Genome-wide Association Study of SPAD Values Using Diverse Soybean Germplasm
Received: 16-Jan-2024 / Manuscript No. jpgb-24-125101 / Editor assigned: 18-Jan-2024 / PreQC No. jpgb-24-125101 (PQ) / Reviewed: 23-Jan-2024 / QC No. jpgb-24-125101 / Revised: 27-Jan-2024 / Manuscript No. jpgb-24-125101 (R) / Published Date: 31-Jan-2024 DOI: 10.4172/jpgb.1000187
Abstract
Leaf photosynthesis and biological nitrogen fixation are two of the most important metabolic processes for soybean growth and development. Since they are considered closely linked, measuring chlorophyll content using non-destructive tools, such as a Soil Plant Analysis Development (SPAD) chlorophyll meter, may help determine the nodulation and nitrogen fixation status of soybean plants. This study aimed to identify single nucleotide polymorphism (SNP) markers associated with SPAD values in a global panel of 187 diverse accessions. The population structure and genome-wide association analyses were carried out using 1,243 high-quality SNPs and greenhouse data obtained in two consecutive years. The results revealed 14 SNPs significantly related to SPAD values on Chr. 5, 10, 12, 15, 17, and 18. In addition, 33 candidate genes were found in the Glyma.Wm82.a2 within 10 kb flanking regions of each significant SNP. Of these, three candidate genes on Chr. 10 and 12 encoded proteins related to photosynthesis, chlorophyll content, and nitrogen status. Overall, our data may help better understand the underlying molecular mechanisms controlling chlorophyll content in relation to nitrogen fixation in soybean.
Keywords
Chlorophyll meter; Nitrogen fixation; Soybean
Introduction
Soybean (Glycine max Merr. [L.]) is one of the most widely cultivated legumes worldwide due to its high economic value, primarily derived from its high protein (approx. 40%) and oil (approx. 20%) contents (2023 data from ers.usda.gov). For optimal growth and productivity, soybeans require 14 mineral nutrients, of which nitrogen (N) is the most predominant. Thus, inorganic N fertilizers are applied in large amounts annually to increase soybean yields [1], causing significant environmental pollution [2-4]. In the lack of inorganic N, symbiotic fixation, carried out by bacteria living in root nodules, provides the necessary N to the plant [5]. Leaf physiology and metabolism depend on available N since it is the critical component of chlorophyll, photosystems (PS) I and II, and RuBisCO [6]. Therefore, both light and dark photosynthetic reactions occur in the presence of N. Approximately 75% of the total plant N is found in the chloroplast, where it is utilized to synthesize the photosynthetic apparatus [7]. Previous studies in different plant species at local, regional, and global scales showed that the concentration of leaf N is significantly and positively correlated with that of chlorophyll as well as the photosynthetic rate [8,9].
The quantification of chlorophyll using conventional methods is time-consuming, is based on destructive extraction with organic solvents (i.e., acetone, ethanol, dimethyl sulfoxide), and requires expensive equipment for digestion and analysis [10]. On the other hand, the Minolta Soil Plant Analysis Development (SPAD) chlorophyll meter allows the quick and non-destructive measurement of chlorophyll content [11]. The SPAD meter has been previously used in studies to indirectly determine foliar chlorophyll and the N content in situ and, thus, can be a valuable tool to monitor the crop N status and potential N use efficiency or N fixation in legumes [12-14].
In soybeans, the relative leaf chlorophyll content based on SPAD values could allow the identification of quantitative trait loci (QTL) related to high N concentration and N fixation efficiency. The available methods for pinpointing QTL for economically important complex traits are linkage mapping using bi-parental populations and genome-wide association study (GWAS) using diverse germplasm accessions. Of these, GWAS allows the exploitation of historical linkage disequilibrium in diverse populations and the mining of QTL with relatively higher accuracy [15]. Previous studies for SPAD values in soybeans based on different genetic populations, experimental locations, and mapping strategies reported multiple putative loci on 16 different chromosomes [16]; however, research on QTL for soybean leaf chlorophyll-content using SPAD is still limited.
