Genome Wide Sequencing Compared to Candidate Gene Association Studies for Predisposition to Substance Abuse a Subset of Reward Deficiency Syndrome (RDS): Are we throwing the Baby Out with the Bathwater?
Received: 25-Mar-2014 / Accepted Date: 27-Apr-2014 / Published Date: 30-Apr-2014 DOI: 10.4172/2161-1165.1000158
Abstract
In 1990 Blum, Noble and associates utilized a candidate approach to associate the first genetic polymorphic association with severe alcoholism that was published in JAMA. This experiment was based on a “blue print” of the reward circuitry proposed as the “brain Reward Cascade”. Impairment of this system leads to aberrant substance seeking behavior. Over the last five years newer and more sophisticated techniques have been developed, including whole genome sequencing, as well as exome sequencing. While there are different schools of thought regarding appropriate approaches to dissecting a very complex disorder known as Reward Deficiency Syndrome (RDS) as an umbrella term for all addictions, future approaches may combine both genome sequencing with gene candidates. Importantly, GWAS/WES generally provides the most value for highly penetrant, rare alleles and as such may not currently be as informative as the candidate gene approach. However, since there is convergence of GWAS and neurotransmitter clusters including specific genes (e.g. DRD1, DRD2, DAt1, etc.) albeit small contributions for each gene, thousands of studies have elucidated risk alleles to RDS behaviors. Thus, we are proposing herein that we should not hasten to “throw out the baby with the bathwater”, because genetic addiction risk stratification depends upon the current candidate gene analysis.
Keywords: Candidate genes; Neurotransmitters; Dopaminergic
161901Abbreviations
Genome Wide Sequencing Association Studies (GWAS); Whole Exome Sequencing (WES); Reward Deficiency Syndrome (RDS)
Genome Wide Association Studies vs. Candidate Gene Studies: Controversy
The need for genetic testing as a way of understanding or pinpointing therapeutic targets is certainly the wave of the future. It is well established that many polymorphisms since the earliest study from our laboratory on the DRD2 gene has morphed into a remarkable list of gene polymorphism associations in the field known today as Psychiatric Genetics. It is no longer acceptable simply to utilize Diagnostic and Statistical Manual (DSM) criteria as the only method of diagnosis of psychiatric disorders [1,2]. We are hereby proposing that coupling known standard pencil and paper tests (e.g., Addiction Severity Index among others) along with DSM and Genetic Addiction Risk Score (GARS) should enhance our information on each patient presenting for example to an addiction clinic for treatment. In fact it will provide a stratification risk profile removing current quessing, as well as DNA targeted therapeutic opportunities. It is further proposed that since we are beginning to understand the power of Genome Wide Association Studies (GWAS) [1]; Whole- Exome Sequencing (WES) and Epigentic Wide Association Studies (EWAS) studies, and especially epigenetics on gene expression via mRNA transcription, knowing these hot spots will pave the way for either highly specific pharmaceuticals directed at select neuronal sites (reduce toxic side effects) or nutrigenomic solutions. In either case, we strongly recommend additional studies to provide the recovering addict with an epigenetic [3-4] tool to activate DA D2 receptors while attenuating the anti-reward effects of DA D1 and possibly D3 receptors, respectively. Finally, we as neuroscientists should begin to perform studies that control for possible comorbid medical and psychiatric conditions in their research (dual diagnosis) this work supports earlier non-genetic concepts of addiction in psychiatric medicine [5-9].
We are cognizant that there is controversy related to the issue of the importance of both GWAS and WES analytical approaches relative to candidate association and linkage studies to unravel contributions of specific genes and associated polymorphisms to addiction liability. Our laboratory in the late 1980’s, realizing the complexity of the problem of both vulnerability and resilience for risk of drug abuse and other behavioral addictions, decided to analyze candidate genes based on a theoretical model we developed and subsequently published and identified as the “Brain Reward Cascade” [10]. Our initial approach utilizing this “blue print of reward” involved association rather than linkage analysis because Lander et al. [11] argued against linkage analysis for complex disorders like drug addiction instead of linkage analysis as previously accomplished with one gene – one disease (OGOD).
