Research Article Open Access
Genome-Wide Comparative Transcriptional Analysis of Developing Seeds among Seven Oryza sativa L. Subsp. Japonica Cultivars Grown near the Northern Limit of Rice Cultivation
| Sho Takano1, Shuichi Matsuda1, Yuji Hirayama2, Takashi Sato2, Itsuro Takamure3 and Kiyoaki Kato1* | ||
| 1Department of Agro-Environmental Science, Obihiro University of Agriculture and Veterinary Medicine, Nishi 2-11 Inada, Obihiro, Hokkaido, 080-8555, Japan | ||
| 2Rice Breeding Group, Kamikawa Agricultural Experiment Station, Local Independent Administrative Agency Hokkaido Research Organization, Minami 1-5, Pippu, Hokkaido 078-0397, Japan | ||
| 3Graduate School of Agriculture, Hokkaido University, Kita 9 Nishi 9, Kita-ku, Sapporo, Hokkaido 060-811, Japan | ||
| Corresponding Author : | Kiyoaki Kato Department of Agro-Environmental Science Obihiro University of Agriculture and Veterinary Medicine Nishi 2-11 Inada, Obihiro, Hokkaido, 080-8555, Japan Tel: +81-155-49-5477 Fax: +81-155- 49-5593 E-mail: kiyoaki@obihiro.ac.jp |
|
| Received November 11, 2014; Accepted December 05, 2014; Published December 08, 2014 | ||
| Citation: Takano S, Matsuda S, Hirayama Y, Sato T, Takamure I, et al. (2015) Genome-Wide Comparative Transcriptional Analysis of Developing Seeds among Seven Oryza sativa L. Subsp. Japonica Cultivars Grown near the Northern Limit of Rice Cultivation. J Rice Res 3: 130. doi: 10.4172/2375-4338.1000130 | ||
| Copyright: © 2015 Takano S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. | ||
Related article at Pubmed Pubmed  Scholar Google Scholar Google |
||
Visit for more related articles at Rice Research: Open Access
Abstract
Improved grain quality is a major breeding target in rice (Oryza sativa L. subsp. japonica), owing to market demand. Rice cultivars grown in Hokkaido (42-45°N), the northernmost region of rice paddy cultivation in Japan and near the northern limit of rice cultivation, have been bred for over 100 years for adaptation to low temperature together with high yield and grain quality. In this study, for seven closely related rice cultivars released in Hokkaido in the last 70 years, we investigated the transcriptome profiles of developing seeds 8 days after flowering (DAF, middle stage) and 15 DAF (late stage) under natural conditions in Hokkaido (42.52°N) using a whole-genome oligonucleotide microarray. The transcriptome profiles were divided into two groups depending on stage and were more variable at the late than that at the middle stage. The genome-wide transcriptome data revealed the features of differentially expressed genes in two first-generation cultivars compared to that in the parental cultivars. Starch properties were varied and correlated with the expression of starch biosynthesis genes among Hokkaido cultivars. Apparent amylose content was positively correlated with Waxy gene expression at the late stage. The expression of starch biosynthesis genes and starch properties were varied among Hokkaido cultivars. The transcriptomes of the most recent cultivars reveal the expression of ideal genes for the development of cultivars with high grain yield, grain quality, and grain traits essential for rice production near the northern and southern limits of rice cultivation.
| Keywords | |
| Grain quality; Rice; Transcriptome; Cultivar; Grain filling | |
| Introduction | |
| Rice (Oryza sativa L.) is a staple food for nearly one-half of the world’s population. Rice productivity has been greatly improved during the Green Revolution since the 1960s, and scientists and breeders now focus on improving grain quality for different purposes and markets. Rice grain quality is determined mainly by four constituents: grain appearance, nutritional content, and the cooking and eating quality [1]. Grain quality involves many characteristics ranging from physical to biochemical and physiological properties [2]. Starch and protein are two main components of rice endosperm and, consequently, key components of grain quality. Rice grain is also an important source of micronutrients and vitamins, whose storage plays crucial roles in grain quality and nutritional value. Consumer preferences for grain quality vary worldwide based on food habits. Therefore, the improvement of rice grain cooking quality has become the most important research component of almost all rice improvement programs worldwide. For instance, people in the Far East, including Japan, prefer sticky and soft rice, whereas those in India prefer non-sticky rice. Consumers in developed countries demand mainly for grain with good cooking quality and eating characteristics, but in many developing regions, nutritional value is crucial. | |