In this study, we aimed to use GWAS to identify loci for foliar chlorophyll as an indicator of N fixation potential [17] using SPAD values in a global panel of 186 diverse soybean accessions. Our findings may assist in elucidating the underlying molecular relation of leaf chlorophyll content and N fixation and create markers for developing high-yielding soybean varieties with minimum requirements in inorganic N.
Materials and Methods
Germplasm panel
A panel of 186 diverse soybean accessions was used in this study. Of these, 133 maturity group (MG) IV and 18 MG V represented the most genetically diverse accessions in previously published mapping studies, whereas 19 MG 00-III and 16 MG VI-X were selected from the USDAARS Germplasm Resources Information Network (GRIN)-Global, assessing the provided phenotypic information and breeding values. The accessions were originated from 21 different countries, including Argentina, Brazil, China, Colombia, Costa Rica, France, Georgia, India, Japan, Morocco, Nepal, South and North Korea, Russian Federation, Serbia, South Africa, Taiwan, Uganda, United States, Vietnam, and Zambia (Table S1).
kindly go through this link for Table S1 Reference
https://www.omicsonline.org/supplementary-files/genome-wide-association-study-of-SPAD-values-using-diverse-soybean-germplasm.pdf
Phenotypic data
Two greenhouse experiments were conducted at the Horticulture Research Center, Southern Illinois University, Carbondale, IL, which has a north-south orientation, from early September to late November in 2022 and 2023. The assays started with planting all accessions in sixinch plastic nursery pots filled with Berger BM1 growing medium. The plants were grown under ambient conditions with no supplemental lighting and watered based on the environmental needs (approx. every two days), using tap water. Both experiments were arranged as randomized complete block designs with two blocks and three repetitions per block (Figure 1). Chlorophyll content was measured using a portable chlorophyll meter (SPAD-502DL, Minolta, Tokyo, Japan). SPAD-502DL measures light transmittance of the leaf in the red and infrared wavelengths at 650 nm and 940 nm, generating a numerical output that indicates leaf greenness and chlorophyll content [11]. Assessing chlorophyll content with SPAD-502DL is a fast, costeffective, and straightforward approach to collecting phenotypic data for soybean leaf chlorophyll contents. SPAD values were collected from the top, middle, and bottom sites of each leaf at the vegetative (V1-V3) and reproductive (R1-R6) growth stages around 10: 00 am. Data analysis, including descriptive statistics, univariate distribution, analysis of variance, Tukey-Kramer post-hoc test, and hierarchical clustering, were performed using JMP Pro 17 (SAS Institute Inc., Cary, NC, USA).
Genotypic data
Two leaf disks were collected from each accession at the V1 growth stage and placed in two 96-well PCR plates. DNA extraction using the HotSHOT method and single nucleotide polymorphism (SNP) genotyping with PlexSeq™ were carried out by AgriPlex Genomics (Cleveland, OH, USA). After filtering markers with missing data, minor allele frequency lower than 5%, and heterozygosity higher than 10% [18,19], we used 1,243 high-quality SNPs for population structure and association analysis.
Population structure
STRUCTURE 2.3.4, which is a Bayesian model-based software, was used to generate population structure and create a Q-matrix for GWAS [20]. The burn-in iteration was 10,000, followed by 10,000 Markov chain Monte Carlo replications after burn-in using an admixture and allele frequencies correlated model. A hypothetical number of subpopulations (k) ranging from 1 to 10 was set to perform structure analysis. The statistical value delta K was calculated as described previously [21]. STRUCTURE HARVESTER was used to determine the optimal K [22]. Consequently, all 186 soybean accessions were assigned to a subpopulation based on the optimum k (k = 4), and the population structure matrix (Q) was generated for further association analysis.
