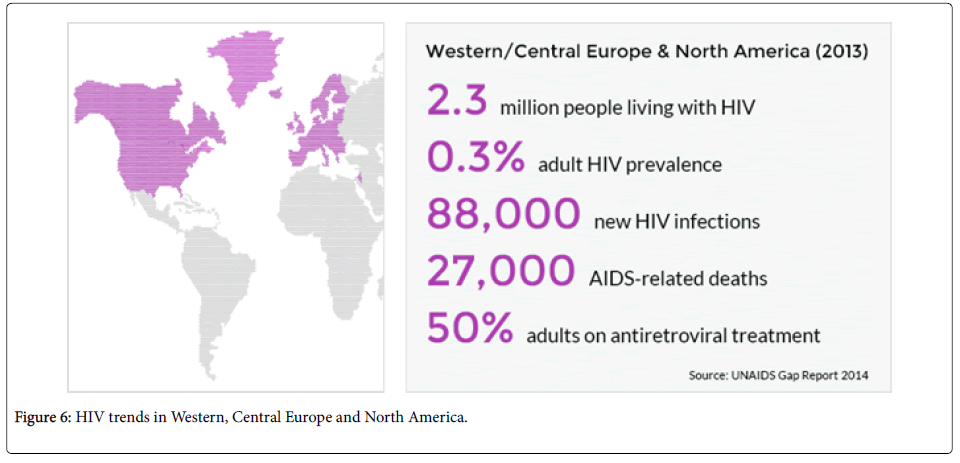
Across Western, Central Europe and North America, HIV prevalence is very low because their nations are capable of reaching to over 80% of those who are in need treatment.
HIV transmission can be prevented through various means. These may involve limiting the number of sexual partners, using condoms, never sharing needles as well as using Pre and Post exposure prophylaxis [17-47].
Influenza
Influenza is a misunderstood disease. Episodes of seasonal and inter-pandemic influenza are confronted each and every year. Seasonal influenza is capable of killing almost 500,000 individuals every year throughout the world [19]. In the US, 36,000 people die of influenza every year, over 90% of who are over 65 years20. Due to the frequent antigenic shifts and drifts in the influenza genome, there is a danger of an influenza pandemic. Pandemics occur when there is an exposure to a microbe with no baseline immunity in a population. The influenza pandemic of 1918 to 1919 was the worst in history with over 40 million deaths throughout the world. The seasonal influenza kills majorly older people but the 1919 pandemic killed both young and old. The young deaths were because exposure was to a highly virulent virus that had no background immunity [19].
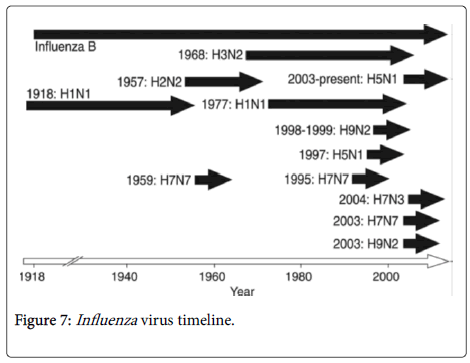
The designation of influenza viruses is by the composition of their haemagglutinin and neuraminidase proteins. The major influenza strain in 2005 was the H3N2 strain. Each and every year, the prevalent strains mutate due to the antigenic drift. For example, a drift from H3N2 Panama strain to H3N2 Fujian strain occurred in the 2003-2004 influenza season. This was a minor mutation thus if one was immune to panama strain, they would be immune the Fujian strain due to immune cross reactions.
An antigenic shift occurs when an influenza strain emerges which is different from what the population had been previously exposed to. The figure below illustrates the influenza pandemics in 20th century.
Specific Influenza strains:
1. H1N1 (Spanish flu): The virulent 1918 flu pandemic was a placed in category 5 and it was caused by Influenza A virus strain of the H1N1 subtype. It lasted from 1918 to 1920 [20,21]. Older estimates say 40-50 million people were killed while currently the estimates are at 50 to 100 million people [22,23].
Many of the deaths from H1N1 were from bacterial pneumonia which was a secondary infection, haemorrhages and edema in the lungs due to the viral infection [24].
H1N1 was global, spreading to the Arctic and to the remote Pacific islands. The virus killed 2-20% of those infected against the flu epidemic mortality rate of 0.1% [23,24].
2. H2N2 (Asian flu): This was a category two outbreak of avian influenza with its origins in China from the year 1956 to 1958. Its emergence is linked to a mutation in a wild ducks strain that was combined with a human strain that was already in existence [25]. Epidemiologically, the virus originated from Guizhou before it spread to Singapore in February 1957, to Hong Kong in April and finally to the United States in June [25,26]. The estimates of death varied worldly.
3. H3N2 (Hong Kong Flu): Occurred within the years 1968 and 1969. The H3N2 flu emanated from H2N2 due to antigenic shift. Genes in different multiple subtypes re-assorted to a new virus. The flu killed about 1 million people in the whole world [27,28]. Majority of the death occurred in those who were over 65 years of age [29].
