Research Article Open Access
Genomewide Array Comparative Genomic Hybridization in 55 JapaneseNormokaryotypic Patients with Non-Syndromic Intellectual Disability
Miki Asahina1*, Yusaku Endoh2, Tomoko Matsubayashi1, Koichi Hirano2, Tokiko Fukuda1 and Tsutomu Ogata1
1Department of Pediatrics, Hamamatsu University School of Medicine, Hamamatsu, Japan
2Department of Pediatrics, Hamamatsu City Welfare and Medical Center for Development, Hamamatsu, Japan
- *Corresponding Author:
- Miki Asahina
Department of Pediatrics
Hamamatsu University School of Medicine,
Hamamatsu 431-3192, Japan
Tel: +81-53-435-2310
Fax: +81-53-435-2310
E-mail: miki.a@hama-med.ac.jp
Received date: October 05, 2016; Accepted date: October 28, 2016; Published date: November 07, 2016
Citation: Asahina M, Endoh Y, Matsubayashi T, HiranoK, Fukuda T, et al. (2016) Genomewide Array Comparative Genomic Hybridization in 55 Japanese Normokaryotypic Patients with Non-Syndromic Intellectual Disability. J Pediatr Neurol Disord 2: 108. doi: 10.4172/2572-5203.1000108
Copyright: © 2016 Asahina M, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Pediatric Neurological Disorders
Abstract
Background: Genomewide array comparative genomic hybridization (aCGH) has widely been utilized as the diagnostic tool in patients with non-syndromic intellectual disability (ID). Indeed, aCGH has identified pathogenic copy number variants (pCNVs), as well as variants of uncertain clinical significance (VsUS) and benign CNVs (bCNVs), in such patients. Aims: To examine the frequencies of various CNVs and clinical findings in patients with non-syndromic ID. Patients and methods: We studied 55 Japanese normokaryotypic patients (35 males, 20 females) with apparently non-syndromic ID. Genomewide aCGH was performed using leukocyte genomic DNA samples. Clinical findings were compared among patients with pCNVs (group 1), those with VsUS (group 2), and those with bCNVs or no CNVs (group 3). Results: Nine patients had pCNVs: one had 5p deletion syndrome, two had 22q11.2 deletion syndrome, one had 17q23.1q23.2 microdeletion syndrome, three had CNVs involving known pathogenic genes, and the remaining two had CNVs overlapping with previously described CNVs in patients with ID (one with duplication at 1q36 and the other with deletion at 12q42). Furthermore, 11 patients had VsUS, and nine patients had bCNVs. Clinical findings were grossly comparable among groups 1-3. Conclusions: The results provide further support for the usefulness of aCGH in the identification of underlying genetic factor(s) for ID, although there was no clinical finding indicative of the presence of pCNVs or VsUS. Furthermore, our data are expected to serve to identify pathogenic genes on chromosomes 1q36 and 12q42, as well as those on several VsUS.
Keywords
Array comparative genomic hybridization; Intellectual disability; Copy number variants; Pathogenic gene; Clinical finding
Introduction
Intellectual disability (ID) is a highly heterogeneous condition occurring in 1–3% of the general population [1]. ID is divided into syndromic and non-syndromic forms, with the former accounting for roughly one-third of affected patients [2]. The syndromic form is associated with a constellation of clinical features characteristic of known syndromes, and is usually caused by specific genetic factors such as mutations of causative genes, copy-number variants (CNVs) involving relevant genes, and aneuploidies [3]. By contrast, nonsyndromic form, though it may be accompanied by non-specific multiple congenital anomalies (MCA), is free from diagnostic clinical manifestations, and is usually subject to multiple (epi) genetic and environmental factors such as mutations of genes for non-specific ID, various usually non-recurrent CNVs, central nervous infections, and environmental chemicals [4].
Recently, genomewide array comparative genomic hybridization (aCGH) has widely been utilized as the diagnostic tool in patients with non-syndromic ID. Indeed, aCGH has identified pathogenic copy number variants (pCNVs) in 5-35% of such patients (average 12.2%) [5]. In addition, aCGH has also detected multiple variants of uncertain clinical significance (VsUS) that could be relevant to ID, and benign CNVs (bCNVs) that are irrelevant to ID [6].
Here, we examined the frequencies of various CNVs and clinical finding in patients with apparently non-syndromic ID.
Patients and Methods
Patients
This study consisted of 55 Japanese patients (35 males, 20 females) with apparently non-syndromic ID with or without non-specific MCA. The ages at examination ranged from 0.8 to 42 years (median, 4.0 years). All patients had normal karyotype in the 50 lymphocytes examined by the conventional 400–550 level G-banding analysis. The ID was assessed as extremely severe (developmental quotient (DQ)/ intelligence quotient (IQ), < 20) in 21 patients, severe (DQ/IQ, 21-34) in 11 patients, moderate (DQ/IQ, 35-49) in six patients, and mild (DQ/ IQ, 50-–69) in 17 patients, by the DSM-IV method [7].