| Starch comprises two major components: amylose and amylopectin. Amylose is a linear molecule containing α((1,4)-linked glucose units with few branches, whereas amylopectin is a highly branched biopolymer containing linear chains of α(1,4)-linked glucose residues joined by α(1,6)-linkages. Within the granule, the chains are considered to be arranged in clusters at intervals of 9 nm within which chains are associated to form double helices. These helices are packed in ordered arrays to form concentric crystalline lamellae, and the distribution of branch lengths in amylopectin is important in determining the crystalline nature of the starch granule and the physical properties of the starch [3-6]. The percentage of amylose in total starch, measured as Apparent Amylose Content (AAC), is the key determinant of rice cooking properties. The suggested classification of amylose content (AC) identifies classes as waxy (0-5%), very low (5- 12%), low (12-20%), intermediate (20-25%), or high (25-33%), whereas rice is commercially classified by amylose content as low (<20% amylose), medium (21-25%), or high (26-33%) [7,8]. Rice grains with high AC (≥26%), like risotto varieties, cook dry and become less tender and hard upon cooling, whereas rice grains with low AC (<20%) cook moist and become sticky, glossy, and cohesive after cooking. In most rice-growing or consuming regions of the world, rice with intermediate AC is preferred. In contrast, low-AC japonicas are preferred in Japan. | |
| In addition to AAC (AC), alteration of amylopectin content and structure affects cooking quality and consumption traits [9,10]. The high degree of organization within amylopectin, a non-random distribution of linear chains, the clustered positioning of branch linkages, and the double helical conformation between adjacent parallel side chains contribute to the crystalline features of amylopectin molecules. In the current model of the multiple cluster structure of amylopectin, A chains, which lack other chains, are linked to other chains at their reducing ends, whereas B chains comprise one or more chains belonging to a cluster. Amylopectins exhibit polymodal chain-length distributions with periodic waves of varying Degrees of Polymerization (DP). The chains are grouped into four different fractions with DP ranging in the intervals 6−12 (A), 13−24 (B1), 25−36 (B2), and >37 (B3) [11]. Based on the ratio of the number of short chains of DP ≤ 10 to the short and intermediate chains of DP ≥ 24 (short chain ratio, SCR), amylopectin with lower SCR is called L-type and that with a higher SCR is called S-type [12] Cooked rice with the S-type amylopectin of japonica cultivars is much softer after cooling than rice with the L-type amylopectin of indica cultivars [13]. Among japonica rice cultivars, a higher content of short chain amylopectin is also positively correlated with softness and stickiness of cooked rice [14]. | |
| The variations in starch structures arise from a differential expression of various isoforms of starch biosynthesis enzymes. Starch biosynthesis in higher plants, including rice, is catalyzed by four classes of enzymes: ADP-Glc pyrophosphorylase (AGPase), starch synthase (SS), starch branching enzymes (SBE), and starch Debranching Enzymes (DBE). Several studies have led to the identification of various SS isoforms via gene sequencing in several plant genomes. In rice, there are 10 SS isoforms, which can be grouped into five types: GBSS (I, II); SSI; SSII (SSIIa, SSIIb, and SSIIc); SSIII (SSIIIa and SSIIIb); and SSIV (SSIVa and SSIVb) [15-17]. All of these isoforms of starch-synthesizing enzymes coordinate and form a regulatory network to control starch synthesis in rice endosperm, affecting grain cooking and eating quality [18-20]. Amylose content is regulated primarily by the Waxy (Wx) gene, which encodes the granule-bound starch synthase I (GBSSI) enzyme, and the level of grain amylose is directly associated with the amount of GBSSI in the endosperm [21-23]. Amylopectin biosynthesis is controlled by SS, SBE, and DBE [8,16,17,23-25]. The gene identified as being responsible for amylopectin structure and processing suitability is the starch synthase IIa gene (SSIIa), known as the Alk (alkali disintegration) gene [26]. SSIIa is responsible for the elongation of short chains with DP 5−10 to form longer A and B1 chains with DP 11−24 within the cluster of amylopectin. Therefore, the difference in allele of the SSIIa is responsible for the structural alteration of amylopectin between indica- and japonica-type rice varieties having active SSIIa and inactive SSIIa, respectively [8,27]. SBEIIb plays a specific role in the formation of A chains of the amylopectin cluster [28], and the levels of SBEIIb activity affect the extent of structural changes in the cluster [25]. Isoamylase (ISA1), one of the DBE, is essential for the formation of the highly organized cluster structure of amylopectin [29], and the level of ISA1 expression influences both the fine structure of amylopectin and the thermal properties of starch [30,31]. In addition, Quantitative Trait Locus (QTL) analysis has revealed that many grain quality traits are controlled by multiple regions in the rice genome [32-36]. | |
| Rice seed development can be divided into four stages: the initiation stage [1-3 days after flowering (DAF)], during which starch is synthesized exclusively in the pericarp; the early developmental stage (3-5 DAF), indicated by endosperm starch accumulation with a marked increase in seed weight; the middle stage (5-10 DAF), with a rapid increase in starch deposition and grain weight; and the late stage (10 DAF and beyond), in which seed maturation occurs [37]. The expressions of genes involved in starch biosynthesis are differentially regulated depending on the developmental stage of the seed [15,38,39]. The challenge lies in understanding the genetic basis and identification of genomic regions controlling these isoforms in order to develop cultivars with desirable combinations of alleles that control these isoforms depending on the developmental stage of the seed. | |