4. H1N1 (Russian Flu): This outbreak occurred within the years 1977 to 1978. The H1N1 flu was a benign pandemic, and affected those born after 1950 as the earlier generation had protective immunity due to prior exposure to the virus [30,31].
5. H1N1/09: In 2009-2010, there was a H1N1/09 disease outbreak. The outbreak started in the US, Mexico, Canada and spread to Spain. The disease spread through the spring, and in May 2009, it was confirmed in the whole world [32]. In 2009, the A/H1N1 was declared by WHO as the first influenza pandemic in the 21st century. The A/H1N1 is a re-assortment of four strains of Influenza A virus subtype H1N1. The pandemic killed more than 18,000 people in over 199 countries by the end of November 2009. However, 284,500 people were killed by the pandemic [32-35].
From the discussions above, future influenza pandemics are likely to be caused by an avian virus with a hemagluttinin surface protein that lacks immunity in humans.
Flu can be prevented by avoiding close contact, staying at home when sick, covering the mouth and nose, cleaning the hands as well as vaccination.
Marburg haemorrhagic fever
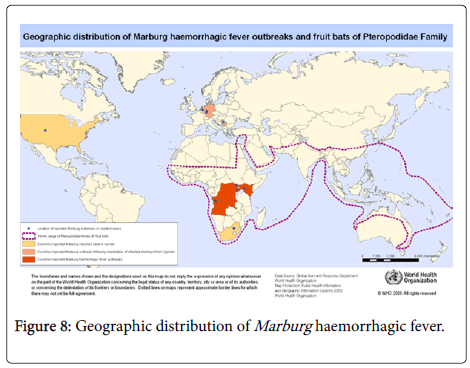
Marburg haemorrhagic fever is caused by Marburg virus. The disease has a high fatality ratio due to its virulent nature. Its name is derived from the German town Marburg where it was first detected in 1967. It was also detected in Frankfurt, and in Belgrade. The virus has no treatment or vaccine and is transmitted to people through fruit bats then human-human contact for human transmission [36].
The disease simultaneously occurred in the year 1967 in Marburg and Frankfurt in Germany and Belgrade in Serbia where 31 cases were reported with seven of the reported cases dying [37,38]. In 1975, Marburg haemorrhagic fever was reported in Rhodesia and South Africa where three people were infected [39]. Two cases of the haemorrhagic fever were reported in Kenya in the year 1980 while one case was reported in 1987 [23]. Since the late 1990s, the haemorrhagic fever outbreaks have been mainly concentrated in the Sub-Saharan Africa. During the Marburg outbreak of the years 1998 to 2000 in Congo, 154 subjects were infected with 128 of those infected dying [23,31,40]. The largest outbreak recently was however in Angola in the years 2004-2005 where 252 individuals were infected with 227 of those passing away [41-44]. The years 2007, 2008, 2012 and 2014 have seen the disease being reported in Uganda, East Africa with about 20 cases being reported.
In case of a Marburg outbreak, it is likely to occur in West Africa or in East Africa. The disease can be prevented by avoiding fruit bats, and sick non-human primates in central Africa, is one way to protect against infection [45-47].
Measures for prevention of secondary, or person-to-person, transmission are similar to those used for other hemorrhagic fevers. If a patient is either suspected or confirmed to have Marburg hemorrhagic fever, barrier nursing techniques should be used to prevent direct physical contact with the patient. These precautions include wearing of protective gowns, gloves, and masks; placing the infected individual in strict isolation; and sterilization or proper disposal of needles, equipment, and patient excretions.
In conclusion, the above diseases highlight the balance of environmental as well as genetic factors that can impact on the evolutionary patterns of RNA viruses while also illustrating the complexity of these systems. Prediction of future disease emergencies due to RNA viruses is difficult due to the above factors. However, as efforts to improve the capacity to respond to these pathogens are improved, there is the likelihood that these emerging and reemerging diseases can be dealt with in the future.
References
- Smolinski MS, Hamburg MA, Lederberg J (2003) Microbial Threats to Health: Emergence, Detection, and Response. The National Academy Press: 1-367.
- Bruce AW, Rita RC (2005) Emerging and Reemerging Infectious Diseases: Biocomplexity as an Interdisciplinary Paradigm. EcoHealth 2: 244-257.
- Horwitz P, Wilcox BA (2005) Parasites, ecosystems and sustainability: an ecological and complex systems perspective. Int J Parasitol 35: 725-732.
- Morse SS (1995) Factors in the emergence of infectious diseases. Emerg Infect Dis 1: 7-15.
- Wilcox BA, Gubler DJ (2005) Disease ecology and the global emergence of zoonotic pathogens. Environ Health Prev Med 10: 263-272.