Ethical approval and samples
This study was approved by the Institutional Review Board Committees of Hamamatsu University School of Medicine, and was performed after obtaining written informed consent for the molecular analysis and the publication of genetic and clinical data after removing information for personal identification.
Genomewide aCGH analysis: Genomewide aCGH was performed with a catalog human array (4×180K format, ID G4449A) (Agilent Technologies) using leukocyte genomic DNA samples of all the patients, the parents who agreed to genetic analysis, and sex-matched control subjects. The procedure was as described in the manufacturer’s instructions. For autosomes and female X chromosomes, log2 signal ratios of around –1.0 and around + 0.5 were regarded as indicative of heterozygous deletions and duplications, respectively. For male sex chromosomes that appear in a heterogametic condition, log2 signal ratios of – ∞ and around + 1.0 were interpreted as hemizygous deletions and duplications, respectively. When ≥three consecutive probes showed abnormal log2 ratios, the corresponding region was regarded as CNVs. Minimum and maximum sizes of CNVs were obtained as the regions between two distal ends of signals indicative of deletions or duplications and those between two proximal ends of signals indicative of normal copy numbers. The genomic position was based on human GRCh37/hg19 (http://genome.ucsc.edu/).
CNVs were regarded as pCNVs, when they were identical to those of established ID-positive syndromes with causative or candidate genes, they included known pathogenic genes in which intragenic mutations or CNVs involving the genes alone have been identified in patients with ID, or they shared an overlapping region with previously described plural CNVs in patients with ID [1-3]. By contrast, CNVs were interpreted as bCNVs, when (1) they were inherited from either of the healthy parents, or (2) they have been registered as normal variants in the public databases such as Database of Genomic Variants (http:// dgv.tcag.ca/dgv/app/home) and ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/) [1,2]. Other CNVs were regarded as VsUS, most of which contained candidate genes for ID or were formed as de novo events.
Clinical assessment: Multiple clinical features were examined in all the patients by two clinicians (M.A. and Y.E.), except for ophthalmologic, cardiac, and renal features which were evaluated by professional doctors in each field. We summarized clinical findings in patients with pCNVs (group 1), those with VsUS (group 2), and those with bCNVs or no CNVs (group 3), and compared them among different groups. Statistical significance of the median was examined by the Mann-Whitney′s U-test, and that of the frequency by the Fisher’s exact test. P < 0.05 was considered significant.
Results
Genomewide aCGH analysis
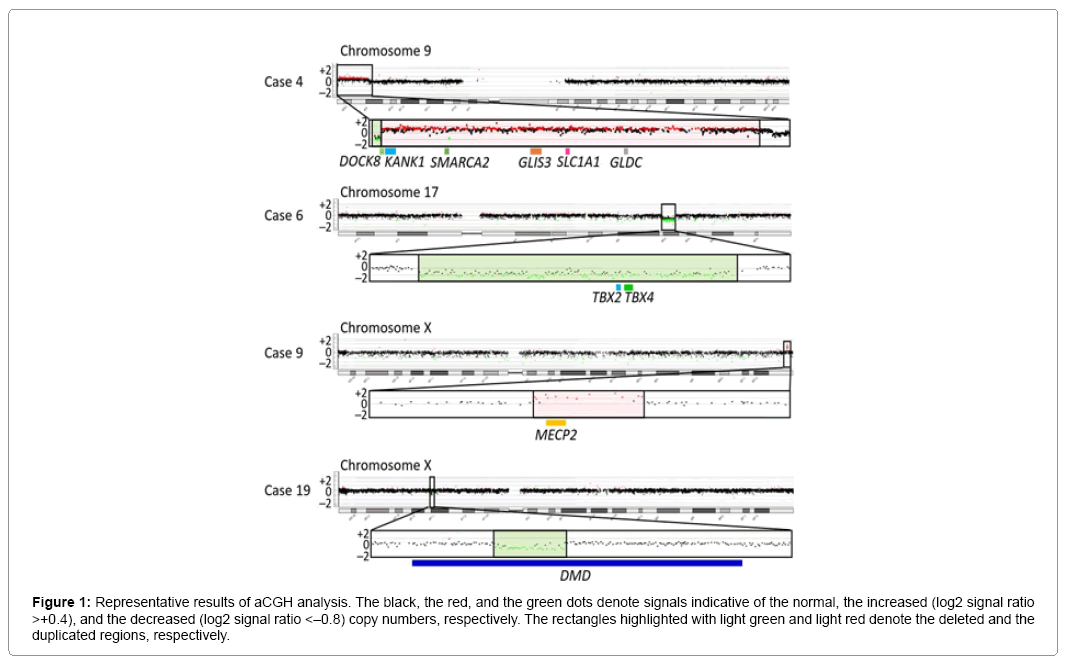
Representative aCGH findings are shown in Figure 1. The data of groups 1 and 2 are summarized in Table 1, and the pathogenic and candidate genes on the identified pCNVs and VsUS are shown in Supplementary Table 1. CNVs were identified in 29 of the 55 patients. Of the 29 patients, nine patients (cases 1–9) were assessed to have pCNVs, because: (1) case 2, and cases 7 and 8, had deletions for 5p deletion syndrome (Cri Du Chat syndrome) and 22q11.2 deletion syndrome (Di George syndrome) with known pathogenic genes, respectively; (2) case 6 had a deletion typical of 17q23.1q23.2 microdeletion syndrome with candidate genes; (3) cases 3, 4, and 9 had CNVs involving known pathogenic genes, as well as candidate genes; and (4) cases 1 and 5 had CNVs partially overlapping with previously described CNVs in patients with ID (Figures 2 and 3) [8-10]. Furthermore, of case 1-9, cases 1–3, 5, 6, and 9 had de novo CNVs. By contrast, nine of the 22 patients were evaluated to have bCNVs, because they were present in either of the healthy parents. The remaining 11 patients (cases 10-20) were assessed to have VsUS with or without candidate genes. Of cases 10-20, cases 10, 14, 17, and 18 had de novo CNVs. Notably, cases 4, 10, and 12-14 had plural CNVs. In particular, the co-existing two VsUS in cases 10 and those in case 14 were found to be generated as de novo CNVs, although parental samples were not available in cases 4, 12, and 13.
