Research Article Open Access
Genotyping of Bacillus anthracis Strains Circulating in Albania
Ardi Peculi1, Emanuele Campese2, Luigina Serrecchia2, Leonardo Marino2, JonidaBoci1, Bizena Bijo1, Alessia Affuso2, Valentina Mercurio2, Luigi Giangrossi2 and Antonio Fasanella2*
1Instituti I SigurisëUshqimoredhe Veterinarisë, Rruga Aleksandër Moisiu 82, Tiranë, Albania, Italy
2Istituto Zoo profilattico Sperimentale of Puglia and Basilicata, Anthrax Reference Centre of Italy, via Manfredonia 20, Foggia, Italy
- *Corresponding Author:
- Dr. Antonio Fasanella
Istituto Zoo profilattico Sperimentale of Puglia and Basilicata
Anthrax Reference Centre of Italy
via Manfredonia 20, Foggia, Italy
Tel: +39 080 405 78
E-mail: a.fasanella@izsfg.it
Received Date: November 26, 2015; Accepted Date: January 31, 2015; Published Date: February 08, 2015
Citation: Peculi A, Campese E, Serrecchia L, Marino L, Boci J, et al. (2015) Genotyping of Bacillus anthracis Strains Circulating in Albania. J Bioterror Biodef 7:131. doi: 10.4172/2157-2526.1000131
Copyright: © 2015 Peculi A, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited
Visit for more related articles at Journal of Bioterrorism & Biodefense
Abstract
Anthrax in Albania is an endemic disease characterized by few outbreaks involving a very low number of animals. Nineteen samples of soil coming from burial sites and 11 strains of Bacillus spp isolated from died animals from different districts of Albania were examined. The analysis of soil samples revealed that 11 of them were contaminated with anthrax spores, while only 8 strains were confirmed as Bacillus anthracis. The analysis of CanSNPs showed that all isolates belong to lineage A major subgroup A.Br.008/009 (Trans-Eurasian or TEA strains). The MLVA test at 15 loci showed three different genotypes: Albania GT/1, Albania GT/2 and Albania GT/3. However all the genotypes are genetically very similar to each other which confirm the hypothesis that all of them are the result of the evolution of a local common ancestral strain. Two distinct genotypes (Albania GT/2 and Albania GT/3) were found in the same burial site in the district of Kukës. Although in the scientific literature there have been cases in which two different genotypes were found in the same carcass, the authors believe that in this case the isolation of two different genotypes could be justified by the presence in the same burial sites of two or more animals that died in different times and as a consequence of different outbreaks.
Keywords
Albania; Molecular epidemiology; Anthrax; CanSNP; MLVA; Bacillus anthracis
Introduction
Anthrax, whose causative agent is Bacillus anthracis, is a non-contagious infectious disease that affects several animal species, the human one included. Domestic and wild ruminants represent the most susceptible categories [1]. The bacterial agent has the characteristic of producing spores that can survive in the environment for several decades [2]. In susceptible animals anthrax generally has a fatal outcome characterized by sudden death and leakage of blood coming out from the natural openings. In human beings such a disease develops in three ways and forms, depending on the route of penetration of the bacterium: cutaneous, pulmonary and gastrointestinal [3]. Recently, a fourth fatal form has been reported in drug users as a result of injection of heroin contaminated with anthrax spores [4]. Thanks to the considerable ability of spores to maintain viability and pathogenicity for many decades and thanks to its low costs of production, B. anthracis is considered one