Research Article Open Access
Genome-Wide Identification and Characterization of Light Harvesting Chlorophyll a/b Binding Protein Genes in Barley (Hordeum vulgare L.)
Dandan Qin1,2, Jing Dong1,2, Fuchao Xu1,2, Shuangtao Ge1,2, Qing Xu1,2 and Meifang Li1,2*
1Institute of Food Crops, Hubei Academy of Agricultural Science, Hubei Wuhan, China
2Hubei Key Laboratory of Food Crop Germplasm and Genetic Improvement, Hubei Wuhan, China
- *Corresponding Author:
- Meifang Li
Nanhu Avenue No 3, Hongshan District
Wuhan City, Hubei Province, 430064 China
Tel: 086-13237178679
E-mail: limeifang100@126.com
Received Date: August 10, 2017 Accepted Date: August 21, 2017 Published Date: September 02, 2017
Citation: Qin D, Dong J, Xu F, Ge S, Xu Q, et al. (2017) Genome-Wide Identification and Characterization of Light Harvesting Chlorophyll a/b Binding Protein Genes in Barley (Hordeum vulgare L.). Adv Crop Sci Tech 5: 301. doi:10.4172/2329-8863.1000301
Copyright: © 2017 Qin D, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Advances in Crop Science and Technology
Abstract
Light harvesting chlorophyll a/b binding proteins (LHC) play an important role in photosynthesis and development of plant. However, limited data is available for this important gene family in barley, especially for their biological functions. In this study, sequences of rice and Arabidopsis LHCs were used as queries to identify the respective orthologues from available barley genomic database. Total 17 non-reduntant HvLHCs were identified. Genomic sequences of these genes were ranged from 780 bp to 2779 bp in length, and ORF sequence of 738 bp to 933 bp. The phylogenetic relationship of members from barley, rice and Arabidopsis revealed that most of them were common to the three species, while some of them were specific to Arabidopsis or the poaceae family. Cis-element analysis showed that along with the universal transcription initiation and enhancement relevant elements, there were also light as well as biotic and abiotic stresses responsive elements in promoter regions of these HvLHCs. Additionally, these genes exhibited quite similar expression pattern during the development of barley plant according to the public normalized RNA-seq and barley array datasets, but responded distinctively to different treatments including dark, heat, salinity and drought, which were confirmed by quantitative real-time PCR analysis. This is the first report of identification of HvLHCs at the genome level, and results presented here would be useful not only for determining the precise role of HvLHCs during barley development and abiotic stress responses, but also for using them in molecular breeding of barley varieties with high yield or high tolerance to stresses.
Keywords
Barley; Light harvesting chlorophyll a/b; Binding protein; Genome level; Gene family
Introduction
Photosynthesis is one of the most important physiological activities in the plant kingdom. Many studies in plants, including barley, suggested that there was a significant correlation between photosynthesis efficiency and yield. Plants with higher leaf area and net photosynthesis rate often perform well under normal or controlled conditions [1-3].
However, these previous studies were mainly focused on the effects of different cultivation methods or physiological treatments on photosynthesis and yield of plant, and also on the relationship between photosynthesis relevant characteristics to yield. Very few studies were carried out to apply photosynthesis related genes into molecular breeding of crops with the aim of increasing efficiency of photosynthesis and yield potential [4].
Release of genome sequences of many crop plants enabled us to identify key genes involved in the process of photosynthesis of plants. Over expression of some photosynthesis related genes improved the efficiency of photosynthesis, such as HvRACB [5], OsRCA [6], ZmPEPC and ZmPPDK [7]. OsHYR’s over-expression in rice not only increased the net photosynthesis rate under normal condition as well as in drought and high temperature exposures, but also the yield of transgenic plants was more [8].