Association analysis and candidate gene identification
Associations between phenotypic and genotypic data were identified using TASSEL 5.0 [23] based on the general linear model (GLM) jointly with population structure (Q) as a covariate. The results were visualized with quantile-quantile (Q-Q) and Manhattan plots. SNPs with-log10(P) (LOD) > 3 were considered significantly associated with SPAD values at the 5% level [24]. Flanking regions within 10 kb of each significant SNP were screened for discovering candidate genes using the Glyma.Wm82.a2 reference in SoyBase (https: //www. soybase.org) [25]. The Arabidopsis Information Resource (TAIR) was the primary source of information for gene description, followed by PANTHER and KEGG Orthology databases.
Results and Discussion
Phenotypic data
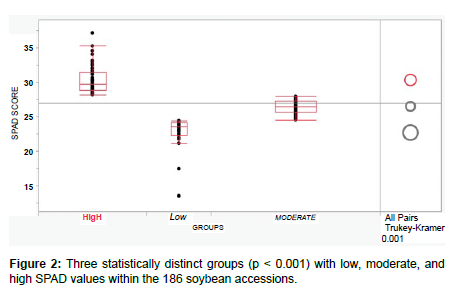
All 186 accessions from the global soybean panel were analyzed using data of SPAD values collected in 2022 and 2023 under greenhouse conditions. Descriptive statistics revealed that data were of normal distribution (mean: 27; median 27; mode 26.4; skewness -0.33; kurtosis 3.09) with a minimum value of 13.5 (PI548427, Wilson, China) and a maximum value of 37.1 (PI385942, Enrei, Japan). The accessions were classified into three statistically distinct groups (p < 0.001) based on their SPAD values (low, moderate, and high; Figure 2). The low group (22.6 ± 2.65) consisted of 35 accessions (18.8%), including the two non-nodulating PI573285 (D68-0099, USA) and PI642732 (Nitrasoy, USA); the moderate group (26.4 ± 0.95) consisted of 92 accessions (49.5%); and the high group (30.3 ± 2.12) consisted of 59 accessions (31.7%), including PI96171 (466, North Korea) and PI385942 (Enrei, Japan) that were previously reported to have no significant differences in terms of N fixation traits including the SPAD values from the super-nodulating line SS2-2 [17]. Our results were in accordance with previously published data that showed significant differences in chlorophyll content among nodulating and non-nodulating soybean accessions, especially at the reproductive (R1-R6) growth stages [26]. Photosynthesis and symbiotic N fixation are probably the most important metabolic processes in soybean growth and development [27]. Previous studies supported that N availability, either via fixation or inorganic fertilization, was positively correlated with the net leaf photosynthetic rate [28,29]. Thus, the collection of SPAD values appears to be a straightforward and economically efficient technique for phenotyping the status of biological N fixation in soybean.
Population structure