of the pathogens of greatest interest as a bacteriological weapon in a possible bioterrorist attack [5]. However, the knowledge of the disease, the causative agent, the mode of transmission, the development of an effective vaccine and especially the correct management of the infected carcasses have contributed to the nearly disappearance of this disease. In agricultural areas of the industrialized countries, sporadic outbreaks of animal anthrax still tend to occur in places where infected animals were previously buried inappropriately or where they were placed in waste tanneries. Outbreaks are often reported as a result of the introduction of contaminated feed. Despite the standard of the developing countries, anthrax is still a serious health problem for both animals and humans [1,6]. Currently in European anthrax has almost entirely disappeared, except for some Eastern Europe and Mediterranean countries such as Italy, Albania, Greece, Romania, Georgia, Russia and Turkey in particular. The population of B. anthracis in Europe is phylogenetically heterogeneous and the most prevalent subgroups are A.Br.008/009, Br.CNEVA and A.Br.001/002 [7]. In Albania anthrax is an endemic disease characterized by few outbreaks involving a small number of animals. The disease was widespread in the past, but now the number of outbreaks has been drastically reduced thanks to the control programs carried out in accordance with the guidelines of WHO and OIE. For the prophylaxis against animal anthrax, the live attenuated anthrax Sterne vaccine is used, produced in Albania. The prophylaxis program provides for the vaccination of animals for at least ten years since the last outbreak. From 2009 to 2011, 36 anthrax outbreaks have been confirmed and during the outbreaks were also reported human cases of cutaneous anthrax [8]. From 2009 to 2011, 3.008.278 doses of vaccine were administrated covering the 25,03% of the bovine population, 2,33% of the equine, 34,08% of the sheeps/goats and 0,29% of the pigs [9]. An emergence of the disease was observed in 2012, when 54 anthrax outbreaks were notified to the OIE [8]; the peak was recorded between July and October and the disease has mainly affected goats and sheeps. Regarding the 2012, the OIE data report that the most affected sites were in the districts of Gjirokastër, Vlorë and Kukës. In Albania, in case of a suspected anthrax outbreak, the veterinarians are obliged to inform the Ministry of Agriculture which alert the veterinary services of the area that implement the appropriate security procedures and control. The isolation and the biomolecular tests that permit to obtain a certain identification of the agent are not always used for diagnosis. The diagnosis is often based on the observation under the microscope of the slides prepared with blood from dead animals and stained with methylene blue. Considering that this is not a diagnosis of certainty, there are doubts on the number of animal died because of anthrax; it is over estimated and tends to define an epidemiological picture that does not correspond to the real situation. This paper reports the results of genotype analysis of Bacillus anthracis strains isolated from dead animals and from soil samples collected in areas of Albania considered at high risk of anthrax.
Materials and Methods
Soil samples
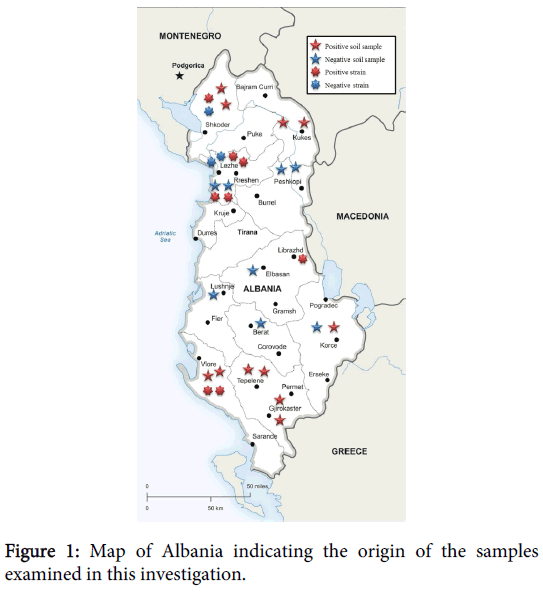
Soil samples were collected in 19 burial sites of dead animals with suspicion of anthrax in the districts of: Shkodër, Vlorë, Lushnjë, Kurbin, Tepelenë, Dibër, Kukës, Elbasan, Korçë Berat and Gjirokastër (Figure 1).