During the photosynthesis process, most of the photons that are converted to biochemical energy and biomass are harvested by the major light-harvesting chlorophyll a/b binding antenna complex (light-harvesting complex II, LHCII), which is one of the most abundant proteins on earth [9]. Except for their roles as antenna proteins, some members of this gene family have also been proved to be involved in development of plants. For instance, six LHCB members of Arabidopsis (Lhcb1.1, Lhcb 2.2, Lhcb 3, Lhcb 4.4, Lhcb 5 and Lhcb 6) were positively involved in ABA signaling in stomatal movement and response to drought [10], and also in seed germination and postgermination growth in response to ABA [11]. Down-regulation of Lhcb1 by microRNA in Arabidopsis slowed down the growth as compared to wide type, and leaves of these transgenic lines were slightly smaller and paler [9]. In rice, starch content in leaf sheath was clearly increased in CRCT over-expression lines and decreased in knockdown lines, which was a CONSTANS, CONSTANS-like, and Time of Chlorophyll a/b Binding Protein 1 (CCT) Protein [12]. Similar to this, expression level of major chlorophyll a/b binding protein (Cab) in wheat was up-regulated at anthesis and found to be associated with higher photosynthetic capacity and antioxidant activities during both winter and spring night-warming treatments. Both of these two treatments profoundly affected the vegetative growth and post-anthesis grain productivity of wheat [13]. In barley, SNPs in HvLhcb1 were significantly associated with agronomic traits, such as plant height, spike length, number of grains per spike, thousand grain weights, flag leaf area [14].
There are 17 and 23 chlorophyll a/b-binding proteins in rice and Arabidopsis, respectively [15]. The release of the genome of barley [16] will help us a lot in the cloning and characterization of this gene family in barley. The present study focused on extensive genome-wide analysis of barley light harvesting chlorophyll a/b binding proteins, which will be the foundation for functional characterization of HvLHCs and using them in molecular breeding of barley.
Materials and Methods
Plant material and growth conditions
Barley variety Edamai No 934 was used during the present study. 10-day’s old seedlings growing in pots with soil under normal conditions were subjected to the four treatments, viz . exposure to 40°C, 150 mM NaCl, 20% PEG (w/v) and dark to mimic the heat, salinity, drought stresses and dark stimulus, respectively. After three hours’ treatment, leaves from each treatment were harvested and immediately frozen into liquid nitrogen, and then were stored at -80°C till further use. Seedlings without any treatment were used as a control.
In silico identification of chlorophyll a/b binding proteins from barley
Arabidopsis and rice LHCs sequences [15] were used as queries in BLASTN searches at the International Barley Sequencing Consortium (IBSC) website (http://www.public.iastate.edu/~imagefpc/IBSC%20Webpage/ IBSC%20Template-home.html) [17]. Non-redundant HC genes CDS_Seq and Morex_Contigs with high similarity to AtLHCs and OsLHCs were picked up as a candidate cDNA and putative genomic sequence of HvLHCs, respectively. Open reading frame of HcLHCs was identified according to their counterparts from Arabidopsis and rice, and chloroa/b-bind domain (PLN00170) in amino acid sequences was identified through blastP on NCBI (https://www.ncbi.nlm.nih.gov).
Chromosomal locations of HvLCHs were retrieved according to the Morex_Contigs containing them. Exon/intron distributions were constructed using Gene Structure Display Server (GSDS) 2.0 (http:// gsds.cbi.pku.edu.cn) by aligning the sequence from the start codon to the stop codon of cDNA and genomic DNA.
Multiple sequence alignment and construction of phylogenetic tree
deduced amino acid sequences of HvLHCs and homologous family members from Arabidopsis and rice were aligned using MEGA 6 the multiple sequence alignment program [18] with the following parameters: poisson correction, pairwise deletion, uniform rates and bootstrap (1000 replicates). Phylogenetic trees were constructed using the neighbor-joining (NJ) method and the output generated was used for further analysis.
Prediction of cis-acting regulatory element from the promoter regions of HvLHCs
Promoter regions comprising approximately 1500 bp upstream of these HvLHCs were downloaded from the barley genome database. The conserved cis-acting regulatory elements present in the promoter regions were computationally predicted using PlantCARE database [19].
Digital expression profiles of HvLHCs
Expression values of HvLHCs in eight different organs at different developmental stages were obtained from the barley genome database (http://apex.ipk-gatersleben.de/apex/f?p=284:10) [20]. Those include 4-day old embryos dissected from germinating grains, roots and shoots from the seedlings (10 cm shoot stage), young developing inflorescences (5 mm), developing inflorescences (1-1.5 cm), developing tillers at sixleaf stage (the third internode), 5 and 15 days post-anthesis developing grain (DPA) (bracts removed), which were selected for deep RNA sequencing [17]. Heat map was constructed using R heatmap 2 based on the absolute expression values of each gene, which was indicated by FPKM (Fragments Per Kilobase of transcript per Million mapped reads).