Figure 1: Representative results of aCGH analysis. The black, the red, and the green dots denote signals indicative of the normal, the increased (log2 signal ratio >+0.4), and the decreased (log2 signal ratio <–0.8) copy numbers, respectively. The rectangles highlighted with light green and light red denote the deleted and the duplicated regions, respectively.
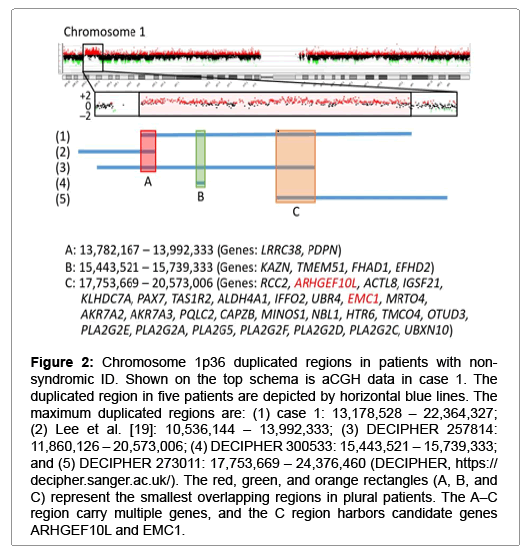
Figure 2: Chromosome 1p36 duplicated regions in patients with nonsyndromic ID. Shown on the top schema is aCGH data in case 1. The duplicated region in five patients are depicted by horizontal blue lines. The maximum duplicated regions are: (1) case 1: 13,178,528 – 22,364,327; (2) Lee et al. [19]: 10,536,144 – 13,992,333; (3) DECIPHER 257814: 11,860,126 – 20,573,006; (4) DECIPHER 300533: 15,443,521 – 15,739,333; and (5) DECIPHER 273011: 17,753,669 – 24,376,460 (DECIPHER, https:// decipher.sanger.ac.uk/). The red, green, and orange rectangles (A, B, and C) represent the smallest overlapping regions in plural patients. The A–C region carry multiple genes, and the C region harbors candidate genes ARHGEF10L and EMC1.
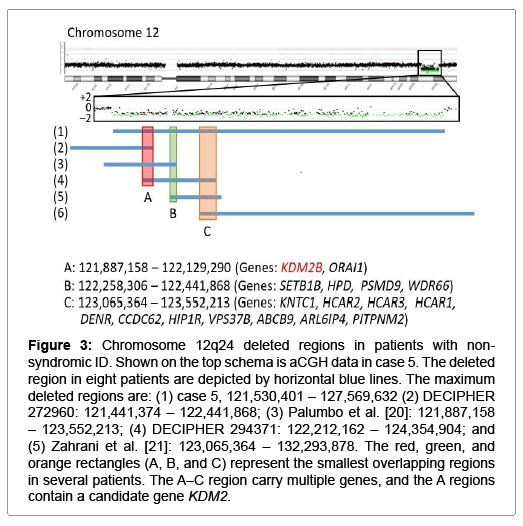
Figure 3: Chromosome 12q24 deleted regions in patients with nonsyndromic ID. Shown on the top schema is aCGH data in case 5. The deleted region in eight patients are depicted by horizontal blue lines. The maximum deleted regions are: (1) case 5, 121,530,401 – 127,569,632 (2) DECIPHER 272960: 121,441,374 – 122,441,868; (3) Palumbo et al. [20]: 121,887,158 – 123,552,213; (4) DECIPHER 294371: 122,212,162 – 124,354,904; and (5) Zahrani et al. [21]: 123,065,364 – 132,293,878. The red, green, and orange rectangles (A, B, and C) represent the smallest overlapping regions in several patients. The A–C region carry multiple genes, and the A regions contain a candidate gene KDM2.