Bacterial strains
From the Instituti i Sigurisë Ushqimore dhe Veterinare of Tirana eleven strains of Bacillus spp isolated from sheeps died in several districts of the Albanian territory were sent to the Italian Reference Centre for Anthrax at the IstitutoZooprofilattico of Puglia and Basilicata (Foggia - Italy). The districts from which the strains came were: Vlorë, Librazhd, Shkodër, Lezhë, Kurbin (Figure 1). All shipments were made in accordance with the international standards of biosafety.
Isolation of B. anthracis from soil samples
The soil samples were analyzed by the Ground Anthrax Bacillus Refined Isolation method [10]. Briefly to 7.5grams of ground were added 22.5 ml of a 0.5% aqueous solution of Tween 20. After washing in magnetic stirrer for 30 minutes, the suspension was centrifuged for 5 minutes at 2000 rpm and the supernatant incubated for 20 minutes at 64°C. After incubation the suspension was diluted1:1 with Tryptose Phosphate Broth containing125 µg/ml of Fosfomycin and 1ml of the diluted suspension was seeded in TSMP plates and incubated at 37°C for 24 hours. After this period, the dishes were subjected to the reading.
Extraction of DNA
Each suspect colony was transferred to a blood agar plate and after 24 hours of incubation was taken adequate rate of bacterial patina and subjected to DNA extraction using the DNeasy Blood and Tissue Kit - Qiagen.
PCR
DNAs were subjected to Polymerase Chain Reaction (PCR) using anthrax-specific primers for the sequences located on plasmids pXO1 and pXO2 and primers specific for the chromosome [11].
CanSNP analysis
We utilized 13 TaqMan-Minor Groove Binding (MGB) allelic discrimination assays with oligos and probes as described by VanErt [12] for each of 13CanSNPs. Each10µl reaction contained 1× TaqMan Genotyping Master Mix (Applied Biosystems), 250nMofeachprobeand600nMeachofforward and reverse primers and approximately 10ng of template DNA. For all assays, thermal cycling parameters were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Endpoint fluoscent data were measured on the AB17900HT. CanSNPs profiles were compared to the 12 recognized worldwide sublineages and subgroups [12].
SNP discrimination by HRM
We used High Resolution Melting (HRM) assays for specific A.Br.011 CanSNP [13] using Precision Melt Analysis Software. The position based on the Ames Ancestor genome (NC_007530.2) is 2552486. Amplification was performed on CFX Connect Real-Time System (BIORAD) using the Precision Melt supermix BIORAD. The reaction mixture consisted of 0.2 µM of each primer, 1x Precision Melt supermix BIORAD in a 20µl final volume. The following parameters were used: 2 min at 95°C were followed by 35 cycles consisting of 10 s at 95°C and 30 s at 60°C. Samples were next heated to 95°C for 30 s, cooled down to 60°C for 1 min and heated from 65°C to 95°C at rate of 0,5°C/s. HRM data were analyzed by Precision Melt Analysis Software.