| Grain quality is determined by a variety of developmental processes, which are affected not only by the genetic constitution but also by environmental conditions such as temperature, water availability, fertilizer application, and drought and salinity stresses [2]. Strong genotype × environment effects have been found for several quality traits in rice cultivars grown at different locations and during different seasons [12,40-42]. Wx gene expression and Wx protein were increased when rice plants were exposed to low temperature (18°C) [21,42]. The G−T polymorphism in the first intron of the Wx gene (Wxb) is associated with a different efficiency of RNA splicing and processing under low and high temperature during grain development [43]. In addition, lower temperatures increased the molar and weight ratios (A + B1)/(B2 + B3) of amylopectin unit chains in japonica cultivars [44] and thus in Hokkaido (japonica) cultivars leading, under cool conditions during grain filling, to low eating quality [45]. The molecular basis of the increased number of short chains at low temperature remains to be elucidated. Nonetheless, for the past 30 years, rice breeders have selected genotypes optimizing the activity of Wx and other genes, affecting amylose content and amylopectin chain length, to adapt to environmental conditions during grain filling in Hokkaido. | |
| The rice cultivars growing in Hokkaido (42-45°N), the northernmost region of rice paddy cultivation in Japan and one of the northern limits of rice cultivation in the world, have been improved to be adapted to low temperatures during the grain-filling stage along with high yield and good eating quality. There is little information about transcriptional profiles in developing seeds of Hokkaido cultivars under natural conditions. In the present study, we investigated the gene expression in developing seeds of seven very closely related cultivars released in Hokkaido during the last 70 years using Agilent 44K rice oligoarrays. Using genome-wide transcriptome profiling, we addressed the breeding history of these seven cultivars in the transcriptome, transmission of transcriptome profiles to two firstgeneration cultivars, and the transcriptional profile unique to the most recent Hokkaido cultivar. These genome-wide transcriptome data provide a foundation for deep exploration of gene-trait relationships, and for rice improvement near the northern and southern limits of world rice cultivation. | |
| Materials and Methods | |
| Plant materials and growth conditions | |
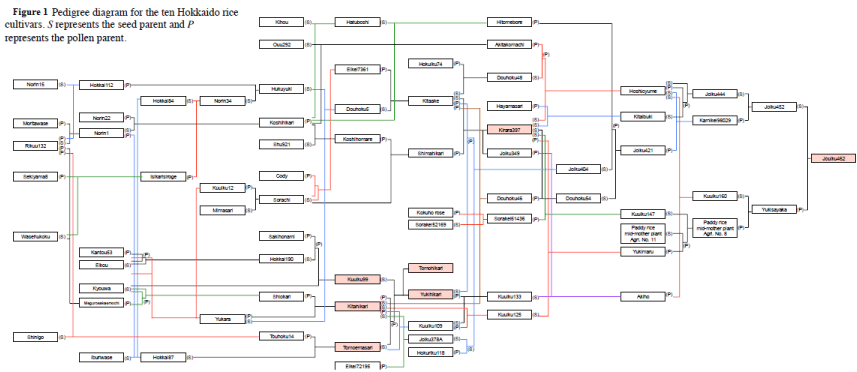
| Rice (Oryza sativa L. ssp. japonica) plants of seven Hokkaido cultivars, Tomoemasari, Kitahikari, Kuuiku99, Tomohikari, Yukihikari, Kirara397, and Jouiku462 (Figure 1), that carry Wxb were grown in 3-L pots filled with 2 kg of soil including 1.2 g each of N, P2O5 and K2O per pot under natural conditions at Obihiro, Hokkaido (42.52°N) in 2013. Immature seeds of each cultivar were collected at 8 DAF (middle stage), and 15 DAF (late stage). Depending on flowering time, immature seeds of Kuuiku99, Tomohikari, Yukihikari, Kirara397, and Jouiku462 were collected on August 19 (8 DAF) and August 26 (15 DAF), immature seeds of Kitahikari were collected on August 23 (8 DAF) and August 30 (15 DAF), and Tomoemasari seeds were collected on August 29 (8 DAF) and September 5 (15 DAF). In all, 50 immature seeds of each cultivar were collected from 10-12 plants of each cultivar and bulked for comprehensive gene expression analysis (Figure 2). For amylose and amylopectin analysis, >300 plants of each cultivar were grown on the experimental farm of the Hokkaido University Field Science Centre for Northern Bio-Research (Sapporo, Hokkaido, 43.074°N) in 2013. | |
| RNA isolation and microarray analysis | |
| Agilent 44 K rice oligoarrays (Agilent Technologies), which contain 44,000 features, were used as one-color oligoarrays. Each feature comprises a 60-mer oligonucleotide corresponding to a fulllength cDNA of rice. Total RNA was extracted from the endosperm of all cultivars at 8 and 15 DAF using the RN easy plant mini kit (QIAGEN). Microarray experiments were performed according to the manufacturer’s manual. Feature Extraction software 10.7.3.1 (Agilent Technologies) was used to delineate and measure the signal intensity of each spot on the array. The resulting data were normalized by a 75 percentile shift [46]. Poor quality features that were either non uniform or saturated were flagged and not used for further analysis. | |
| Cluster analysis of SNPs/InDels between seven cultivars | |