| Case | Age (y) |
Sex | Chromosomal location |
CNV | Inheritance | Start (max) * Start (min) * |
End (max) * End (min) * |
Size (max) * Size (min) * |
Protein coding genes | Pathogenic gene(s) for ID | Candidate gene(s) for ID | Clinical features | ID (DQ/IQ) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <Patients with pathologic copy number variations (pCNVs)> | |||||||||||||
| 1 | 18 | F | 1p36.21-p36.12 | Duplication | De novo | 13,178,528 13,791,307 |
22,364,327 22,352,713 |
9,185,799 8,561,406 |
65 | – | ARHGEF10L, EMC1 | Epilepsy, Pulmonary artery stenosis, Syndactyly, Congenital dislocation of the hip joint, Hyperphosphatasia ¶, Strabismus, Amenorrhea | Most severe |
| 2 | 5 | F | 5p15.33-p15.2 † | Deletion | De novo | 1 26,142 |
13,897,399 13,882,896 |
13,897,398 13,856,755 |
40 | CTNND2 TERT |
– | Sleep disorder, Bushy eyebrow, Astigmatism | Moderate (40) |
| 3 | 4 | F | 5q14.3 | Duplication | De novo | 87,556,055 87,574,384 |
88,110,678 88,100,645 |
554,623 526,261 |
3 | MEF2C | – | Motor delay, Microcephaly, Choreoathetosis, Hyperopia, Sleep disorder, Eating disorder | Most severe |
| 4 | 2 | M | 9p24.2 | Deletion | Unknown | 1 20,4193 |
373,868 356,138 |
373,867 151,945 |
4 | DOCK8 | – | Short stature, Macrocephaly, Deafness, Hypertelorism, Epicanthus, Low-set ears, Sleep disorder, Eating disorder | Mild (61) |
| 9p23-p24.2 | Duplication | Unknown | 356,138 373,868 |
9,781,684 9,766,294 |
9,425,546 9,392,426 |
31 | – | KANK1, SLC1A1 SMARCA2 |
|||||
| Xp 22.2 | Duplication | Unknown | 16,997,317 16,997,258 |
17,733,918 17,725,454 |
736,601 728,196 |
2 | – | NHS | |||||
| ãÂ?Â? | ãÂ?Â? | ãÂ?Â? | Xp 21.3 | Duplication | Unknown | 28,817,193 28,829,765 |
28,919,846 28,902,121 |
102,653 72,356 |
1 | IL1RAPL1 # (Intragenic duplication) | – | ãÂ?Â? | |
| 5 | 6 | F | 12q24.31-q24.32 | Deletion | De novo | 121,496,723 121,530,401 |
127,569,632 127,537,641 |
6,072,909 6,007,240 |
49 | – | KDM2B | Motor delay, Short stature, Epilepsy, Tetralogy of Fallot | Most severe |
| 6 | 2 | M | 17q23.1-q23.2 ‡ | Deletion | De novo | 58,066,851 58,120,809 |
60,316,690 60,251,568 |
2,249,839 2,130,759 |
12 | – | TBX2, TBX4 | Motor delay, Short stature, Deafness, Camptodactyly, Congenital aural atresia, Cryptorchidism, Sleep disorder | Mild (60) |
| 7 | 4 | M | 22q11.21§ | Deletion | Unknown | 18,651,673 18,894,835 |
20,719,112 20,311,763 |
2,067,439 1,416,928 |
28 | TBX1 | – | Motor delay, Epicanthus, Low-set ears, Sleep disorder | Mild (56) |
| 8 | 6 | M | 22q11.21§ | Deletion | Unknown | 18,651,673 18,894,835 |
20,719,112 20,311,763 |
2,067,439 1,416,928 |
28 | TBX1 | – | Motor delay, Hypertelorism, Epicanthus, Low-set ears | Mild (56) |
| 9 | 4 | M | Xq28 | Duplication | De novo | 153,190,720 153,197,498 |
153,630,137 153,609,163 |
439,417 411,665 |
13 | MECP2 | – | Motor delay, Autism, Sleep disorder | Severe (27) |
| <Patients with variants of uncertain clinical significance (VsUS)> | |||||||||||||
| 10 | 7 | M | 3q29 | Duplication | De novo | 193,201,737 193,220,394 |
193,353,202 193,345,836 |
151,465 125,442 |
2 | – | ATP13A4, OPA1 | Motor delay, Short stature, Low-set ears, Macrocephaly | Severe (30) |
| ãÂ?Â? | ãÂ?Â? | ãÂ?Â? | 21q21.1 | Duplication | De novo | 17,196,538 17,205,639 |
17,385,656 17,360,796 |
189,118 155,157 |
1 | – | – | ãÂ?Â? | |
| 11 | 39 | M | 5q23.3-q31.1 | Duplication | Unknown | 130,594,844 130,616,397 |
130,740,811 130,728,922 |
145,967 112,525 |
1 | – | CDC42SE2 | Motor delay, Epilepsy, Hypertelorism, Epicanthus, Bushy eyebrow, Down-slanted palpebral Fissure, Wide mouth, Extrapyramidal symptoms | Most severe |
| 12 | 42 | M | 5q31.1 | Duplication | Unknown | 130,405,344 130,476,155 |
130,808,141 130,797,583 |