15-loci MLVA and SNR analysis
We utilized 5'fluorescent-labeled oligos, deprotected and desalted, specifically selected for the Variable Number Tandem Repeats (VNTRs) and Single Nucleotide Repeats (SNRs) used. The 15 specific primer pairs for the Multilocus Variable Tandem Repeat Analysis (MLVA) were selected as described by Van Ert [12]. The four specific primer pairs were selected for SNR reactions following Garofolo, et al. [14]. MLVA PCRs were performed in two single plex and five multiplex reactions in a final volume of 15 µl. The reaction mixture contained: 1x PCR reaction buffer (Qiagen), 1 U of Hot StarTaq Plus DNA polymerase (Qiagen), dNTPs (0.2 mMfor each), 3mM MgCl2, appropriate concentrations of each primer (singleplex 1: vrrC1 0.20μM; singleplex 2: vrrC2 0.20μM; multiplex 1: vrrA 0.25μM, vrrB1 0.20μM, CG3 0.40μM; multiplex 2: vrrB2 0.25μM, pXO2 0.60μM, pXO1 0.25μM; multiplex 3: VNTR12 0.20μM, VNTR19 0.15μM and VNTR35 0.20μM; multiplex 4: VNTR16 0.20μM and VNTR23 0.30μM; multiplex 5: VNTR170.10μM and VNTR32 0.15μM) and approximately 10ng of template DNA. The thermocycling conditions were the same for single plex 1 and 2 and multiplex 1 and 2 as follows: 95°C for 5 min; 35 cycles at 94°C for 30s, at 60°C for 30s, and at 72°C for 30s; and finally, 72°C for 5 min. The conditions for multiplex 3 were: 95°C for 5 min; 35 cycles at 94°C for 30 s, at 54°C for 30 s and at 72°C for 45s; and finally 72°C for 5 min; for multiplex 4: 95°C for 5 min; 35 cycles at 94°C for 30 s, at 56°C for 45 s and at 72°C for 1 min; and finally 72°C for 5 min; for multiplex 5: 95°C for 5 min; 35 cycles at 94°C for 30 s, at 59°C for 45 s and at 72°C for 1 min; and finally 72°C for 5 min. The SNR was performed in a multiplex PCR reaction [15] in a final volume of 15 µl containing 1× PCR reaction buffer (Qiagen), 1 U of Hot Star Taq Plus DNA polymerase (Qiagen), dNTPs (0.2 mM each), 3.5 mM MgCl2, appropriate concentrations of forward and reverse primers (0.2 µM HM1, 0.2 µM HM2, 0.1µM HM6, 0.2 µM HM13)and approximately 10ng of template DNA. The thermocycling conditions were as follows: 95°C for 5 min; 35 cycles at 94°C for 30 s, 60°C for 30 s and 72°C for 30 s; and finally 72°C for 5 min.
Automated genotype analysis
The MLVA PCR products were diluted 1:80 and subjected to capillary electrophoresis on ABI Prism 3130 genetic analyzer (Applied Biosystems) with 0.25μl of GeneScan 1200 and sized by Gene Mapper 4.0 (Applied Biosystems Inc.). SNR amplified PCR products were diluted 1:80 and subjected to capillary electrophoresis on ABI Prism 3130 genetic analyzer (Applied Biosystems) with 0,25 μl of Gene Scan 120 Liz and sized by Gene Mapper 4.0 (Applied Biosystems Inc.). In all the analyzes the samples were processed in triplicate to allow the correct sizing of the fragments.
Results
The analysis of soil samples of 19 suspected burial sites showed that 11 were contaminated with anthrax spores (Table 1), while only 8 out of 11 strains of Bacillus spp were confirmed as Bacillus anthracis (Table 2).
| District | N° of collected soil samples | Result | CanSNP | Genotype |
|---|---|---|---|---|
| BERAT | 1 | 1 negative | -- | -- |
| DIBËR | 2 | 2 negative | -- | -- |
| ELBASAN | 1 | 1 negative | -- | -- |
| GJIROKASTËR | 2 | 2 positive | A.Br.008/009 | Albania GT/1-GT/2 |
| KORÇË | 2 | 1 positive | A.Br.008/009 | Albania GT/3 |
| 1 negative | -- | -- | ||
| KUKËS | 2 | 1 positive | A.Br.008/009 | Albania GT/2 |
| 1 positive* | A.Br.008/009 | Albania GT/2-GT/3 | ||
| KURBIN | 2 | 2 negative | -- | -- |
| LUSHNJË | 1 | 1 negative | -- | -- |
| SHKODËR | 2 | 2 positive | A.Br.008/009 | Albania GT/2 |
| TEPELENË | 2 | 2 positive | A.Br.008/009 | Albania GT/1 |
| VLORË | 2 | 2 positive | A.Br.008/009 | Albania GT/1 |
Table 1: Brief description of the results obtained on soil samples: Sample in which two different genotypes were found.