| Whole-genome sequencing and detection of SNP/InDel positional data between Nipponbare (Pseudomolecules build 5.0, http:// rapdblegacy.dna.affrc.go.jp/) and seven Hokkaido cultivars were performed using our previous data [47], yielding 83,738 unique SNP/ InDel sites. A dendrogram was created using complete linkage in Cluster 3.0 [48] (http://bonsai.ims.u-tokyo.ac.jp/∼mdehoon/software/ cluster/), and the result was visualized with Java TreeView software 1.1.6r4 [49] (http://jtreeview.sourceforge.net/). The colors used in the clustergram representation for each of the seven cultivars were based on a comparison with the Nipponbare genome. If the SNP/InDel type matched that of Nipponbare, it was colored blue; otherwise, it was colored yellow. | |
| Apparent amylose content analysis | |
| According to the protocol described by [50] with modifications [51] by the AAC of milled grain was measured with an auto-analyzer (AA-III, Bran Luebbe), which is based on a flow injection of a 0.09% NaOH solution into the sample, addition of an iodine solution, and spectrometric determination of the absorbance at 620 nm was determined by a spectrometer. Calibration was performed using the absorbance of standard rice samples carrying 21.1% AAC. ACC analysis was performed in triplicates for each genotype. | |
| Branch chain-length distribution of α-glucan | |
| Isolation of starch and analysis of the branch chain-length distribution of α-glucan were performed as described by [52]. Mature kernels from each cultivar were dehulled and the endosperm was separated from the germ and milled in a coffee mill. The ground endosperm powder (7 g) was suspended in 100 mL of distilled water and stirred for 1 h at 4°C. The suspension containing starch was centrifuged at 700 ×g for 10 min at 4°C. The precipitate was washed five times with 100 mL of distilled water, followed by centrifugation at 700 ×g for 10 min at 4°C and a final centrifugation at 10,000 ×g. The resulting starch pellet was suspended by stirring in 100 mL of 2% SDS for 2 h at room temperature, and was then centrifuged at 700 ×g for 10 min at room temperature to remove protein, and the supernatant was discarded. The procedure for removing protein was repeated three times. The precipitate was then washed six times with 100 mL of distilled water, followed by centrifugation at 700 ×g for 10 min at room temperature and final centrifugation at 10,000 ×g. Starch samples were dried in vacuo. | |
| The starch was suspended in 5 mL of methanol and heated in a boiling water bath for 10 min. The suspension was centrifuged at 1,000 ×g for 10 min, and the precipitate was collected and washed twice with 5 mL of 90% (v/v) methanol and then re-suspended in 300 mL of 0.25 M NaOH. The suspension was heated in a boiling water bath for 5 min to gelatinize the starch. The pH and concentration of α-glucan sample solutions were adjusted by the addition of 9.6 μL of 100% acetic acid, 100 μL of 600 mM Na-acetate buffer (pH 4.4), 15 μL of 2% NaN3, and 1.09 mL of distilled water. The α-glucan sample was debranched with Pseudomonas amyloderamosa isoamylase (354 units, Seikagaku- Kogyo, Tokyo) at 37°C for 24 h. The debranched sample was heated in a boiling water bath for 20 min to stop enzyme activity, and was then centrifuged. The supernatant was deionized with ion exchange resin [Bio-Rad AG 501-X8(D)] in a microtube. An aliquot containing the equivalent of 5 nmol of reducing sugar, determined by the modified Park Johnson method [53], was evaporated to dryness in a centrifugal vacuum evaporator (Taitec, Tokyo). Fluorescence labeling of α-1,4- glucan at the reducing end with 8-amino-1,3,6-pyrenetrisulfonic acid (APTS) was performed according to the method described by [54]. The APTS-labeled α-1,4-glucan was analyzed using an eCAP N-linked oligosaccharide profiling kit and a P/ACE System 5000 high-resolution capillary electrophoresis system equipped with a laser-induced fluorescence detector (Beckman Instruments, Coulter, CA, U.S.A.). | |
| Statistical analysis | |
| Tukey’s test was applied to determine any significant differences between the means of starch properties. | |
| Results | |
| Gene expression profiles of seven Hokkaido rice cultivars | |
| Genes involved in adaptation to cool summers in Hokkaido during grain filling, or possibly unknown genetic factors that are essential for grain filling in Hokkaido, may be positively selected in flanking chromosomal regions. To assess the correlation between expression profile and genomic structure in seven Hokkaido cultivars, we compared the dendrograms based on hierarchical clustering of transcriptome profiles of seven cultivars at 8 and 15 DAF and of the previous comprehensive SNPs/InDel data set [47] (Figure 1). The dendrogram from hierarchical clustering revealed that the transcriptome profiling was divided into two large clusters depending on the seed development stage (Figure 1). The branch length of the dendrogram of 15 DAF was longer than that of 8 DAF, suggesting that the transcriptome of each cultivar at 15 DAF varied as compared to those at 8 DAF. At both stages, six cultivars formed similar dendrograms with respect to four features; (i) Tomoemasari was the farthest from the other six cultivars, (ii) Tomohikari, Yukihikari, and Kuuiku99 formed the primary group, (iii) Yukihikari and Tomohikari formed a sister pair, and (iv) Kirara397 and Jouiku462 formed a sister pair. Kitahikari was positioned differently depending on the stage; Kitahikari clustered in the primary group with Kirara397 and Jouiku462 at 15 DAF, but was separate from these cultivars at 8 DAF. | |