Important advances have been made over the last two decades concerning “Psychatric Genetics.” Certainly, substantial genetic contributions to addiction liability are now supported by earlier twin studies and more recently linkage, candidate association and GWAS studies. Animal studies initially focused upon genes that targeted major drugs of abuse mechanism of action. Many of these studies were successful in the identification of quantitative trait loci including association of chromosomal 9 (DRD2 gene) and ethanol behavioral responses [12] as well as other reward genes including serotonin, opioids and GABA [13]. Most of these and many other studies have identified gene/proteins that affect responses to drugs of abuse [14]. Parallel to these animal studies, human genetic research exploded over the last five years, to inexpensively detect genetic variation in the human genome. The era of genome sequencing began with the identification of single-nucleotide polymorphisms (SNPs) utilizing gene chips. Very recently we are also utilizing WES which utilizes high-throughput sequencing to identify the molecular arrangement of DNA base pairs specifying the coding regions of a person’s genome also referred to as the exome. While this is exciting, it may not have clinical utility, because the exome only comprises about 1% of the entire genome [15]. However, while the exome represents ~1-2% of the genome, 85% of currently described disease alterations are located within the coding regions (this 1-2%). The prudent argument here is that psychiatric disease may over-represented within the ~15% of alterations outside of the coding region (regulatory elements, epigenetic factors which are more often associated with multi-factorial diseases.
Compared to an enormous literature on candidate gene analysis (6120 studies) there is currently a paucity of GWAS/ WES studies relevant to addiction liability (239 studies). A commentary by Hall et al. [16] from NIDA and others have argued that candidate gene analysis may be wrong. However, they do suggest that from many GWAS studies the following: (1) many genes play roles in addiction; variants of genetic alleles additively contribute to risk for addiction; (2) classes of genes (such as reward circuitry based genes) are paramount in explaining both risk and resilience in all addictions; (3) considerable genetic differences exist, yet GWAS signal convergence occurs on particular candidate genes.
It is well –known that the action of psychoactive drugs primarily affects synaptic neurotransmission. Reynolds et al. [17] correctly suggests that specific genes for neurotransmitter receptors and transporters have provided strong candidates in pharmacogenetic research in psychiatry. Moreover, there are many inconsistencies between candidate gene and GWAS studies. Reynolds further suggests that consistencies have accumulated through candidate gene studies involving the dopamine D2 receptor; serotonin transporter; and GABA dysfunction in mental illness. Moreover, Han et al. [18] reported that following GWAS, functional enrichment analysis revealed specific genes to underlie alcohol risk such as cation transport, synaptic transmission, and transmission of nerve impulses representing meaningful biological processes. In agreement Uhl et al. [19] suggest that following GWAS, clusters of SNPs within selected genes display 10(-2) > p>10 (-8) associations with dependence in many independent studies. Importantly, along these lines specific candidate genes associated with substance dependence phenotypes, for example, in Native Americans include: OPRM1,CRN1, COMT, GABA2, MAOA, and HTR3-B [20]. In a cross species GWAS study to access risk genes in alcoholism it was found that 47 genes associated including GABA, signaling pathway and cell communication [21]. In one study using GWAS and factor analysis Agrawal et al. [22] found a high loading (0.89) for alcohol craving and convergence resulted in an association of SNPs in DRD3 and craving.
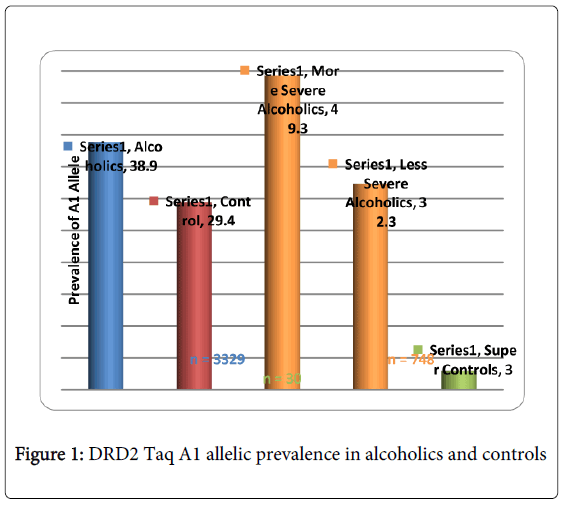
While there are many more examples showing promising convergence between GWAS and candidate gene analyses, albeit others showing no convergence, Li et al. [23] conducted a meta-analysis of GWAS and candidate gene analyses to interpret genetic risk for drug addiction and consider potential mechanisms associated with this risk. When they compared the lists of drug-related genes identified by molecular biological studies and those of association studies, they observed significantly higher participation in similar genetic interaction networks than expected by chance. This work is underscored by Li’s earlier work revealing that KARG analysis evaluated 1500 human genes regulating addictive behaviors and found that these genes significantly impact glutaminergic and dopaminergic pathways [24] (Figure 1).