- Burroughs T, Knobler SL, Lederberg J (2002) The Emergence of Zoonotic Diseases: Understanding the Impact on Animal and Human Health. The National Academy Press.
- Knobler SL, Lemon SM, Najafi M, Burroughs T (2003) The Resistance Phenomenon in Microbes and Infectious Disease Vectors: Implications for Human Health and Strategies for Containment. The National Academy Press.
- Morens DM, Folkers GK, Fauci AS (2004) The challenge of emerging and re-emerging infectious diseases. Nature 430: 242-249.
- (1981) Pneumocystis pneumonia-Los Angeles. Morb Mortal Wkly Rep 30: 250-252.
- Friedman-Kien A, Laubenstein L, Marmor M, Hymes K, Green J, et al. (1981) Kaposi’s sarcoma and pneumocystis pneumonia among homosexual men-New York City and California. MMWR. Morbidity and Mortality Weekly Report 30: 305-308.
- WHO (2015) Global summary of the aids epidemic: 2014.
- WHO (2015) Global Health observatory data.
- WHO (2015) Global HIV and AIDS statistics.
- WHO (2015) Overview: HIV/AIDS.
- WHO (2015) HIV disease burden in Sub-Saharan Africa by December 2013.
- UNAIDS (2015) HIV in Asia and The Pacific.
- UNAIDS (2015) AIDS related deaths in Asia and The Pacific.
- UNAIDS (2015) HIV trends in Western, Central Europe and North America.
- WHO (2005) Avian influenza: assessing the pandemic threat.
- CDC (2004) Key Facts about the flu and flu vaccine.
- Andrew Price-Smith (2009) Contagion and Chaos: Disease, Ecology, and National Security in the Era of Globalization. Cambridge, MA: MIT Press.
- Patterson KD, Pyle GF (1991) The geography and mortality of the 1918-1919 influenza pandemic. Bull Hist Med 65: 4-21.
- Knobler SL, Mack A, Mahmoud A, Lemon SM (2005) The Threat of Pandemic Influenza: Are We Ready? The National Academies Press.
- Taubenberger J, Morens DM (2006) 1918 Influenza: the Mother of All Pandemics. Emerging Infectious Diseases (Centers for Disease Control and Prevention) 12: 15-22.
- Greene J, Moline K (2006). The Bird Flu Pandemic.
- Goldsmith, Connie (2007) Influenza: The Next Pandemic? 21st century publishing.
- Paul WE (1993) Fundamental Immunology. New York: Plenum Press. p.1273.
- Mandel M, Toronto S (2009) No need to panic. yet Ontario officials are worried swine flu could be pandemic, killing thousands.
- BBC News (2009) Asia on high alert for flu virus.
- Hilleman M (2002) Realities and enigmas of human viral influenza: pathogenesis, epidemiology and control. Vaccine 20: 3068-3087.
- Potter CW (2006) A History of Influenza. J ApplMicrobiol 91: 572-579.
- The Associated Press (2009) WHO: Swine flu pandemic has begun, 1st in 41 years.
- Enserink M(2010) WHO Declares Official End to H1N1 'Swine Flu' Pandemic. Science Insider.
- Dawood FS, Iuliano AD, Reed C, Meltzer MI, Shay DK, et al. (2012) Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis 12: 687-695.
- WHO (2012) Marburg Haermorrhagic fever.
- Avian influenza ("bird flu") fact sheet. WHO.
- Bonin O (1969) The Cercopithecus monkey disease in Marburg and Frankfurt (Main), 1967. Actazoologica et pathologicaAntverpiensia 48: 319-331.
- CDC (2013) Pandemic Influenza Overview.
- Valleron AJ, Cori A, Valtat S, Meurisse S, Carrat F,et al. (2010). Transmissibility and geographic spread of the 1889 influenza pandemic. ProcNatlAcadSci USA 107: 8778-8781.
- Mills CE, Robins JM, Lipsitch M (2004) Transmissibility of 1918 pandemic influenza. Nature 432: 904-906.
- Donaldson LJ, Rutter PD,Ellis BM, Greaves FEC, MyttonOT,et al. (2009) Mortality from pandemic A/H1N1 2009 influenza in England: public health surveillance study. BMJ 2009: 339.
- CDC (2012) First Global Estimates of 2009 H1N1 Pandemic Mortality Released by CDC-Led Collaboration.
- Geographic distribution of Marburg haemorrhagic fever outbreaks and fruit bats of pteropodidae family. WHO.
- HIV/AIDS prevention. CDC.
- WHO (2014) Consolidated guidelines on HIV prevention, diagnosis, treatment and care for key populations.
- Preventing the Flu: Good Health Habits Can Help Stop Germs. CDC.
- Marburg Haemorrhagic Fever. Prevention. CDC.
Relevant Topics
Recommended Journals
Article Tools
Article Usage
- Total views: 13011
- [From(publication date):
March-2016 - Apr 07, 2025] - Breakdown by view type
- HTML page views : 12071
- PDF downloads : 940