RNA isolation and expression analysis
High quality RNA was extracted using RNAiso Plus reagent (TAKARA, Japan) as per the manufacturer’s instructions. Two microgram of total RNA was used to synthesize first-strand cDNA using oligo (dT) 18 primer with M-MLV reverse transcriptase (Promega, USA). Real-time PCR was performed by using a Premix EX TaqTM kit (TaKaRa, Japan) to analyze the relative expression of HvLHCs under different treatments. HvACT was used as internal control for gene expression analysis [21]. The primers used for these HvLHCs as well as HvACT are listed in Table 1. The threshold cycle (Ct) values of the triplicate PCRs were averaged and relative quantification of transcript levels were performed using the comparative Ct method [22]. Three biological replicates were performed for each sample.
| Gene | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| RT_MLOC_81109 | TCAAGGACCTGCCGTCG | CAGTGGATCACCTCCAGC |
| RT_MLOC_17002 | AGAATCACCATGGGCAAC | CACTTCTGGAGCACCTCG |
| RT_MLOC_60073 | AGAGCTTGCGGTGGAAC | AGCCTGGCTTGATGATGTC |
| RT_MLOC_65066 | AGTACCTGCAGTACGACGTG | TCCACCAGCTCCACCTTG |
| RT_MLOC_44990 | TGTTCGGCAAGTCCAAGAC | CGGTCTCAAGGTTCAGCTG |
| RT_MLOC_57061 | GACGACGAACTCGCCAAG | GTCTTGAACCAAACGGCCT |
| RT_MLOC_63821 | GGTTCGATATAGCAGGCCT | GATGACGATGATGCCTTGAC |
| RT_MLOC_22471 | GAGATCCTGTCCAAGAACG | GAGCTCCTTGACCTTGAGCT |
| RT_MLOC_49483 | GCTCCAGGACTGGTACAACC | GAGGTGGTCGAGGAGGTTC |
| RT_MLOC_12844 | TGATCCGTGTGTCAGGAATC | GCTTCACTTCATTCGCATTC |
| RT_MLOC_56051 | AGGTGATCCACTGTCGATG | TCAACAATCTCGCCAAGTG |
| RT_MLOC_20487 | TCAAGGAGTCCGAGATCTACC | AACTCGATCGCCAGGATG |
| RT_MLOC_67012 | AGAACCGTGAGCTGGAGG | GTCGACTATCTCGCCGAG |
| RT_MLOC_11961 | AGCGATGCCGAGTTCATC | CATGAGCAGCAGCTGGGT |
| RT_MLOC_44755 | ACGCTCTTCGTCATCGAGT | TGCTGCAGGTTCTCGAAC |
| RT_MLOC_69498 | CAAGAACCGTGAGCTGGA | CGACGATCTCTCCGAGTG |
| RT_MLOC_18354 | CGCACCAAGGAGATCAAGAA | GTGAATGCCGAGAAGATGTTG |
| HvACT | GCGAGTTGTCTGGGTCTTCT | ACATGGCAAGGACTTGAGAAA |
Table 1: Specific primers for real-time PCR analysis.
Results
Identification of 17 LHC family genes in barley
For identification of LHC gene family members in barley, BLAST against the barley genome databases was performed with genes from Arabidopsis and rice. In silico analysis revealed that there were 17 unique members of LHC gene family in barley (Table 2 and Figure 1). The average length of coding sequence of these genes was around 800 bp in length, with the shortest and longest one being 738 bp and 933 bp, respectively. However, length of the genomic sequence varied significantly among these genes, ranging from less than 780 bp to about 2779 bp. Gene structure of these genes was also variable. There was no intron for four of them, while the other 13 members have one to five introns in the coding regions of the genes. Further analysis of deduced amino acid showed that all of the coding sequences of these genes had a conserved domain of chlorophyll a/b binding protein, suggesting that they belonged to LHC gene family.