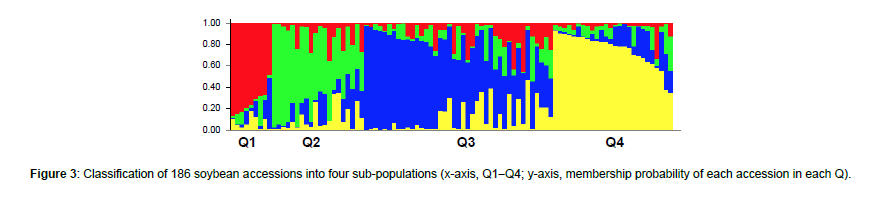
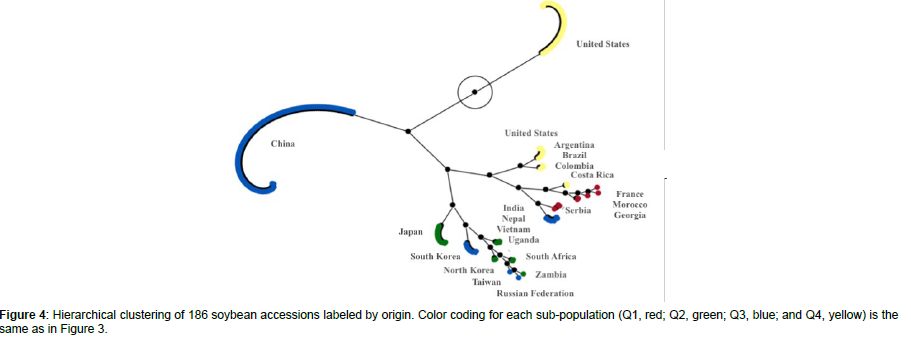
Using STRUCTURE analysis and STRUCTURE HARVESTER, the 186 accessions in the global panel were grouped into four subpopulations, Q1 (red), Q2 (green), Q3 (blue), and Q4 (yellow), since the maximal delta K value was identified at K = 4 (Figure 3). The results generated from STRUCTURE were in accordance with those yielded from hierarchical clustering using JMP Pro 17. Cluster I (red) was comprised of 12 accessions originated from France, Georgia, Morocco, and Serbia; Cluster II (green) was comprised of 18 accessions originated from Japan, North Korea, South Africa, Uganda, and Zambia; Cluster III (blue) was comprised of 108 accessions originated mainly from China as well as from India, Nepal, Russian Federation, South Korea, Taiwan, and Vietnam; and Cluster IV (yellow) was comprised of 48 accessions originated mainly from the United States as well as from Argentina, Brazil, Colombia, and Costa Rica (Figure 4).
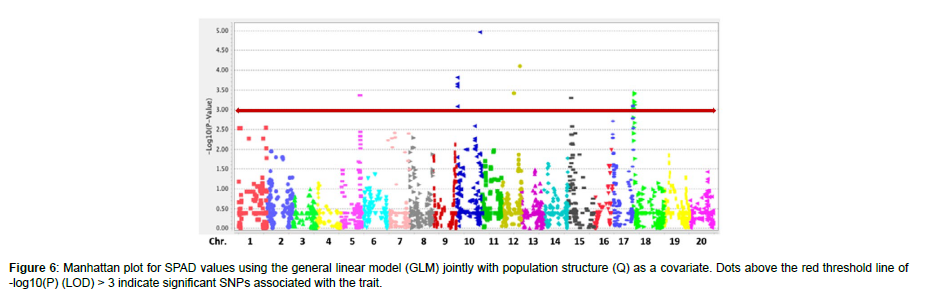
Association analysis
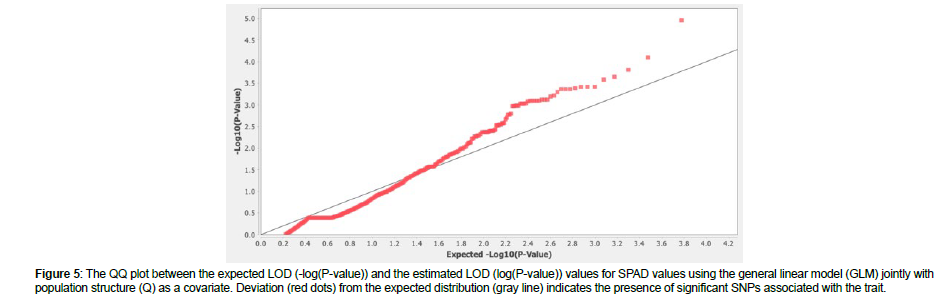
The QQ plot for SPAD values using the GLM + Q revealed deviation from the expected distribution and sufficient control of Type I and II errors, indicating the presence of significant SNPs associated with the trait (Figure 5). In total, 14 SNPs (p < 0.001, LOD > 3) were identified to be associated with SPAD values in the panel of 186 soybean accessions, according to (Figure 6). Of these, two SNPs (ss715607235 and ss715613440) displayed significant association at LOD > 4; three SNPs (ss715605997, ss715607272, and ss715608386) displayed significant association at LOD > 3.5; and the rest nine SNPs (ss715591991, ss715606900, ss715613242, ss715621452, ss715627640, ss715628823, ss715630330, ss715632280, and ss715631618) displayed LOD > 3 that was above the suggested threshold. Chromosome (Chr.) 10 contained five associations, including the one with the highest LOD, followed by Chr. 18 with four associations, Chr. 12 with two associations, and Chr. 5, 15, and 17 with one association each. A previous linkage map study on leaf chlorophyll content reported 59 SPAD QTLs distributing on Chr.1-18, excluding 12, in the advanced recombinant inbred line population Zhonghuang 24 × Huaxia 3 [30]. Besides, we found 24 published SPAD QTLs on Chr. 1, 2, 3, 6, 7, 9, 10, 11, 13, 17, 18, and 20 [31-34]. Compared with previous studies, our SPAD-associated regions were on the same chromosomes with various distances from the reported QTLs or alleles, except for the two associations on Chr. 12 that can be considered novel.