| Based on genome-wide SNPs/InDels, the genome structures of the seven cultivars were divided into two groups, the first containing Tomoemasari, Kuuiku99, Tomohikari, and Yukihikari and the second containing Kitahikari, Kirara397, and Jouiku462 (Figure 1). In the first group, Yukihikari and Tomohikari formed a sister pair. In the second group, Kirara397 and Jouiku462 formed a sister pair. Thus, the shape of the dendrogram based on comprehensive SNPs/InDels was similar to that of the dendrogram based on the transcriptome profiles at 15 DAF. These findings, taken together, suggest that breeders have positively selected genotypes associated with the transcriptome profiles at 15 DAF to improve grain-filling traits in Hokkaido in the past 70 years. | |
| Transmission of gene expression in the first generation cultivars | |
| Yukihikari and Tomohikari were first-generation cultivars and were developed from (Kitahikari/Tomoemasari)/Kuuiku99 (Figure 1). We assessed the transmission of gene expression from each parental cultivar to the first generation cultivars. As described above, the dendrogram of hierarchical clustering based on the transcriptome showed that Yukihikari and Tomohikari formed a sister pair and clustered with Kuuiku 99 at both stages, and that Tomoemasari was the farthest from Yukihikari and Tomohikari at both stages. These features were very similar to those of clustering based on genome-wide SNPs/ InDels (Figure 1). | |
| In Yukihikari, 4,714 genes (8 DAF) and 2,348 genes (15 DAF) were similar to at least one parental line (showing between 1/1.2 and 1.2 times the Yukihikari reference expression). Among them, 3,651 (8 DAF) and 1,341 (15 DAF) were conserved between Yukihikari and all three parental cultivars. Thirty (8 DAF) and 42 (15 DAF) genes were conserved with Kitahikari, 30 (8 DAF) and 75 (15 DAF) with Tomoemasari, and 157 (8 DAF) and 285 (15 DAF) with Kuuiku99. Like Yukihikari, 4,310 (8 DAF) and 2,893 (15 DAF) genes in Tomohikari were similar in at least one parental line (between 1/1.2 and 1.2 times Tomohikari reference expression). Among these, 3,367 (8 DAF) and 1,231 (15 DAF) genes were conserved between Tomohikari and the three parental cultivars. Thirty-seven (8 DAF) and 57 (15 DAF) genes were conserved with Kitahikari, 25 (8 DAF) and 40 (15 DAF) with Tomoemasari, and 173 (8 DAF) and 328 (15 DAF) with Kuuiku99. | |
| Some genes were found to be differentially expressed in Yukihikari and/or Tomohikari compared to either parental cultivar, and were therefore considered “transgressive.” In Yukihikari, 63 (8 DAF) and 45 (15 DAF) genes were upregulated at least two-fold compared to either parental cultivar, while 50 (8 DAF) and 41 (15 DAF) were downregulated at least two-fold compared to either parental cultivar. In Tomohikari, 61 (8 DAF) and 344 (15 DAF) genes were upregulated at least two-fold compared to either parental cultivar, while 95 (8 DAF) and 80 (15 DAF) were downregulated to less than 1/2 times compared to either parental cultivar. Among these differentially expressed genes, 6 (8 DAF) and 16 (15 DAF) genes were consistently upregulated in Yukihikari and Tomohikari compared to either parental cultivar. In contrast, 10 (8 DAF) and 18 (15 DAF) genes were consistently downregulated in Yukihikari and Tomohikari compared to either parental cultivar. | |
| Transgressive gene expression and flanking genomic regions | |
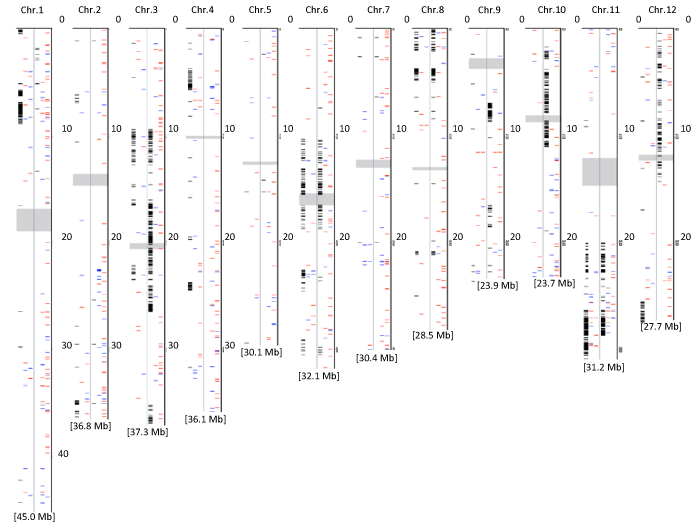
| Our previous study of genome-wide SNPs/InDels revealed that Yukihikari and Tomoemasari have clusters of unique SNPs/InDels (haplotype blocks) compared to the parental cultivars [47] (Figure 2). Each unique haplotype block was thought to be transmitted from the parental accession used originally for the breeding of the cultivars but not the parental accession used in the genomic sequencing study [46] and the present transcriptome study. To assess the possibility that differentially expressed genes are present in these unique genomic regions, we compared the SNPs/InDel sites unique to Yukihikari and/ or Tomohikari with the positions of differentially expressed genes in Yukihikari and/or Tomohikari (Figure 2). The within-chromosome distributions of differentially expressed genes in Yukihikari and Tomoemasari were nonuniform (Figure 2). Among 113 (8 DAF) and 86 (15 DAF) differentially expressed genes in Yukihikari, 17 (15.0% of 113 genes, 8 DAF) and 21 (24.4% of 86 genes, 15 DAF) were located in a unique genomic region. As with Yukihikari, 22 (14.1% of 156 genes, 8 DAF) and 56 (13.2% of 424 genes, 15 DAF) differentially expressed genes were located in a unique genomic region in Tomohikari. Differentially expressed genes located in unique genomic regions in the first generation cultivars may be controlled by cis-regulatory elements from the parental accession used originally for breeding the cultivars. In contrast, 96 (85.0% of 113 genes, 8 DAF) and 65 genes (75.6% of 86 genes, 15 DAF) in Yukihikari and 134 (85.9% of 156 genes, 8 DAF) and 368 (86.8% of 424 genes, 15 DAF) in Tomohikari may be controlled by trans-regulatory elements and/or novel combinations of cis-regulatory elements. | |