402,797 321,428 |
1 | – | CDC42SE2 | Motor delay, Epilepsy, Hypertelorism, Epicanthus, Bushy eyebrow, Down-slanted palpebral Fissure, Wide mouth, Extrapyramidal symptoms | Most severe |
| ãÂ?Â? | ãÂ?Â? | ãÂ?Â? | 9p23 | Duplication | Unknown | 9,679,865 9,762,182 |
9,972,017 9,957,044 |
292,152 194,862 |
1 | – | PTPRD | ãÂ?Â? | |
| 13 | 1.5 | F | 6q25.1 | Duplication | Unknown | 150,244,525 150,263,131 |
150,294,629 150,281,545 |
50,104 18,414 |
1 | – | – | Motor delay, High arched palate, Short stature, Strabismus, Eating disorder, Sleep disorder | Mild (50) |
| ãÂ?Â? | ãÂ?Â? | ãÂ?Â? | 20q11.21 | Deletion | Unknown | 29,842,845 29,857,590 |
30,167,165 30,157,286 |
324,320 299,696 |
8 | – | – | ãÂ?Â? | |
| 14 | 1.8 | M | 8p23.3 | Duplication | De novo | 1,507,333 1,522,999 |
1,833,763 1,815,984 |
326,430 292,985 |
3 | – | CLN8, ARHGEF10 | Microcephaly, Highly arched eyebrow, Short fingers, Macrotia, Eating disorderãÂ?Â? | Mild |
| ãÂ?Â? | ãÂ?Â? | ãÂ?Â? | 8p23.2 | Duplication | De novo | 2,701,555 2,725,834 |
3,006,205 2,993,737 |
304,650 267,903 |
1 | – | CSMD1 | ãÂ?Â? | |
| 15 | 15 | M | 18q12.2 | Duplication | Unknown | 34,439,213 34,447,971 |
34,685,037 34,662,565 |
245,824 214,594 |
1 | – | KIAA1328 | Motor delay, Scoliosis, Dandy-walker variants, Hypercholesterolemia | Most severe |
| 16 | 3 | M | 14q23.1 | Deletion | Unknown | 61,503,826 61,518,515 |
61,543,918 61,536,213 |
40,092 17,698 |
1 | – | SLC38A6 | Short stature, Autism, Ventricular septal defect, Blepharophimosis | Moderate (42) |
| 17 | 2 | F | 20q11.21 | Deletion | De novo | 29,638,422 29,652,452 |
30,212,322 30,193,658 |
573,900 541,206 |
8 | – | – | Epicanthus, Blepharophimosis, Protruding ear, Brachydactyly of the hand, Strabismus | Mild |
| 18 | 1 | M | 21q22.11 | Deletion | De novo | 34,911,504 34,925,956 |
35,109,693 35,103,544 |
198,189 177,588 |
4 | – | ITSN1 | Motor delay, Epilepsy, Down-slanted palpebral Fissure, Right macrotia, Plagiocephaly, Heterotopic gray matter, Eating disorder | Most severe |
| 19 | 16 | F | Xp21.1 | Deletion | Unknown | 31,575,837 31,608,431 |
31,980,038 31,979,979 |
404,201 371,548 |
1 | – | DMD (Intragenic deletion) | Motor delay, Scoliosis | Moderate |
| 20 | 3 | M | Xq25 | Deletion | Unknown | 127,083,788 127,144,318 |
127,232,612 127,222,241 |
148,824 77,923 |
1 | – | ACTRT1 | Motor delay, Epilepsy, Hypertelorism, Blepharoptosis, Cryptotia, High-arched palate, Microcephaly, Overlapping finger, Short stature, Inguinal hernia, Cryptorchidism, Microphthalmia, Eating disorder | Most severe |
According to GRCh37/hg19.
†The 5p deletion syndrome (Cri Du Chat syndrome).
‡The 17q23.1q23.2 microdeletion syndrome.
§The 22q11.2 deletion syndrome (Di Gorge syndrome).
¶Hyperphosphatasia may be due to duplication of ALPL.
#This intragenic duplication is predicted to have a loss-of-function effect.
Abbreviations: y, years, F, female; M, male; CNV, copy number variant; ID, intellectual disability; DQ, developmental quotient; and IQ, intelligence quotient.
Table 1: Summary of array CGH data and clinical findings in 20 patients with pCNVs or VsUS.
Clinical assessment
Clinical features in patients of groups 1 and 2 are described in Table 1. Clinical features were highly variable with no pathognomonic features. Indeed, while case 2 had 5p deletion for Cri Du Chat syndrome, she showed no characteristic mewing cry. Similarly, while case 7 and 8 had 22q11.2 deletion for Di George syndrome, they were free from cardiovascular anomalies, abnormal calcium metabolism, and immune deficiency. In addition, while case 6 had a deletion typical of 17q23.1q23.2 microdeletion syndrome, his overall clinical features remained rather non-specific.