| District | N° of collected strains | Animal | Result | CanSNP | Genotype |
|---|---|---|---|---|---|
| KURBIN | 2 | sheep | 2 positive | A.Br.008/009 | Albania GT/3 |
| LEZHË | 2 | sheep | 2 positive | A.Br.008/009 | Albania GT/3 |
| LEZHË | 2 | sheep | 2 negative | -- | -- |
| LIBRAZHD | 1 | sheep | 1 positive | A.Br.008/009 | Albania GT/2 |
| SHKODËR | 1 | sheep | 1 negative | -- | -- |
| SHKODËR | 1 | sheep | 1 positive | A.Br.008/009 | Albania GT/2 |
| VLORË | 2 | sheep | 2 positive | A.Br.008/009 | Albania GT/1 |
Table 2: Brief description of results obtained on bacterial strains
The analysis of CanSNPs showed that all isolates belong to lineage A major subgroup A.Br.008/009 which is widely represented in Europe and Asia: Trans-Eurasian or TEA strains.
The HRM analysis showed that all the strains belong to the subgroup A.Br.008/011 [16]. The MLVA test at 15 loci showed three different genotypes: Albania GT/1, Albania GT/2and Albania GT/3 (Figure 2 and Table 3).
| Fragment size (bp) for each locus | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | size | vrrA | vrrB1 | vrrB2 | vrrC1 | vrrC2 | CG3 | pXO1 | pXO2 | VNTR12 | VNTR16 | VNTR17 | VNTR19 | VNTR23 | VNTR32 | VNTR35 |
| Ba001/1FG *Reference strain |
obtained expected |
306 314 |
223 229 |
154 162 |
620 616 |
594 604 |
148 153 |
130 135 |
133 137 |
110 115 |
262 273 |
382 386 |
90 96 |
193 197 |
501 485 |
109 110 |
| Albania GT/1 | obtained expected |
306 314 |
223 229 |
154 162 |
620 616 |
594 604 |
148 153 |
130 135 |
133 137 |
110 115 |
262 273 |
390 394 |
93 99 |
193 197 |
501 485 |
109 110 |
| Albania GT/2 | obtained expected |
283 290 |
223 229 |
154 162 |
620 616 |
594 604 |
148 153 |
130 135 |
133 137 |
110 115 |
262 273 |
390 394 |
93 99 |
193 197 |
501 485 |
109 110 |
| Albania GT/3 | obtained expected |
306 314 |
223 229 |
145 153 |
620 616 |
594 604 |
148 153 |
130 135 |
133 137 |
110 115 |
262 273 |
390 394 |
93 99 |
193 197 |
501 485 |
109 110 |
Table 3: Results of 15-loci MLVA analysis on B. anthracis strains circulating in Albania.
We included the 15-loci MLVA typing panel of each genotypes in the MLVAbank (https://mlva.u-psud.fr/) and the results reveal that these three genotypes have never been reported in the literature. The strains isolated from the burial sites of the districts of Vlorë, Gjirokastër and Tepelenë belong to genotype Albania GT/1.
The strains of Bacillus anthracis isolated from soil samples collected in burial sites of the districts of Shkodër, Kukës and Gjirokastër showed a mutation at the locus vrrA in comparison to the genotype Albania GT/1 and for this reason they belong to a different genotype that was called Albania GT/2 in this work.
Finally, from the soil samples collected on the burial sites of the area of Kukës and Korçë, Bacillus anthracis belonging to a genotype different from the previous was isolated, characterized by a mutation at the locus vrrB2 from the previous genotypes.
It was interesting finding two different genotypes in one soil sample collected in a burial site of the Kukës district.
With regard to the strains sent by InstitutiiSigurisëUshqimoredheVeterinarisë of Tirana, all those sent from the district of Vlorë were found to belong to genotype Albania GT/1, while the strains from the districts of Shkodër and Librazhd belong to the Albania GT/2 and the strains from the
The SNR test performed on all isolates showed the existence of five sub genotypes within the Albania GT/1, two sub genotypes within Albania GT/2 and four sub genotypes within the Albania GT/3 [17].