GWAS studies in psychiatry frequently fail to explain a large proportion of variance and non-replication of individual SNPs. Derringer et al. [25] utilizing a “selective scoring” whereby variants (273 SNPs) from eight dopamine-related genes for association with cocaine dependence were considered. They identified a four SNP score significantly associated with the variance. They suggested that (1) if only SNPs of genes with theoretical importance are investigated, than replicable effects are more likely to be identified; furthermore, (2) if a scoring approach evaluating the aggregate of the highest associated SNPs is utilized, than more replicable effects can be identified than would be revealed in individual SNP level studies. Along with these precautions, we also propose the utilization of “super controls” to eliminate cross contamination of having a control phenotype that also carries the associated disorder, especially when considering the complex nature of RDS. In one study, we found that while the risk allele of the DRD2 Taq A1 in over 3,000 non-screened population controls was 32%, super controls (screened for all RDS behaviors in the proband and family) was only 3% [26]. Also in developing GARS we are cognizant that the proposed test only provides risk stratification and not actual diagnosis of any disorder including RDS. Most importantly, full comprehension of “psychiatric pharmacogenomics” will undoubtedly involve epigenetic factors, such as DNA histone modifications (e.g. methylation and or deacetylation) that can affect responses to drugs and polymorphic antecedents for either vulnerability or resilience to reward behaviors like RDS [27-36]. Finally, GWAS/WES generally provides the most value for highly penetrant, rare alleles and as such may not currently be as informative as the candidate gene approach.
Acknowledgements
The writing of this paper was supported in part by funds from the National Institutes of Health, NIAAA (RO1-AA07112 and K05-AA00219) and the Medical Research Service of the US Department of Veterans Affairs (MOB). The authors appreciate the editorial assistance of Margaret A. Madigan and Paula J. Edge. Kenneth Blum is the recipient of a grant to PATH FOUNDATION NY, by Life Extension Foundation, Ft/ Lauderdale, Florida.
Conflicts
Kenneth Blum, PhD holds certain US and foreign patents related to genetic testing. Dr. Blum is also an officer and stock holder of IGENE, LLC and is a paid consultant of Dominion Diagnostics, LLC., IGENE, Malibu Recovery Center, and RDSolutions, LLC. Both Drs. Blum is a member of the scientific advisory board of Dominion Diagnostics, LLC and has a title of Chief Scientific Advisor.
References
- Teo YY (2008) Common statistical issues in genome-wide association studies: a review on power, data quality control, genotype calling and population structure. CurrOpinLipidol 19: 133-143.
- Insel TR (2014) The NIMH Research Domain Criteria (RDoC) Project: Precision Medicine for Psychiatry. Am J Psychiatry 171: 395-397.
- Archer T, Oscar-Berman M, Blum K, Gold M (2013) Epigenetic Modulation of Mood Disorders. J Genet Syndr Gene Ther 4.
- Archer T, Oscar-Berman M, Blum K (2011) Epigenetics in Developmental Disorder: ADHD and Endophenotypes. J Genet Syndr Gene Ther 2.
- Dackis CA, Gold MS (1988) Biological aspects of cocaine addiction. In: Volkow ND, Swann AC (edn) Cocaine in the brain. Rutgers University Press, New Brunswick, NJ.
- Neill DB, Peay LA, Gold MS (1978) Identification of a subregion within rat neostriatum for the dopaminergic modulation of lateral hypothalamic self-stimulation. Brain Res 153: 515-528.
- Miller NS, Gold MS (1991) Dual diagnoses: psychiatric syndromes in alcoholism and drug addiction. Am Fam Physician 43: 2071-2076.