Figure 5: The QQ plot between the expected LOD (-log(P-value)) and the estimated LOD (log(P-value)) values for SPAD values using the general linear model (GLM) jointly with population structure (Q) as a covariate. Deviation (red dots) from the expected distribution (gray line) indicates the presence of significant SNPs associated with the trait.
Candidate genes
We found a total of 33 candidate genes from the Glyma.Wm82.a2 within 10 kb flanking regions of each significant SNP (Table 1). There were three on Chr. 5 (Glyma.05g216600, Glyma.05g216700, and Glyma.05g216800), 14 on Chr. 10 (Glyma.10g044300, Glyma.10g044400, Glyma.10g044500, Glyma.10g044600, Glyma.10g044700, Glyma.10g029700, Glyma.10g047800, Glyma.10g047900, Glyma.10g065000, Glyma.10g065100, Glyma.10g065200, Glyma.10g199100, Glyma.10g199200, and Glyma.10g199300), five on Chr. 12 (Glyma.12g079000, Glyma.12g078900, Glyma.12g091900, Glyma.12g092000, and Glyma.12g092100), three on Chr. 15 (Glyma.15g034100, Glyma.15g034200, and Glyma.15g034300), one on Chr. 17 (Glyma.17g239700), and seven on Chr. 18 (Glyma.18g018100, Glyma.18g041100, Glyma.18g066200, Glyma.18g066100, Glyma.18g000200, Glyma.18g000300, and Glyma.18g000400). Candidate gene models belonged to various metabolic and biosynthetic pathways. Of these, Glyma.12g092000 on Chr. 12 encoded the PS I subunit O, which is known to balance the Chl a/b ratio and the excitation pressure between PSI and PSII in Arabidopsis [35]. Glyma.10g199100 within the 10-kb flanking region of the SNP with the highest LOD on Chr. 10 encodes a leghemoglobin-related protein, a strong indicator of symbiotic nitrogen fixation [36]. These findings support the previously reported hypothesis that the chlorophyll content might be related to the nitrogen supply in soybean leaves [37,38]. Glyma.10g029700 encoded a serine/threonine-protein kinase, which was annotated to be associated with the chlorophyll catabolic process. A previous study showed that Arabidopsis plants overexpressing a serine/threonine-protein kinase gene from a wild soybean had higher chlorophyll contents under salt-stress conditions [39].