Detailed clinical findings in groups 1-3 are summarized in Table 2. Male-dominant sex ratio was common to groups 1-3 as well as total patients, and the examined age and the degree of ID were similar among groups 1-3. Furthermore, the frequencies of clinical features were similar among groups 1-3, except for significantly high frequency of eating disorder in group 2 and that of sleep disorder in group 1.
| Total | pCNV | VOUS | bCNV and no CNV | |
|---|---|---|---|---|
| Group 1 | Group 2 | Group 3 | ||
| (n=55) | (n=9) | (n=11) | (n=35) | |
| Male | 35 | 5 | 8 | 22 |
| Female | 20 | 4 | 3 | 13 |
| Examined age (median [range]) (years) | 4.0 [0.8–18] | 4.0 [1.9–18] | 3.0 [1.0–42] | 4.0 [0.8–30] |
| Paternal age at birth (median [range]) (years) | 33 [21–42] | 33 [24–40] | 32 [21–42] | 32.5 [23–42] |
| Maternal age at birth (median [range]) (years) | 30 [21–44] | 31 [25–37] | 28 [21–33] | 30 [21–44] |
| Assisted reproductive technology | 5/55 | 0/9 | 2/11 | 3/35 |
| Premature birth | 8/55 | 0/9 | 3/11 | 5/35 |
| Small for gestational age | 8/55 | 0/9 | 1/11 | 7/35 |
| Short stature | 24/55 | 3/9 | 5/11 | 16/35 |
| Intellectual disability | 55/55 | 9/9 | 11/11 | 35/35 |
| Most severe (DQ/IQ <20) | 21/55 | 4/9 | 3/11 | 14/35 |
| Severe (DQ/IQ, 21–34) | 11/55 | 1/9 | 2/11 | 8/35 |
| Moderate (DQ/IQ, 35–49) | 6/55 | 1/9 | 1/11 | 4/35 |
| Mild (DQ/IQ, 50–69) | 17/55 | 3/9 | 5/11 | 9/35 |
| Speech delay | 51/55 | 8/9 | 10/11 | 33/35 |
| No single word | 31/55 | 4/9 | 5/11 | 22/35 |
| Motor delay | 42/55 | 6/9 | 8/11 | 28/35 |
| Unable to walk without support | 19/55 | 2/9 | 6/11 | 11/35 |
| Hypotonia | 29/55 | 5/9 | 5/11 | 19/35 |
| Nuerodevelopmental disorders | 10/55 | 1/9 | 1/11 | 8/35 |
| Autism spectrum disorder | 10/55 | 1/9 | 1/11 | 8/35 |
| Attention-deficit/hyperactivity disorder | 1/55 | 0/9 | 0/11 | 1/35 |
| Eating disorders | 12/55 | 2/9 | 5/11* | 5/35* |
| Sleep disorders | 16/55 | 6/9† | 1/11 | 9/35† |
| Magnetic resonance imaging abnormalities | 19/55 | 1/9 | 6/11 | 12/35 |
| Epilepsy | 25/55 | 2/9 | 4/11 | 19/35 |
| Craniofacial features | 33/55 | 7/9 | 8/11 | 18/35 |
| Macrocephaly | 5/55 | 1/9 | 1/11 | 3/35 |
| Microcephaly | 9/55 | 1/9 | 2/11 | 6/35 |
| Hypertelorism | 7/55 | 2/9 | 3/11 | 2/35 |
| Strabismus | 13/55 | 2/9 | 2/11 | 9/35 |
| Microphthalmus | 2/55 | 0/9 | 1/11 | 1/35 |
| Palpebral fissure deformity | 17/55 | 3/9 | 6/11 | 8/35 |
| Eyebrow deformity | 6/55 | 1/9 | 3/11 | 2/35 |
| Ear deformity | 16/55 | 4/9 | 5/11 | 7/35 |
| Deafness | 6/55 | 2/9 | 0/11 | 4/35 |
| Cleft palate | 3/55 | 0/9 | 0/11 | 3/35 |
| High-arched palate | 6/55 | 0/9 | 2/11 | 4/35 |
| Abnormal teeth | 3/55 | 0/9 | 1/11 | 2/35 |
| Congenital heart defect | 8/55 | 2/9 | 1/11 | 5/35 |
| Congenital renal anomaly | 1/55 | 0/9 | 0/11 | 1/35 |
| Skeletal abnormality | 18/55 | 3/9 | 5/11 | 10/35 |
| Ambiguous genitalia (male) | 5/35 | 1/5 | 1/8 | 3/22 |
Each feature has been examined in all the patients.
*P=0.043.
†P=0.044.
Table 2: Summary of clinical findings.
Discussion
This study identified pCNVs in nine of 55 patients with apparently non-syndromic ID. The frequency (16%) is grossly similar to that reported previously [5]. The results provide further support for the usefulness of aCGH in the clarification of underlying genetic factor(s) for ID.