Discussion
The analysis of burial sites and the isolated strains indicate that in the investigated areas of Albania there are 3 different genotypes of Bacillus anthracis that were referred as Albania GT/1, Albania GT/2 and Albania GT/3.
districts of Kurbin and Lezhë were found to belong to genotype Albania GT/3. and at the locus vrrA from the Albania GT/2 and it in this work was called Albania GT/3.
The MLVA analysis indicated that all the genotypes are genetically very similar to each other, which confirms the hypothesis that all of them are the result of the evolution of a local common ancestral strain. However, the research should be extended to other parts of Albanian territory and especially in the southern regions more close to Turkey, where the genetic variability of the circulating strains of Bacillus anthracis is very high [18].The genotypes of B. anthracis isolated from the soil samples of burial sites of the regions of Vlorë and Shkodër are identical to those of the strains isolated from animals died of anthrax. This data indicate that the environmental analysis can permit to obtain a very comparable results to those obtained by passive surveillance that is based on analysis of the strains isolated from dead animals. In addition, the improvement of the detection analysis of soil samples could provide very useful indications on the level of contamination and its risk assessment of new outbreaks. Another interesting aspect concerns the discovery of two distinct genotypes (Albania GT/2 and Albania GT/3) in the same burial site in the district of Kukës. Beyer and Turnbull [19] found co-infections with more than one GT in carcasses of animal died for anthrax and suggested that this is probably not especially uncommon. However in our case this result could also be due to the presence in the same burial sites of two or more animals that died at different times and in different outbreaks. Regarding the detection of multiple sub genotypes in the same burial site, it is not rare (data not shown). B. anthracis is a bacterium that from a genetic point of view is highly conserved because it spends most of its existence in the soil under spore form. Nature provides few opportunities to the bacterium for its replication cycle and the development of an extraordinary pathogenicity is the effective strategy to substantially increase the chances of success against the host immune mechanisms.
The rapid and intense multiplication of vegetative cells within the host leads quickly to death. Although many of the new generations of bacteria will be neutralized by putrefactive processes, a good portion will survive and spread into the surrounding soil in the form of spores. This process will ensure the standard of environmental density of the bacteria which is an essential condition for the continuation of the species.
In summary, the few cases of anthrax that occur each year are only the result of a natural ecological balance that through these extraordinary events promote the maintenance of a bacterial species that otherwise would have become extinct a long time ago [20].
Therefore the intense replication can promote the development of more sub genotypes and the research on them, carried out through the analysis of SNRs, can be of great help in the molecular epidemiology studies of anthrax outbreaks because it allows researchers to be able to make a detailed and evolutionary analysis of the infectious episodes. In summary the obvious explanation would be that the development of the variations might have originated during the course of infection. This would agree with the findings of Stratilo and Bader [21], who compared SNR profiles of B. anthracis soil isolates distributed around carcass sites of bison diseased from anthrax. The authors found similar distributions of SNR variations within soil samples of single carcass sites which were proposed by Kenefic et al. [22] to be acquired during host passaging.