| CHR. | SNP MARKER | MAJOR ALLELE | MINOR ALLELE | GLM+Q LOD | GENOMIC LOCATION | GENE NAME | GENE ANNOTATION |
|---|---|---|---|---|---|---|---|
| 5 | ss715591991 | C | A | 3.41 | Intergenic | Glyma.05g216600 | Long-chain acyl-CoA synthetase |
| Glyma.05g216700 | Nucleoside diphosphatase kinase family protein | ||||||
| Glyma.05g216800 | Pleckstrin homology (PH) domain-containing protein | ||||||
| 10 | ss715606900 | C | T | 3.10 | Intergenic | Glyma.10g044300 | Arginine biosynthesis protein |
| Glyma.10g044400 | Unknown | ||||||
| Glyma.10g044500 | Unknown | ||||||
| Glyma.10g044600 | Unknown | ||||||
| Glyma.10g044700 | Pyrophosphorylase 4 | ||||||
| 10 | ss715605997 | G | A | 3.55 | Coding sequence | Glyma.10g029700 | Serine/Threonine-protein kinase |
| 10 | ss715607272 | G | A | 3.61 | Intergenic | Glyma.10g047800 | Ypt/Rab-GAP domain of gyp1p superfamily protein |
| Glyma.10g047900 | Protein kinase superfamily | ||||||
| 10 | ss715608386 | T | G | 3.80 | Coding sequence | Glyma.10g065000 | Pyruvate kinase |
| Glyma.10g065100 | Gamma-glutamyl kinase | ||||||
| Glyma.10g065200 | Leucine-rich repeat protein kinase family | ||||||
| 10 | ss715607235 | G | A | 4.95 | Intergenic | Glyma.10g199100 | Leghemoglobin-related |
| Glyma.10g199200 | Unknown | ||||||
| Glyma.10g199300 | Coatomer, α-subunit | ||||||
| 12 | ss715613242 | A | G | 3.46 | 3′ untranslated regions | Glyma.12g079000 | Transmembrane protein |
| Glyma.12g078900 | Nitrate transporter | ||||||
| 12 | ss715613440 | C | T | 4.18 | Intron | Glyma.12g091900 | Unknown |
| Glyma.12g092000 | Photosystem I subunit O | ||||||
| Glyma.12g092100 | Viral movement protein | ||||||
| 15 | ss715621452 | A | C | 3.35 | Coding sequence | Glyma.15g034100 | Acyltransferase |
| Glyma.15g034200 | RNA-binding family protein | ||||||
| Glyma.15g034300 | Origin recognition complex 1 | ||||||
| 17 | ss715627640 | T | C | 3.12 | Intergenic | Glyma.17g239700 | Subunit of serine palmitoyltransferase |
| 18 | ss715628823 | A | G | 3.11 | Intergenic | Glyma.18g018100 | Trehalose-phosphatase |
| 18 | ss715630330 | A | G | 3.21 | Intron | Glyma.18g041100 | Glutamine synthetase 1,4 |
| 18 | ss715632280 | T | G | 3.25 | Intergenic | Glyma.18g066200 | Nucleotide-sugar transporter |
| Glyma.18g066100 | Unknown | ||||||
| 18 | ss715631618 | G | A | 3.48 | Intron | Glyma.18g000200 | Ring/Ubox superfamily protein |
| Glyma.18g000300 | α/β-hydrolases family | ||||||
| Glyma.18g000400 | Calmodulin-binding family protein |
Table 1:Candidate gene models and descriptions within 10 kb flanking regions of SNPs significantly associated with SPAD values using Wm82.a2.v1.
Conclusion
In the present study, we created a global panel of 186 soybean accessions to collect SPAD value under greenhouse conditions and genotype them with SNP markers. Population structure was assessed and revealed the existence of four subpopulations. GWAS using the GLM + Q model allowed the detection of 14 SNPs significantly related to SPAD values. We also found 33 candidate genes, three of which encoded proteins important for photosynthesis, chlorophyll content, and nitrogen status. Overall, our data provide helpful information for further elucidating the underlying mechanism connecting chlorophyll content and nitrogen status in soybeans and favorable alleles for improving photosynthesis rates and biological N fixation ability.
Acknowledgments
We thank United Soybean Board for fully funding the project.
References
- Cassman KG, Dobermann A (2022) Nitrogen and the future of agriculture: 20 years on: This article belongs to Ambio's 50th Anniversary Collection. Theme: Solutions-oriented research. Ambio 51: 17-24.
- Good AG, Beatty PH (2011) Fertilizing nature: a tragedy of excess in the commons. PLoS Biol 9: e1001124.