Of cases 1-9 with pCNVs, cases 2-4 and 7-9 had pathogenic genes for ID on the pCNVs (Supplementary Table 1). Indeed, intragenic mutations of CTNND2, DOCK8, IL1RAPL1, and TBX1, and loss of only TERT, are associated with ID, as are duplication of only MECP2 or overexpression of MEF2C [11-17]. Thus, copy number alterations of these genes would have played a major role in the development of ID in the six cases, while the relevance of candidate genes on the pCNVs in case 4, and probably other non-specified genes as well, would remain tenable. In addition, since the CNV in case 2 and the CNVs in cases 7 and 8 are known to cause 5p deletion syndrome and 21q22.11 deletion syndrome, respectively, this confirms their pathogenicity [8,9]. Furthermore, since the CNVs in cases 2, 3, and 9 were formed as de novo events, this would also support their pathogenicity.
The remaining cases 1, 5, and 6 were also interpreted as having pCNVs. Actually, they had de novo CNVs, although there were no definite pathogenic genes on the identified CNVs. In particular, the ~2.2 Mb deletion in case 6 is identical to that reported as chromosome 17q23.1-q23.2 microdeletion syndrome with ID, and TBX2 and TBX4 have been regarded as candidate genes for ID because of their biological functions and expression pattern [18]. Since this 17q23.1-q23.2 microdeletion is known to be generated by non-allelic homologous recombination mediated by low-copy repeats [10], this would explain the recurrence of the same deletion in multiple unrelated subjects. For the ~9 Mb duplication on chromosome 1p36.21–p36.12 in case 1 and the ~6 Mb deletion on chromosome 12q24.31–q24.32 in case 5, previous studies have revealed similar duplications and deletions in patients with ID (Figures 2 and 3) [19-21]. Notably, although there is no segment shared by all the duplications and deletions, the duplicated region in case 1 and the deleted region in case 5 encompass three different smallest regions of overlaps (SROs-A–C) common to plural patients, and SRO-C on chromosome 1p36 and SRO-A on chromosome 12q24 harbor candidate genes for ID. Thus, it is likely that the duplication in case 1 and the deletion in case 5 are pathogenic, and that different genes are involved in the development of ID in patients with duplications involving 1p36 and in those with deletions affecting 12q24.
Cases 10-20 were evaluated to have VsUS rather than pCNVs. Indeed, since similar CNVs have not been reported in patients with ID, their pathogenicity remains uncertain. However, the VsUS in cases 10, 14, 17, and 18 were de novo CNVs, and those in cases 10–13, 14-16, and 18-20 harbor candidate genes (Supplementary Table 1). Thus, some of the VsUS would actually be pCNVs that harbor hitherto unrecognized pathogenic genes for ID. One may argue that the intragenic deletion of DMD identified in female case 19 is unlikely to explain her phenotype, although it could have phenotypic effects depending on the X-inactivation patterns in target tissues. However, since it is a disease-causing pCNV and could lead to ID in affected males [22], we categorized this intragenic deletion in female case 19 as a VUS rather than a pCNV or a bCNV.
Unexpectedly, plural pCNVs or VsUS were found in cases 4, 10, and 12–14. In particular those in cases 10 and 14 were generated as de novo abnormalities. Such co-existence of plural CNVs has been reported previously [23]. This would imply that de novo CNVs can occur with a certain frequency.
Several findings are noteworthy with regard to the clinical findings. First, the patient number was larger in males than in females. This would primarily be due to the presence of a large number of genes for X-linked non-syndromic ID [11]. It is predicted that a substantial fraction of male patients have hidden mutations of such genes. Furthermore, identification of pCNVs involving such X-linked genes in cases 4 (IL1RAPL1) and 9 (MECP2) suggests that X-chromosomal pCNVs are also more frequent in males than in females. Second, the age at examination, the degree of ID, and the frequencies of various features were grossly similar among groups 1-3. This suggests lack of a clinical indication for pCNVs as well as VsUS in patients with nonsyndromic ID.
In summary, we performed aCGH in 55 patients with non-syndromic ID. The results provide further support for the usefulness of aCGH in the identification of underlying genetic factor(s) for ID, although there was no clinical finding indicative of the presence of pCNVs or VsUS. Furthermore, our data are expected to serve to identify pathogenic genes on chromosomes 1q36 and 12q42, as well as those on several VsUS.
References
- Roeleveld N, Zielhuis GA, Gabreëls F (1997) The prevalence of mental retardation: a critical review of recent literature. Developmental Medicine & Child Neurology 39: 125-132
- Hunter AG (2000) Outcome of the routine assessment of patients with mental retardation in a genetics clinic. Am J Med Genet 90: 60-68.
- Rauch A, Hoyer J, Guth S, Zweier C, Kraus C, et al. (2006) Diagnostic yield of various genetic approaches in patients with unexplained developmental delay or mental retardation. Am J Med Genet 140: 2063-2074.
- Kaufman L, Ayub M, Vincent JB (2010) The genetic basis of non-syndromic intellectual disability: a review. J Neurodev Disord 2: 182-209.
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, et al. (2010) Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 86: 749-764.
- Choucair N, Ghoch JA, Corbani S, Cacciagli P, Mignon-Ravix C, et al. (2015) Contribution of copy number variants (CNVs) to congenital, unexplained intellectual and developmental disabilities in Lebanese patients. Mol Cytogenet 8: 26
- American Psychiatric Association (1994) DSM-IV: Diagnostic and Statistical Manual of Mental Disorders. American Psychiatric Association Publishing, Arlington.