Chromosomal locations of HvLHCs
After searching for homologous sequences from genome sequence of barley cv. Morex, the 17 HvLHCs could be located to the different chromosomes of Morex genome. They were distributed on all the seven chromosomes of barley according to the position of these Morex_contigs, with 1, 4, 1, 1, 5, 2 and 3 member (s) for chromosome 1H to 7H, respectively (Table 2). Remarkably, we observed that the four and three members on chromosome 2H and 7H were concentrated in the interval of 56 cm to 59 cm and 67 cm to 70 cm of these two chromosomes, respectively (Table 2).
| Gene Information | Morex_contig_Information | Chromosome Information | Gene Annotation | ||||
|---|---|---|---|---|---|---|---|
| Accesion No. | Length of CDS (bp) | Length of genomic sequence (bp) | Morex_Contig | Gene Position on Morex_Contig | Chromosome | Chromosome Position (cM) | |
| MLOC_11961 | 768 | 892 | morex_contig_1562233 | 1720-2611 | 2H | 59.348442 | chlorophyll a/b binding protein |
| MLOC_12844 | 774 | 1669 | morex_contig_1564232 | 4667-6335 | 6H | 78.116147 | photosystem I light harvesting complex protein 5 |
| MLOC_17002 | 807 | 909 | morex_contig_1575254 | 1131-2039 | 2H | 56.373938 | chlorophyll a/b binding protein |
| MLOC_18354 | 822 | 1125 | morex_contig_158024 | 3959-5083 | 5H | 43.760478 | chlorophyll a/b-binding protein type II |
| MLOC_20487 | 744 | 1035 | morex_contig_1590103 | 722-1756 | 7H | 67.917847 | chlorophyll a/b binding protein 6a |
| MLOC_22471 | 792 | 917 | morex_contig_160732 | 1384-478 | 5H | NA | chlorophyll a/b binding protein |
| MLOC_44755 | 738 | 933 | morex_contig_275313 | 1181-2110 | 5H | NA | chlorophyll a/b binding protein chloroplastic-like |
| MLOC_44990 | 816 | 2404 | morex_contig_65451 ; Barke_contig_59491*; morex_contig_275924 | 1459-1021;1768-1185 | 3H | 86.3314 | photosystem II subunit |
| MLOC_49483 | 810 | 1004 | morex_contig_342448 | 1691-688 | 6H | 54.745042 | light-harvesting complex I chlorophyll a/b binding protein 3 |
| MLOC_56051 | 801 | 801 | morex_contig_40500 | 3734-2937 | 7H | 65.439093 | chlorophyll a/b binding protein |
| MLOC_57061 | 861 | 1471 | morex_contig_41390 | 5798-7265 | 4H | 51.558074 | chlorophyll a/b binding protein |
| MLOC_60073 | 768 | 1190 | morex_contig_44168 | 2841-1758 | 2H | 56.621813 | chlorophyll a/b binding protein |
| MLOC_63821 | 933 | 2779 | morex_contig_47996 | 4674-7452 | 5H | 6.7361111 | chlorophyll a/b-binding protein |
| MLOC_65066 | 861 | 979 | morex_contig_49702 | 3228-2250 | 2H | 57.8612 | chlorophyll a/b binding protein chloroplastic-like |
| MLOC_67012 | 780 | 780 | morex_contig_52417 | 13960-14740 | 5H | 110.06944 | chlorophyll a/b binding protein |
| MLOC_69498 | 780 | 780 | morex_contig_56519 | 2200-1424 | 7H | 70.538244 | chlorophyll a/b binding protein |
| MLOC_81109 | 801 | 801 | morex_contig_94468 | 2513-1593 | 1H | 121.95467 | chlorophyll a/b binding protein |
Table 2: Chlorophyll a/b binding protein genes in barley.
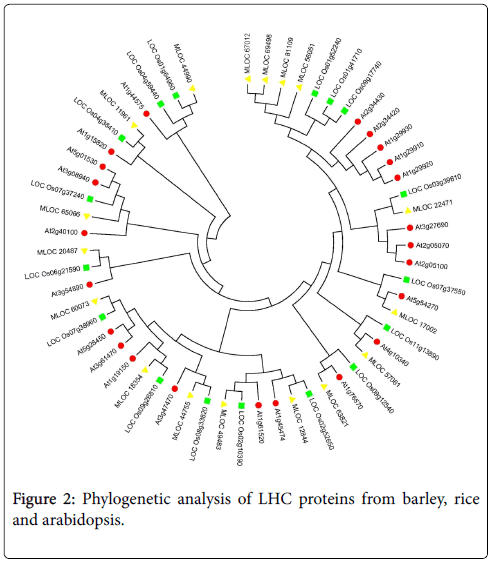
Phylogenetic analysis of LHC proteins from barley, rice and arabidopsis
Phylogenetic analysis of HvLHCs (indicated by yellow triangle), AtLHCs (indicated by red circle) and OsLHCs (indicated by green square) was performed using full length deduced amino acid sequences of these genes to understand the relationships between them (Figure 2). As shown in the figure, most of the HvLHCs could find only one counterpart both in rice and Arabidopsis, while some of them could find more in rice or Arabidopsis. Additionally, some members from the same species showed closer relationship themselves as compared to members from other species. For instance, MLOC_67012, MLOC_69498, MLOC_81109 and MLOC_56051 from barley showed high similarity between each other. They were grouped together by cluster analysis, but separated from relevant genes from rice and Arabidopsis. Similar results were observed for other members from Arabidopsis, such as Lhcb 2.1 (AT2G05100), Lhcb 2.2 (AT2G05070) and Lhcb 2.4 (AT3G27690), which were also clustered together and separated from the relevant genes from the other two species.