| Transcripts unique to the most recent cultivar, Jouiku462 | |
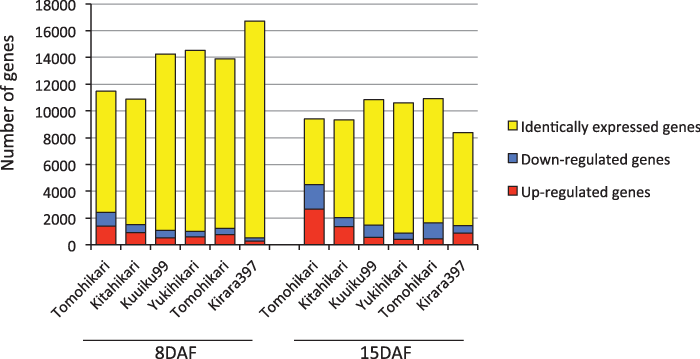
| To characterize changes in gene expression profiles of seed development during >70 years of breeding history in Hokkaido, we compared the gene expression between the most recent cultivar, Jouiku462, and the six older cultivars. A total of 2,594 (8 DAF) and 489 (15 DAF) genes were expressed at similar levels in the seven cultivars (between 1/1.2 times and 1.2 times those of Jouiku462). Compared to Jouiku462, numbers of similarly expressed genes varied among cultivars and fell between 16,214 (Kirara397) and 9,054 (Tomohikari) at 8 DAF and between 9,721 (Yukihikari) and 4,922 (Tomohikari) at 15 DAF (Figure 3). In contrast, numbers of differentially expressed genes fell between 507 (Kirara397) and 2,439 (Tomohikari) at 8 DAF and between 894 (Yukihikari) and 4,474 (Tomohikari) at 15 DAF compared to Jouiku462 (Figure 3). We list the genes unique to Jouiku462 in (8 DAF) and (15 DAF). In Jouiku462, 25 (8 DAF) and 17 (15 DAF) were consistently upregulated compared to the six older cultivars, while 45 (8 DAF) and 31 (15 DAF) were consistently downregulated. As at 8 DAF, 75 genes were differently expressed from all of the other six cultivars by more than 2-fold. Among them, 17 genes were upregulated in Jouiku462 and 31 were downregulated in Jouiku462 compared to all of the other six cultivars. | |
| Expression profiling of genes involved in starch synthesis | |
| AACs of the seven cultivars were varied, ranging from 16.6% (Jouiku462) to 20.2% (Tomoemasari). AACs of the seven cultivars were divided into four groups by statistical analysis. Jouiku462 was classified as the lowest AAC group. Kitahikari (20.1%) and Tomoemasari (20.2%) were classified as the highest AAC group. Kuuiku99 (18.2%) and Yukihikari (18.4%) were classified as a lower AAC group, and Kirara397 (18.8%) and Tomohikari (19.1%) were classified as a higher AAC group, respectively. Thus, breeders have succeeded in reducing the amylose content in rice grain for improvement of the sticky and soft eating quality of cooked rice. | |
| Properties of amylopectin chain length also varied in the seven cultivars. Two older cultivars Tomoemasari (released in 1945) and Kuuiku99 (released in 1972) showed opposite properties of amylopectin chain length. Tomoemasari contained more short chains (Fa; 5 ≤ DP ≤ 12), fewer intermediate-size (Fb2; 25 ≤ DP ≤ 36), and long chains (Fb3; DP ≥ 37). Kuuiku99 contained fewer short chains and more intermediate size and long chains. Thus, Tomoemasari had longer chains (Fb2 + Fb3), and fewer short chains (Fa + Fb1), whereas Kuuiku99 had shorter chains and fewer long chains. The other five cultivars had intermediate properties of amylopectin chain length between Kuuiku99 and Tomoemasari. The most recent cultivar, Jouiku462, was at the midpoint among the seven cultivars. These results suggest that breeders have selected the intermediate type of amylopectin chain length in the current cultivars. | |
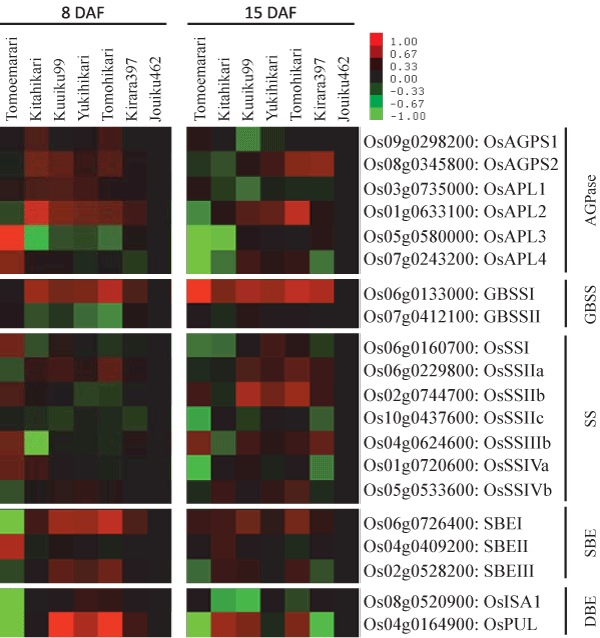
| We assessed the relationship between starch properties and gene expression (Figure 4). The marked reduction of Wx expression (15 DAF) was evident in the recent cultivar Jouiku462 compared to the six older cultivars (Figure 4). The marked reduction of OsAPL3 expression (15 DAF) was evident in Tomoemasari and Kitahikari. The reduced expression of Wx and increased expression of OsAPL3 might thus contribute, at least in part, to the reduction of amylose content in the most recent cultivar, Jouiku462. The increase of OsSSIIa expression (15 DAF) was evident in Kuuiku99 and Tomohikari suggesting that the (Fa + FB1)/(Fb2 + Fb3) ratio was negatively correlated with the expression of OsSSIIa. | |
| Discussion | |
| Possible positive selection for gene expression in Hokkaido rice cultivars | |
| We have shown several lines of evidence that support the preferential selection by breeders, over the past 70 years, of the genotype associated with gene expression at 15 DAF rather than that at 8 DAF. First, the expression profiles in developing seeds of seven rice cultivars were distinct in the two development stages of 8 DAF and 15 DAF (Figure 1). This difference in temporal expression pattern across cultivars between middle (8 DAF) and late (15 DAF) stages seem to be related to the distinct spatial pattern of starch deposition within a caryopsis and embryo development. Second, the expression profile of seven cultivars at 15 DAF was more variable than that at 8 DAF, as shown by the longer branch length at 15 DAF than at 8 DAF (Figure 1). Third, the number of differentially expressed genes between the most recent cultivar, Jouiku462, and the other six cultivars was much greater at 15 than at 8 DAF (Figure 3). In contrast, the number of genes expressed at the same level in the most recent cultivar Jouiku462 and the other six cultivars was lower at 15 than at 8 DAF. Fourth, the clustering dendrogram of genome-wide SNPs/InDels showed high similarity to that from clustering by comprehensive expression profiling at 15 but not at 8 DAF (Figure 1) suggesting that the genome-wide genotype was closely correlated with the transcriptome at 15 DAF. | |