Conclusions
The CanSNPs analysis confirms that B. anthracis strains circulating in Albania belong to A.Br.008/009 which represents the large family of TEA (Trans-Eurasian), while the HRM analysis showed that all the strains belong to the subgroup A.Br.008/011.The MLVA test at 15 loci showed three different genotypes: Albania GT/1, Albania GT/2 and Albania GT/3.On 19 samples (strains and soils), 7 were found to belong to the Albania GT/1 (36,8%) and 7 to Albania GT/2 (36,8%). The Albania GT/2 results the most widespread genotype in the country while the Albania GT/1 results the most geographically restricted and found only in the South. Finally 6 samples were found to belong to the Albania GT/3(31,5%) which is less widespread than the Albania GT/1 and Albania GT/2.All the genotypes are genetically very similar to each other, which confirms the hypothesis that all of them are the results of the evolution of a local common ancestral strain. The uniform geographic distribution of the Albania GT/2 and the Albania GT/3 genotypes is in agreement on the fact that the human activities and the movements of the animals play a very important role in the proliferation and dispersal of the disease. However the Albania GT/1 seems predominantly localized almost exclusively in the southern part of the country. It seems that in the southern part of the country the movements of the animals are much more controlled by health authorities’ compared to the remaining part of the country (ArdiPeculi personal communication). This aspect should justify that this genotype remains located in this part and does not spread in other parts of the country. Albania has an important trade of animals with neighboring countries and it would be very interesting to see the influence of these trades on the epidemiology of anthrax in Balkan region, but unfortunately this kind of data is very fragmentary and not reliable. However it is not excluded that in further investigation strains belonging to other genotypes or lineages can be found, as it has been observed in Italy, where, in addition to the dominant genotype TEA, there are ecological niches of B.Br. CNEVA and A.Br.005/006 regions in Northern Italy [23].Despite every year a vaccination program using a live attenuated vaccine type Sterne takes place, anthrax outbreaks tend to occur with some consistency mainly in areas where the disease has been reported in the past. This could be related to the usual management of died animals which consists to bury the carcasses in a deep hole; this practice tends to favor the maintenance of high levels of environmental contamination (data not shown) which could be considered the reason that maintains endemic the disease in Albania. One aspect that emerges regards the Albanian diagnostic structure and their ability to identify the pathogen agents is that the isolation and the biomolecular tests that permit to obtain a certain identification of the agent is not always used for diagnosis. The diagnosis is often based on the observation under the microscope of the slides prepared with blood from dead animals and stained with methylene blue. Considering that this is not a diagnosis of certainty, there are doubts on the number of animal died for anthrax; it is overestimated and tends to define an epidemiological picture that does not correspond to the real situation. In fact only 8 out of 11strains identified as B. anthracis were confirmed in our investigation. The improvement of the Public Health laboratories as well as the diagnostic procedures could have a positive impact not only on the prevention and control of the disease but also on the trade of animals and their products in order to ensure the standard quality required by European countries in an eventual program providing for the entry of Albania into the European Community.
Fund
Current Research 2010 “IZSPB 04/10 "funded by the Ministry of Health of Italy.
Acknowledgments
We thank Francesco Tolve, MichelaIatarola e Giuseppe Stramaglia for the technical support.
References
- Hugh-Jones M, Blackburn J (2009) The ecology of Bacillus anthracis. Mol Aspects Med 30: 356-367.
- Dragon DC, Rennie RP (1995) The ecology of anthrax spores: tough but not invincible. Can Vet J. 36: 295-301.
- Dixon TC, Meselson M, Guillemin J, Hanna PC (1999) Anthrax. N Engl J Med 341: 815-826.
- Price EP, Seymour ML, Sarovich DS, Latham J, Wolken SR, et al. (2012) Molecular epidemiologic investigation of an anthrax outbreak among heroin users, Europe. Emerg Infect Dis 18: 1307-1313.
- Christopher GW, Cieslak TJ, Pavlin JA, Eitzen EM Jr (1997) Biological warfare. A historical perspective. JAMA 278: 412-417.
- Hugh-Jones M (1999) 1996-97 Global Anthrax Report. J Appl Microbiol 87: 189-191.
- Derzelle S, Thierry S (2013) Genetic diversity of Bacillus anthracis in Europe: genotyping methods in forensic and epidemiologic investigations. Biosecur Bioterror 11 Suppl 1: S166-176.
- OIE reporting history. World Animal Health Information Database (WAHID), France.
- Animal vaccination. World Animal Health Information Database (WAHID), France.