- Overhauser J, Huang X, Gersh M, Wilson W, McMahon J, et al. (1994) Molecular and phenotypic mapping of the short arm of chromosome 5: sublocalization of the critical region for the cri-du-chat syndrome. Hum Mol Genet 3: 247-252.
- Driscoll DA, Budarf ML, Emanuel BS (1992) A genetic etiology for DiGeorge syndrome: consistent deletions and micro deletions of 22q11. Am J Hum Genet 50: 924-933.
- Ballif BC, Theisen A, Rosenfeld JA, Traylor RN, Gastier FJ, et al. (2010) Identification of a recurrent micro deletion at 17q23.1q23.2 flanked by segmental duplications associated with heart defects and limb abnormalities. Am J Hum Genet 86: 454-461.
- Honda S, Hayashi S, Imoto I, Toyama J, Okazawa H, et al. (2010) Copy-number variations on the X chromosome in Japanese patients with mental retardation detected by array-based comparative genomic hybridization analysis. J Hum Genet 55: 590-599.
- Griggs BL, Ladd S, Saul RA, DuPont BR, Srivastava AK (2008) Dedicator of cytokinesis 8 is disrupted in two patients with mental retardation and developmental disabilities. Genomics 91: 195-202.
- Nawara M, Klapecki J, Borg K, Jurek M, Moreno S, et al. (2008) Novel mutation of IL1RAPL1 gene in a nonspecific X-linked mental retardation (MRX) family. Am J Med Genet 146: 3167-3172.
- Chieffo C, Garvey N, Gong W, Roe B, Zhang G, et al. (1997) Isolation and characterization of a gene from the DiGeorge chromosomal region homologous to the mouse Tbx1 gene. Genomics 43: 267-277.
- Nguyen JM, Qualmann KJ, Okashah R, Reilly A, Alexeyev MF, et al. (2015) 5p deletions: Current knowledge and future directions. Am J Med Genet C Semin Med Genet 169: 224-238.
- Ramocki MB, Tavyev YJ, Peters SU (2010) The MECP2 duplication syndrome. Am J Med Genet 152: 1079-1088.
- Le MN, Holder EM, Jaillard S, Goldenberg A, Joriot S, et al. (2010) MEF2C haploinsufficiency caused by either microdeletion of the 5q14.3 region or mutation is responsible for severe mental retardation with stereotypic movements, epilepsy and/or cerebral malformations. J Med Genet 47: 22-29.
- Ballif BC, Theisen A, Rosenfeld JA, Traylor RN, Gastier-Foster J, et al. (2010) Identification of a recurrent microdeletion at 17q23.1q23.2 flanked by segmental duplications associated with heart defects and limb abnormalities. Am J Hum Genet 86: 454-461.
- Lee BH, Kasparis C, Chen B, Mei H, Edelmann L, et al. (2015) Setleis syndrome due to inheritance of the 1p36.22p36.21 duplication: evidence for lack of penetrance. J Hum Genet 60: 717-722.
- Palumbo O, Palumbo P, Delvecchio M, Palladino T, Stallone R, et al. (2015) Microdeletion of 12q24.31: report of a girl with intellectual disability, stereotypies, seizures and facial dysmorphisms. Am J Med Genet 167: 438-444
- Al-Zahrani J, Al-Dosari N, Abudheim N, Alshidi TA, Colak D, et al. (2011) Chromosome 12q24.31-q24.33 deletion causes multiple dysmorphic features and developmental delay: First mosaic patient and overview of the phenotype related to 12q24qter defects. Mol Cytogenet 4: 9.
- Daoud F, Angeard N, Demerre B, Martie I, Benyaou R, et al. (2009) Analysis of Dp71 contribution in the severity of mental retardation through comparison of Duchenne and Becker patients differing by mutation consequences on Dp71 expression. Hum Mol Genet 18: 3779-3794.
- Hayashi S, Imoto I, Aizu Y, Okamoto N, Mizuno S, et al. (2011) Clinical application of array-based comparative genomic hybridization by two-stage screening for 536 patients with mental retardation and multiple congenital anomalies. J Hum Genet 56: 110-124.
Relevant Topics
- Behavioral Psychology
- Chiari malformation
- Chronic Traumatic Encephalopathy
- Congenital Brain Defects
- Duchenne Muscular Dystrophy
- Epilepsy and Seizures
- Genetic and Metabolic Disorders
- Genetic Epilepsies
- Headaches and Migraines
- Movement Disorders
- Neonatal encephalopathy
- Neurodevelopmental Disorders
- Neurogenetic Disorders
- Neurological Complications of AIDS
- Neuromuscular Disease
- Pediatric Brain Tumour
- Pediatric Sleep Disorders
- Stroke and Perinatal Injuries
Recommended Journals
Article Tools
Article Usage
- Total views: 23868
- [From(publication date):
December-2016 - Mar 31, 2025] - Breakdown by view type
- HTML page views : 22863
- PDF downloads : 1005