- Liu L, Zheng X, Wei X, Kai Z, Xu Y, et al. (2021) Excessive application of chemical fertilizer and organophosphorus pesticides induced total phosphorus loss from planting causing surface water eutrophication. Sci Rep 11: 23015.
- Waqas M, Hawkesford MJ, Geilfus C-M (2023) Feeding the world sustainably: efficient nitrogen use. Trends Plant Sci 28: 505-508.
- Oldroy GE, Murray JD, Poole PS, Downie JA (2011) The rules of engagement in the legume rhizobial symbiosis. Annu Rev Genet 45: 119-144.
- Suzuki Y, Ohsaki K, Takahashi Y, Wada S, Miyake C, et al. (2023) Behavior of photosystems II and I is modulated depending on N partitioning to Rubisco in mature leaves acclimated to low N levels and senescent leaves in rice. Plant Cell Physiol 64: 55-63.
- Li Y, Ren B, Ding L, Shen Q, Peng S, et al. (2013) Does chloroplast size influence photosynthetic nitrogen use. PLoS One 8: e62036.
- Dong N, Prentice IC, Wright IJ, Wang H, Atkin OK, et al. (2022) Leaf nitrogen from the perspective of optimal plant function. J Ecol 110: 2585-2602.
- Luo X, Keenan TF, Chen JM, Croft H, Prentice IC, et al. (2021) Global variation in the fraction of leaf nitrogen allocated to photosynthesis. Nat Commun 12: 4866.
- Wood NJ, Baker A, Quinnel RJ, Camargo-Valero MA (2020) A simple and non-destructive method for chlorophyll quantification of Chlamydomonas cultures using digital image analysis. Front Bioeng Biotechnol 8: 746.
- Uddling J, Gelang-Alfredsson J, Piikki K, Pleijel H (2007) Evaluating the relationship between leaf chlorophyll concentration and SPAD-502 chlorophyll meter readings. Photosynth Res 91: 37-46.
- Arregui LM, Lasa B, Lafarga A, Iraneta I, Baroja E, et al. (2006) Evaluation of chlorophyll meters as tools for N fertilization in winter wheat under humid Mediterranean conditions. Eur J Agron 24: 140-148.
- Ziadi N, Brassard M, Belanger G, Claessens A, Tremblay N, et al. (2008) Chlorophyll measurements and nitrogen values for the evaluation of corn nitrogen status. Agron J 100: 1264-1273.
- Yuan Z, Ata-UI-Karim ST, Cao Q, Lu Z, Cao W, et al. (2016) Indicators for diagnosing nitrogen status of rice based on chlorophyll meter readings. Field Crop Res 185: 12-20.
- Yu J, Buckler ES (2006) Genetic association mapping and genome organization of maize. Curr Opin Biotechol 17: 155-160.
- Dhanapal AP, Ray JD, Singh, SK, Hoyos-Villegas A, Smith JR et al. (2016) Genome-wide association mapping of soybean chlorophyll traits based on canopy spectral reflectance and leaf extracts. BMC Plant Biol 16: 174.
- Hamawaki RL, Kantartzi SK (2018) Di-nitrogen fixation at the early and late growth stages of soybean. Acta Sci Agron 40: 36372.
- Akond M, Liu S, Schoener L, Anderson JA, Kantartzi SK, et al. (2013) SNP-Based genetic linkage map of soybean using the SoySNP6K Illumina Infinium BeadChip genotyping array. J Plant Genom Sci 1: 80-89.
- Song Q, Hyten DL, Jia G, Quigley CV, Fickus EW, et al. (2015) Fingerprinting soybean germplasm and its utility in genomic research. G3-Genes Genom Genet 5:1999-2006.
- Porras-Hurtado L, Ruiz Y, Santos C, Phillips C, Carracedo A, et al. (2013) An overview of STRUCTURE: applications, parameter settings, and supporting software. Front Genet 4: 98.
- Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611-2620.
- Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4: 359-361.
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, et al. (2007) TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633-2635.
- Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138: 963-971.
- Song Q, Yan L, Quigley C, Fickus E, Wei H, et al. (2020) Soybean BARCSoySNP6K: An assay for soybean genetics and breeding research. Plant J104: 800-811.
- Zhou XJ, Liang Y, Chen H, Shen SH, Jing YX, et al. (2006) Effects of rhizobia inoculation and nitrogen fertilization on photosynthetic physiology of soybean. Photosynthetica 44: 530-535.
- Vollmann J, Walter H, Sato T, Schweiger P (2011) Digital image analysis and chlorophyll metering for phenotyping the effects of nodulation in soybean. Comput Electron Agric 75: 190-195.
- Caetano-Anolles G (1997) Molecular dissection and improvement of the nodule symbiosis in legumes. Field Crops Res 53: 47-68.
- Wu J, Wang D, Rosen CJ, Bauer ME (2007) Comparison of petiole nitrate concentrations, SPAD chlorophyll readings, and QuickBird satellite imagery in detecting nitrogen status of potato canopies. Field Crops Res 101: 96-103.
- Wang L, Conteh B, Fang L, Xia Q, Nian H, et al. (2020) QTL mapping for soybean (Glycine max L.) leaf chlorophyll-content traits in a genotyped RIL population by using RAD-seq based high-density linkage map. BMC Genom 21: 739.
- Fang C, Ma YM, Wu SW, Liu Z, Wang Z, et al. (2017) Genome-wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genom Biol 18:161.
- Hu ZB, Zhang HR, Kan GZ, Ma DY, Zhang D, et al. (2013) Determination of the genetic architecture of seed size and shape via linkage and association analysis in soybean (Glycine max L. Merr.). Genetica 141: 247-254.
- Li GJ, Li HN, Cheng LG, Zhang YM (2010) QTL analysis for dynamic expression of chlorophyll content in soybean (Glycine max L. Merr.). Acta Agron Sin 36: 242-248.
- Shi XL, Yan L, Yang CY, Yan WW, Moseley DO, (2018) Identification of a major quantitative trait locus underlying salt tolerance in ‘Jidou 12’ soybean cultivar. BMC Res Notes 11: 95.
- Jensen PE, Haldrup A, Zhang S, Scheller HV (2004) The PSI-O subunit of plant photosystem I is involved in balancing the excitation pressure between the two photosystems. J Biol Chem 279: 24212-24217.
- Ott T, vanDongen JT, Gu C, Krusell L, Desbrossess G, et al. (2005) Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr Biol 15: 531-535.
- Fritschi FB, Ray JD (2007) Soybean leaf nitrogen, chlorophyll content, and chlorophyll a/b ratio. Photosynthetica 45:92-98.
- Kaler AS, Abdel-Haleem H, Fritschi FB, Gillman JD, Ray JD et al. (2020) Genome-wide association mapping of dark green color index using a diverse panel of soybean accessions. Sci Rep 10: 5166.
- Sun XL, Yu QY, Tang LL, Ji W, Bai X, et al. (2013) GsSRK, a G-type lectin S-receptor-like serine/threonine protein kinase, is a positive regulator of plant tolerance to salt stress. J Plant Physiol 170:505-515.
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Indexed at, Google Scholar, Crossref
Citation: Bhandari R, Hagen W, Kantartzi SK (2024) Genome-wide AssociationStudy of SPAD Values Using Diverse Soybean Germplasm. J Plant Genet Breed8: 187. DOI: 10.4172/jpgb.1000187
Copyright: © 2024 Bhandari R, et al. This is an open-access article distributedunder the terms of the Creative Commons Attribution License, which permitsunrestricted use, distribution, and reproduction in any medium, provided theoriginal author and source are credited.
Share This Article
Open Access Journals
Article Tools
Article Usage
- Total views: 2182
- [From(publication date): 0-2024 - Apr 05, 2025]
- Breakdown by view type
- HTML page views: 1948
- PDF downloads: 234