Cis-acting regulatory element analysis of promoter region of HvLHCs
After searching for cis-elements by PLANTCARE, we found that those common elements involved in transcription initiation or enhancement, such as TATA-box and CAAT-box, also existed in the promoter region of all these HvLHC genes. Along with these elements, various kinds of light-responsive elements were also present, including ATCT-motif, Box I, C-box, CATT-motif, G-box, GAG-motif, GTGGCmotif, I-box, L-box et al. Another two cis-acting regulatory element ARE and ABRE were also found in most of these genes, which was reported to be essential for anaerobic induction and ABA response, respectively. Moreover, there were also elements involved in other stresses and hormones response in promoter regions of these HvLHCs such as CGTCA-motif, ERE, GARE, MBS, HSE, LTR, for MeJA, ethylene, gibberellin, drought response, high and low temperature stresses, respectively.
Expression analysis of HvLHCs at different developmental stages and their response to different stimulus
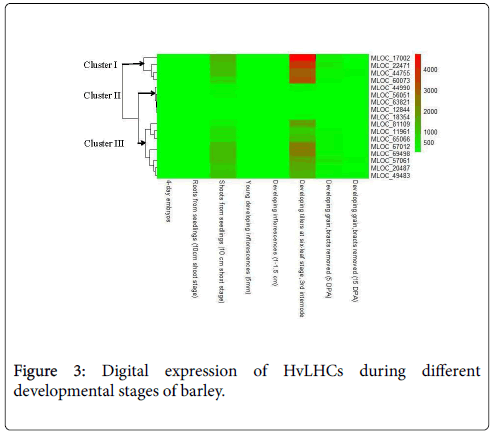
Expression profiling analysis of gene can provide important clues regarding its function. Digital spatio-temporal expression pattern of HvLHCs was detected by RNA sequencing, which was extracted from barley genome database (Figure 3). All of the HvLHCs were undetectable or have very low expression level in 4-day old embryos dissected from germinating grains, roots from the seedlings (10 cm shoot stage), young developing inflorescences (5 mm), developing inflorescences (1-1.5 cm), 5 and 15 days post-anthesis developing grain (DPA) (bracts removed). However, they showed prominently higher expression level in shoots from seedlings (10 cm shoot stage) and developing tillers at six-leaf stage (the third internode) as compared to the six organs mentioned above. They were further classified into 3 clusters according to their expression level in the latter two organs. Cluster I contained four genes, which showed significantly higher expression level in the two organs, especially in the developing tillers. Cluster III contained eight genes showing higher expression to a less extent, while cluster II contained five genes, which showed just a little bit higher expression value in the two organs as compared to the other six organs (Figure 3).
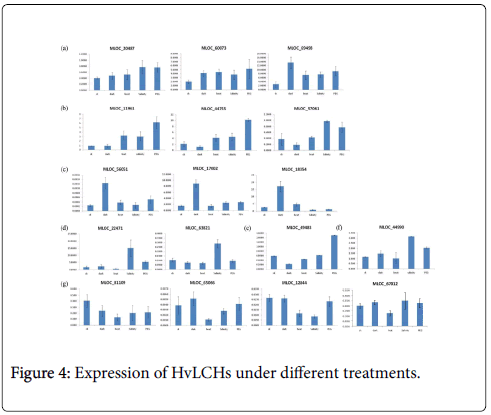
What’s more, dark, heat, drought and salinity stresses were conducted on barley seedlings in the present study. Transcriptional profiles of these genes under the four treatments were investigated using q RT-PCR. The results showed that expression of some of these genes were induced by all the treatments (such as MLOC_60073), some of them responded differently to different treatments (such as MLOC_49483), and some of them were repressed significantly by one of the treatments (MLOC_81109), such as high temperature stress. Further analysis classified these genes into seven groups according to their expression patterns under the treatments studied here (Figure 4a-4g).