| Rice seed development is divided into four stages: the initiation stage (1-3 DAF), during which starch is synthesized exclusively in the pericarp; the early developmental stage (3-5 DAF), indicated by endosperm starch accumulation with a pronounced increase in seed weight; the middle stage (5-10 DAF), with a rapid increase in starch deposition and grain weight; and the late stage (10 DAF and beyond), in which seed maturation occurs [14,35]. Thus, the gene expression profiles at the middle stage were relatively conserved among the seven cultivars studied suggesting that grain traits associated with the increase in starch deposition and grain weight were selected more than 70 years ago and are critical for rice production in Hokkaido. Grain traits associated with seed maturation may have been improved by the change in gene expression profiles during the last 70 years, as shown in the most recent cultivar, Jouiku462 (Figure 3) Further research is needed to clarify the possible association between grain traits and gene expression profiles. | |
| The expression profile of Tomoemasari, the oldest cultivar (released in 1945), was far from that of the other six cultivars at both stages. Because the present study aimed to compare the profiles of gene expression under natural condition, immature seeds of each cultivar were collected at different days (three groups) depending on its flowering time. The flowering date of Tomoemasari was 10 days later than those of Kuuiku99, Tomohikari, Yukihikari, Kirara397, and Jouiku462 and was 6 days later than that of Kitahikari. The distinct expression profile of Tomoemasari might thus represent both the genetic difference and the effects of different temperatures in late summer between August 25 (flowering date) and September 5 (15 DAF) in Hokkaido. | |
| Transgressive gene expression in first-generation cultivars | |
| We showed that some of the gene expression changes in the first generation cultivars are far (by ≥ 2 times) outside the range of gene expression of either parent, and can accordingly be considered transgressive. These differentially expressed genes were classified into three groups: 97 (8 DAF) and 52 (15 DAF) genes were differentially expressed only in Yukihikari (first group), 140 (8 DAF) and 390 (15 DAF) were differentially expressed only in Tomohikari (second group), and 16 (8 DAF) and 34 (15 DAF) were differentially expressed in both cultivars (third group). Among differentially expressed genes, 12 (8 DAF) and 13 (15 DAF) of the first group, 16 (8 DAF) and 50 (15 DAF) of the second group, and 5 (8 DAF) and 6 (15 DAF) of the third group were located in genomic regions unique to each of the first-generation cultivars, which are expected to have been transmitted from the parental accession used originally for breeding the cultivars (Figure 2) In contrast, 85 (8 DAF) and 39 (15 DAF) genes in the first group, 124 (8 DAF) and 340 (15 DAF) in the second group, and 11 (8 DAF) and 28 (15 DAF) in the third group were located outside of unique genomic regions, suggesting that regulation of their expression originated from trans-regulatory factors and/or new combinations of cis-regulatory factors. Recently, expression quantitative trait loci (eQTLs) have been identified as genomic regions that regulate gene expression variation in a genetically segregating population [55], eQTL analysis can detect genetic elements that regulate the expression variation of the e-trait acting in cis if the QTL is located in the vicinity of an e-trait, or in trans if it is located in a distant position. eQTL analysis could suggest the existence of a potential regulator of a given gene [56,57]. Further study is needed to clarify, by eQTL analysis, the genetic basis of the differentially expressed genes identified in the firstgeneration cultivars. In addition, there is some possibility that breeders have selected the genotype associated with the differentially expressed gene to improve grain traits during grain filling. The contributions of the genomic regions harboring regulatory factors for the differentially expressed genes to grain traits during grain filling also remain to be clarified. | |
| Expressed genes unique to the most recent Hokkaido cultivar Jouiku462 | |
| The expression profile of the most recent Hokkaido cultivar, Jouiku462, represents an ideal gene expression profile essential for high grain yield, high grain quality, and grain traits for grain filling under low-temperature conditions in Hokkaido, the northern limit of rice cultivation. In the present study, we identified 39 genes that were differentially upregulated in Jouiku462 and 64 genes that were differentially down regulated in Jouiku462 compared to the six older cultivars. For example, Os07g0689600 (Nicotianamine synthase 3; OsNAS3) was upregulated in Jouiku462 at 15 DAF. Nicotianamine (NA), a chelator of metals, is ubiquitous in higher plants and a key component of metal homeostasis [58]. Thus, manipulation of cellular NA concentrations would seem to be another approach for improving Fe concentrations in planta [58]. NA is synthesized from three molecules of S-adenosyl methionine by nicotianamine synthase (NAS). Transgenic rice seeds expressing HvNAS1 driven by the seed-specific pGluB-1 