- Fasanella A, Di Taranto P, Garofolo G, Colao V, Marino L, et al. (2013) Ground Anthrax Bacillus Refined Isolation (GABRI) method for analyzing environmental samples with low levels of Bacillus anthracis contamination. BMC Microbiol 13: 167.
- Fasanella A, Losito S, Trotta T, Adone R, Massa S, et al. (2001) Detection of anthrax vaccine virulence factors by polymerase chain reaction. Vaccine 19:4214-4218.
- Van Ert MN, Easterday WR, Huynh LY, Okinaka RT, Hugh-Jones ME, et al. (2007) Global genetic population structure of Bacillus anthracis. PLoS One 2: e461.
- Girault G, Thierry S, Cherchame E, Derzelle S (2014) Application of High-Throughput Sequencing: Discovery of Informative SNPs to Subtype Bacillus anthracis. Adv Biosci Biotechnol 5: 669-677.
- Garofolo G, Ciammaruconi A, Fasanella A, Scasciamacchia S, Adone R, et al. (2010) SNR analysis: molecular investigation of an anthrax epidemic. BMC Vet Res 6: 11.
- Kenefic LJ, Beaudry J, Trim C, Huynh L, Zanecki S, et al. (2008) A high resolution four-locus multiplex single nucleotide repeat (SNR) genotyping system in Bacillus anthracis. J Microbiol Methods.73:269-72.
- Marston CK, Allen CA, Beaudry J, Price EP, Wolken SR, et al. (2011) Molecular Epidemiology of Anthrax Cases Associated with Recreational Use of Animal Hides and Yarn in the United States. PLoS ONE, 6: e28274.
- Keim P, Price LB, Klevytska AM, Smith KL, Schupp JM, et al. (2000) Multiple-Locus Variable-Number Tandem Repeat Analysis Reveals Genetic Relationships within Bacillus anthracis. J Bacteriol, 182: 2928-2936.
- Durmaz R, Doganay M, Sahin M, Percin D, Karahocagil MK, et al. (2012) Molecular epidemiology of the Bacillus anthracis isolates collected throughout Turkey from 1983 to 2011. Eur J Clin Microbiol Infect Dis 31: 2783-2790.
- Beyer W, Turnbull PC (2013) Co-infection of an animal with more than one genotype can occur in anthrax. Lett ApplMicrobiol 57: 380-384.
- Fasanella A, Galante D, Garofolo G, Hugh-Jones M (2010) Anthrax undervalued zoonosis. Vet Microbiol. 140:318-311.
- Stratilo CW, Bader DE (2012) Genetic diversity among Bacillus anthracis soil isolates at fine geographic scales. Appl Environ Microbiol. 78:6433-437.
- Kenefic LJ, Beaudry J, Trim C, Daly R, Parmar R, et al. (2008) High resolution genotyping of Bacillus anthracis outbreak strains using four highly mutable single nucleotide repeat markers. Lett Appl Microbiol 46: 600-603.
- Garofolo G, Serrecchia L, Corrò M, Fasanella A (2011) Anthrax phylogenetic structure in Northern Italy. BMC Res Notes 4: 273.
Relevant Topics
- Anthrax Bioterrorism
- Bio surveilliance
- Biodefense
- Biohazards
- Biological Preparedness
- Biological Warfare
- Biological weapons
- Biorisk
- Bioterrorism
- Bioterrorism Agents
- Biothreat Agents
- Disease surveillance
- Emerging infectious disease
- Epidemiology of Breast Cancer
- Information Security
- Mass Prophylaxis
- Nuclear Terrorism
- Probabilistic risk assessment
- United States biological defense program
- Vaccines
Recommended Journals
Article Tools
Article Usage
- Total views: 15245
- [From(publication date):
March-2015 - Apr 11, 2025] - Breakdown by view type
- HTML page views : 10605
- PDF downloads : 4640