Discussion
Characterization of barley LHC gene family
Light harvesting chlorophyll a/b binding protein (LHC) is one of the most abundant gene families in plants. Identifying members of this gene family at the genome level would facilitate a better understanding about the evolution and function of these genes in various plant species. In rice and Arabidopsis, 17 and 23 LHCs or LHC-related genes were identified [15]. Total 17 non-reduntant members from barley genome were identified in the present study. Though all the coding sequences of these HvLHCs had a conserved domain for chlorophyll a/b binding protein, the nucleotide sequences of them showed significant diversity between them, especially the sequences outside the conserved domain (Data not shown). Moreover, gene structures also varied a lot. Number, length and distribution of exon and intron among them were quite different from each other (Figure 1).
After searching for the chromosome locations of HvLHCs, we found that there were two hot regions on chromosome 2H and 7H, which contained four and three members, respectively. Though gene duplication was a common phenomenon in plants with huge genome, our results showed that sequences of genes in the same cluster were quite different, suggesting that the concentration of these genes was not the result of the gene duplication.
Phylogenetic analysis of LHC proteins from barley, rice and arabidopsis
Most of these HvLHCs could find one or more homologous genes both in Arabidopsis and rice, suggesting that they were evolutionarily conserved between dicot and monocot plants. It was interesting that the gene clade on the upper right of Figure 2, in which members from the same species showed closer relationship as compared to members from the other two species. Members of Arabidopsis in this clade were Lhcb1.1 to Lhcb1.5 [15]. The gene clade next to it also showed similar result, including AtLhcb2.1, AtLhcb2.2 and AtLhcb2.4/2.3 in it. These genes from different species may play some different roles as compared to each other. These findings also offered an open challenge to unravel their differentiation and function in barley as well as other relatives of barley.
Expression of HvLHCs during barley development and response to stimulus
Tissue-specific expression data at a given developmental stage is useful for identifying genes involved in defining precise nature of individual tissues. In this study, expression profiling of the 17 HvLHCs using public RNA-seq data showed that they were specifically and highly expressed in shoots from seedlings (10 cm shoot stage) and developing tillers at six-leaf stage (3rd internode) (Figure 3). These two tissues were dominant photosynthesis relevant tissues as compared to the others analyzed. The abundance of LHCs genes in them suggested that these genes played critical roles in photosynthesis of barley.
It was well known that a sudden shift from high light condition to low light conditions during the growth of barley plants in growth chamber resulted in an increase in the chlorophyll a+b content of senescing primary foliage leaves [23]. The abundance of minor chlorophyll a/b binding proteins CP29 and LHCI of barley during leaf senescence was also controlled by light [23]. There were numbers of various kinds of light responsive elements in promoter regions of all the HvLHCs. Accordingly, expression of most of these HvLHCs changed due to the dark treatment, either induced or repressed. Expressions of genes in group c were significantly induced by dark treatment (Figure 4c).
Different from animals, plants must adapt to various biotic and abiotic stresses in their life cycles involving physiological processes as well as molecular activities. Photosynthesis is one of the most sensitive physiological processes when plants are attacked by adverse stimulus, such as heat, drought and fungi. Expression level of CcLHCB1 from pigeon pea was up-regulated by drought stimulation and AM inoculation [24]. In barley, phosphorylation and translocation of HvLhcb1 in thylakoid membranes was induced under Fe deficiency condition, which may be important for balancing the PS/PHenergy distribution during Fe deficiency [25]. A type I chlorophyll a/b binding protein b was identified to be specially expressed in the barley drought tolerant genotype XZ5 [26]. There were also several studies showed that members of LHCB family play an important role in plant adaptation to environmental stresses [10,27,28]. Previous studies also showed that some LHCB members of Arabidopsis were positively involved in ABA dependent processes, such as stomatal movement, response to drought [10] and seed germination [11]. This study found that there were one to five ABREs in the promoter regions of 16 HvLHCs which indicated that expression of them may be responsive to ABA. They may also play some roles in ABA regulated pathways. Additionally and importantly, promoter regions of these HvLHCs also contained various kinds of stress-responsive elements. To further illustrate their expression pattern under stress treatments, responses of them under dark, heat, drought and salinity stresses were investigated in this study. All of the 17 genes were responsive to at least one stimulus, and they were classified into seven groups according to their main features of expression patterns (Figure 4). These genes in the same group may play similar or complementary roles under different conditions, while genes in different groups may play different roles. Over expression and knockdown experiments will needed and be helpful to illustrate the biological function of HvLHCs genes and to make use of them in barley breeding.