promoter also show greater amounts of NA (4-fold) and Fe (1.5-fold) [58,59]. Among the three OsNAS genes present in rice, OsNAS1 and OsNAS2 transcripts are markedly elevated in both roots and leaves in response to Fe deficiency, whereas OsNAS3 expression is induced in roots but suppressed in leaves when Fe supply is inadequate [60]. Seeds from OsNAS3 activation-tagged plants (OsNAS3-D1) in which OsNAS3 expression was increased in seedling leaves, flag leaves, flowers, and immature seeds, grown in a paddy field, contained elevated amounts of Fe (2.9-fold), Zn (2.2-fold), and Cu (1.7-fold) accompanied with a 9.6-fold increase in the NA level [61]. Cu content of grains was significantly correlated with gel consistency and AC. In addition, significant associations were found between protein content, and Zn and Cu content. Jouiku462 was bred for high grain quality of cooked rice but was not directly selected for mineral content. In Jouiku462, improvement of mineral content to accompany its nutritional properties and eating quality by enhancing NAS3 expression remains to be investigated. | |
| Os03g0315400 (OsMYB2) was also upregulated by ≥ 2-fold in Jouiku462 at 15 DAF compared to the six older cultivars. Transgenic rice upregulating OsMYB2 showed enhanced seedling-stage salinitystress tolerance along with drought and cold-stress tolerance without reduced growth rate, compared with a control (Yang et al. 2012). OsLEA3, OsRab16A, and OsDREB2A, were upregulated, suggesting that OsMYB2 encodes a stress-responsive MYB transcription factor that may act as a master switch in stress tolerance [62]. Seed development is initiated by a stage of morphogenesis in which the mature embryo develops from a single fertilized cell. Following morphogenesis, developing seeds enter a maturation stage, during which storage compounds are synthesized [63]. The early and middle phases of maturation are dominated by the action of the phytohormone, abscisic acid (ABA) [64]. Subsequently, ABA levels decline and the seed enters the desiccation phase. This phase is associated with a major loss of water, and the water content of mature dry seeds is generally less than 20% on a fresh weight basis. Such severe desiccation would kill cells in vegetative parts of the plant [65]. After 10 DAF, the water content of the developing seed drops markedly, indicating that this period is the beginning stage of dehydration [66]. However, these severely desiccated seeds germinate and develop into normal seedlings after imbibition, indicating that the developing seeds must acquire sufficient desiccation tolerance before the end of the desiccation phase, when their water content falls below the threshold of tolerance for vegetative cells. The contribution of the upregulation of OsMYB2 to dehydration stress tolerance during grain filling awaits further study. | |
| GBSSII (Wx) expression was reduced in Jouiku462 (Figure 4), although all seven cultivars carry the Wxb allele. Jouiku462 was bred as a low-amylose cultivar by introgression of qAC9.3, which is a lowamylose- content QTL [67], using marker-assisted selection (Sato et al. unpublished data). The molecular mechanism underlying the reduction of AAC by qAC9.3 is still unknown. The possibility that qAC9.3 reduces Wx expression at transcriptional and post-transcriptional levels awaits further study. | |
| Conclusion | |
| Our study provides genome-wide transcriptome data of developing seeds at 8 DAF (middle stage) and 15 DAF (late stage) under natural conditions in Hokkaido (42.52°N) for very closely related Hokkaido rice cultivars released in the past 70 years. These genome-wide transcriptome data reveal the genetic control of differentially expressed genes in first-generation cultivars. The transcriptome of a current cultivar shows possible ideal expressed genes associated with high grain yield and quality and grain traits essential for rice production near the northern and southern limits of rice cultivation. | |
| Acknowledgements | |
| This study was supported in part by Grants-in-Aid for Regional R&D Proposal-Based Program from Northern Advancement Centre for Science & Technology of Hokkaido Japan and the Tojuro Iijima Foundation for Food Science and Technology. Seeds of Hokkaido rice cultivars were provided by the Central Agricultural Experiment Station, Local Independent Administrative Agency Hokkaido Research Organization. We thank Reina Asa, Shoko Yoshikawa, Naomi Shimoda, Hideyuki Kitazawa, Hidetaka Nagasawa, Noriko Kinoshita, and Hiroki Yamamoto for technical support. | |
References |
|
|
|
Figures at a glance
 |
 |
 |
 |
|||
| Figure 1 | Figure 2 | Figure 3 | Figure 4 |
Relevant Topics
- Basmati Rice
- Drought Tolerence
- Golden Rice
- Leaf Diseases
- Long Grain Rice
- Par Boiled Rice
- Raw Rice
- Rice
- Rice and Aquaculture
- Rice and Nutrition
- Rice Blast
- Rice Bran
- Rice Diseases
- Rice Economics
- Rice Genome
- Rice husk
- Rice production
- Rice research
- Rice Yield
- Sticky Rice
- Stress Resistant Rice
- Unpolished Rice
- White Rice
Recommended Journals
Article Tools
Article Usage
- Total views: 16140
- [From(publication date):
January-2015 - Jul 04, 2025] - Breakdown by view type
- HTML page views : 11441
- PDF downloads : 4699