Conclusion
This is the first report about identification of LHCs from barley at the genome level. Results presented here will not only provide a basis for determining the role of HvLHCs during development and stress response. Also this resource will be useful in breeding of new elite barley varieties with high yield or high tolerance to biotic and abiotic stresses.
Acknowledgements
We thank Dr. Liu Gang at Institute of Food Crops, Hubei Academy of Agricultural Sciences and Dr. Guo Ganggang from Institute of Crop Science, Chinese Academy of Agricultural Sciences for their help in the data analysis. This work was financially supported by Natural Science Foundation of Hubei Province (2015CFA108) and China Agriculture Research System (CARS-05).
References
- Cao S, Zhao Y, Wen J, Wang S, Zhang R (2000) Studies on Photosynthesis in Flag Leaves and Its Relation to Grain Filling Course of High-yield Wheat. Scientia Agricultura Sinica 33: 19-25.
- Fu J, Cheng L, Huang Z-h, Wang Z-Q, Yang J-C (2012) Relationship of Leaf Photosynthetic Characteristics and Root Physiological Traits with Grain Yield in Super Rice. Acta Agronomica Sinica 38: 1264-1276.
- He Q, Zhang E, Zhao Y, Zhu Y (2008) Effects of Canopy photosynthetic Characteristics fro Yield of Different Genotype of Malting Barley. Chinese Agricultural Science Bulletin 24: 237-241.
- Lawson T, Kramer DM, Raines CA (2012) Improving yield by exploiting mechanisms underlying natural variation of photosynthesis. Current Opinion in Biotechnology 23: 215-220.
- Schultheiss H, Hensel G, Imani J, Broeders S, Sonnewald U, et al. (2005) Ectopic Expression of Constitutively Activated RACB in Barley Enhances Susceptibility to Powdery Mildew and Abiotic Stress. Plant Physiology 139: 353-362.
- Fukayama H, Ueguchi C Fau - Nishikawa K, Nishikawa K Fau - Katoh N, Katoh N Fau - Ishikawa C, Ishikawa C Fau - Masumoto C, et al. (2012) Overexpression of rubisco activase decreases the photosynthetic CO2 assimilation rate by reducing rubisco content in rice leaves. Plant & Cell Physiology 53: 976-986.
- Zhang H, Xu W, Wang H, Hu L, Li Y, et al. (2014) Pyramiding expression of maize genes encoding phosphoenolpyruvate carboxylase (PEPC) and pyruvate orthophosphate dikinase (PPDK) synergistically improve the photosynthetic characteristics of transgenic wheat. Protoplasma 251: 1163-1173.
- Ambavaram MMR, Basu S, Krishnan A, Ramegowda V, Batlang U, et al. (2014) Coordinated regulation of photosynthesis in rice increases yield and tolerance to environmental stress. Nature Communications 5: 5302.
- Pietrzykowska M, Suorsa M, Semchonok DA, Tikkanen M, Boekema EJ, et al. (2014) The Light-Harvesting Chlorophyll a/b Binding Proteins Lhcb1 and Lhcb2 Play Complementary Roles during State Transitions in Arabidopsis. The Plant Cell 26: 3646-3660.
- Xu Y-H, Liu R, Yan L, Liu Z-Q, Jiang S-C, et al. (2012) Light-harvesting chlorophyll a/b-binding proteins are required for stomatal response to abscisic acid in Arabidopsis. Journal of Experimental Botany 63: 1095-1106.
- Liu R, Xu Y-H, Jiang S-C, Lu K, Lu Y-F, et al. (2013) Light-harvesting chlorophyll a/b-binding proteins, positively involved in abscisic acid signalling, require a transcription repressor, WRKY40, to balance their function. Journal of Experimental Botany 64: 5443-5456.
- Morita R, Sugino M, Hatanaka T, Misoo S, Fukayama H (2015) CO(2)-Responsive CONSTANS, CONSTANS-Like, and Time of Chlorophyll a/b Binding Protein Expression1 Protein Is a Positive Regulator of Starch Synthesis in Vegetative Organs of Rice. Plant Physiology 167: 1321-1331.
- Fan Y, Tian Z, Yan Y, Hu C, Abid M, et al. (2017) Winter Night-Warming Improves Post-anthesis Physiological Activities and Sink Strength in Relation to Grain Filling in Winter Wheat (Triticum aestivum L.). Frontiers in Plant Science 8: 992.
- Xia Y, Ning Z, Bai G, Li R, Yan G, et al. (2012) Allelic Variations of a Light Harvesting Chlorophyll A/B-Binding Protein Gene (Lhcb1) Associated with Agronomic Traits in Barley. PLOS ONE 7: e37573.
- Umate P (2010) Genome-wide analysis of the family of light-harvesting chlorophyll a/b-binding proteins in arabidopsis and rice. Plant Signaling & Behavior 5: 1537-1542.
- Beier S, Himmelbach A, Colmsee C, Zhang X-Q, Barrero RA, et al. (2017) Construction of a map-based reference genome sequence for barley, Hordeum vulgare L. Scientific Data 4: 170044.
- Mayer Kf FWR, Waugh RFBJWS, Brown JwFSA, Schulman AFLP, Langridge PFPM, et al. (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491: 711-716.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution 30: 2725-2729.
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, et al. (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Research 30: 325-327.
- Colmsee C, Beier S, Himmelbach A, Schmutzer T, Stein N, et al. (2015) BARLEX-the Barley Draft Genome Explorer. Molecular Plant 8: 964-966.
- Houston K, McKim SM, Comadran J, Bonar N, Druka I, et al. (2013) Variation in the interaction between alleles of HvAPETALA2 and microRNA172 determines the density of grains on the barley inflorescence. Proceedings of the National Academy of Sciences of the United States of America 110: 16675-16680.
- Qin D, Wang F, Geng X, Zhang L, Yao Y, et al (2015) Overexpression of heat stress-responsive TaMBF1c, a wheat (Triticum aestivum L.) Multiprotein Bridging Factor, confers heat tolerance in both yeast and rice. Plant Molecular Biology 87: 31-45.
- Humbeck K, Krupinska K (2003) The abundance of minor chlorophyll a/b-binding proteins CP29 and LHCI of barley (Hordeum vulgare L.) during leaf senescence is controlled by light. Journal of Experimental Botany 54: 375-383.
- Qiao G, Wen X-P, Zhang T (2015) Molecular Cloning and Characterization of the Light-Harvesting Chlorophyll a/b Gene from the Pigeon pea (Cajanus cajan). Applied Biochemistry and Biotechnology 177: 1447-1455.
- Saito A, Shimizu M, Nakamura H, Maeno S, Katase R, et al. (2014) Fe deficiency induces phosphorylation and translocation of Lhcb1 in barley thylakoid membranes. FEBS Letters 588: 2042-2048.
- Wang N, Zhao J, He X, Sun H, Zhang G, et al. (2015) Comparative proteomic analysis of drought tolerance in the two contrasting Tibetan wild genotypes and cultivated genotype. BMC Genomics 16: 432.
- Andersson J, Walters RG, Horton P, Jansson S (2001) Antisense Inhibition of the Photosynthetic Antenna Proteins CP29 and CP26: Implications for the Mechanism of Protective Energy Dissipation. The Plant Cell 13: 1193-1204.
- Ganeteg U, Kulheim C, Andersson J, Jansson S (2004) Is Each Light-Harvesting Complex Protein Important for Plant Fitness? Plant Physiology 134: 502-509.
Relevant Topics
- Agricultural science
- Agronomy
- Climate impact on crops
- Crop Productivity
- Crop Sciences
- Crop Technology
- Field Crops Research
- Hybrid Seed Technology
- Irrigation Technology
- Organic Cover Crops
- Organic Crops
- Pest Management
- Plant Genetics
- Plant Breeding
- Plant Nutrition
- Seed Production
- Seed Science and Technology
- Soil Fertility
- Weed Control
Recommended Journals
Article Tools
Article Usage
- Total views: 3272
- [From(publication date):
August-2017 - Dec 22, 2024] - Breakdown by view type
- HTML page views : 2500
- PDF downloads : 772