- Miller NS, Dackis CA, Gold MS (1987) The relationship of addiction, tolerance, and dependence to alcohol and drugs: a neurochemical approach. J Subst Abuse Treat 4: 197-207.
- Miller NS, Gold MS (1998) Management of withdrawal syndromes and relapse prevention in drug and alcohol dependence. Am Fam Physician 58: 139-146.
- Blum K, Kozlowski GP (1990) Ethanol and neuromodulator interactions: a cascade model of reward. In: Ollat H, Parvez S, Parvez H (edn) Alcohol and Behavior. VSP Press, Utrecht, Netherlands.
- Lander ES, Schork NJ (1994) Genetic dissection of complex traits. Science 265: 2037-2048.
- Buck K, Lischka T, Dorow J, Crabbe J (2000) Mapping quantitative trait loci that regulate sensitivity and tolerance to quinpirole, a dopamine mimetic selective for D(2)/D(3) receptors. Am J Med Genet 96: 696-705.
- Crabbe JC, Phillips TJ, Harris RA, Arends MA, Koob GF (2006) Alcohol-related genes: contributions from studies with genetically engineered mice. Addict Biol 11: 195-269.
- Gerra G, Somaini L, Leonardi C, Cortese E, Maremmani I, et al. (2014) Association between gene variants and response to buprenorphine maintenance treatment. Psychiatry Res 215: 202-207.
- Feero WG (2014) Clinical application of whole-genome sequencing: proceed with care. JAMA 311: 1017-1019.
- Hall FS, Drgonova J, Jain S, Uhl GR (2013) Implications of genome wide association studies for addiction: are our a priori assumptions all wrong? PharmacolTher 140: 267-279.
- Reynolds GP, McGowan OO, Dalton CF (2014) Pharmacogenomics in psychiatry: the relevance of receptor and transporter polymorphisms. Br J ClinPharmacol 77: 654-672.
- Han S, Yang BZ, Kranzler HR, Liu X, Zhao H, et al. (2013) Integrating GWASs and human protein interaction networks identifies a gene subnetwork underlying alcohol dependence. Am J Hum Genet 93: 1027-1034.
- Uhl GR, Drgonova J, Hall FS (2014) Curious cases: Altered dose-response relationships in addiction genetics. PharmacolTher 141: 335-346.
- Ehlers CL, Gizer IR (2013) Evidence for a genetic component for substance dependence in Native Americans. Am J Psychiatry 170: 154-164.
- Zhao Z, Guo AY, van den Oord EJ, Aliev F, Jia P, et al. (2012) Multi-species data integration and gene ranking enrich significant results in an alcoholism genome-wide association study.BMC Genomics13:S16.
- Agrawal A, Wetherill L, Bucholz KK, Kramer J, Kuperman S, et al. (2013) Genetic influences on craving for alcohol. Addict Behav 38: 1501-1508.
- Li CY, Zhou WZ, Zhang PW, Johnson C, Wei L, et al. (2011) Meta-analysis and genome-wide interpretation of genetic susceptibility to drug addiction. BMC Genomics 12: 508.
- Li CY, Mao X, Wei L (2008) Genes and (common) pathways underlying drug addiction. PLoSComputBiol 4: e2.
- Chen TJ, Blum K, Mathews D, Fisher L, Schnautz N, et al. (2005) Are dopaminergic genes involved in a predisposition to pathological aggression? Hypothesizing the importance of "super normal controls" in psychiatricgenetic research of complex behavioral disorders. Med hypotheses 65:703-707.
- Derringer J, Krueger RF, Dick DM, Aliev F, Grucza RA, et al. (2012) The aggregate effect of dopamine genes on dependence symptoms among cocaine users: cross-validation of a candidate system scoring approach. Behav Genet 42:626-635.
- Jordi E, Heiman M, Marion-Poll L, Guermonprez P, Cheng SK, et al. (2013) Differential effects of cocaine on histone posttranslational modifications in identified populations of striatal neurons. ProcNatlAcadSci U S A 110:9511-9516.
- Byrnes JJ, Johnson NL, Carini LM, Byrnes EM (2013) Multigenerational effects of adolescent morphine exposure on dopamine D2 receptor function. Psychopharmacology (Berl) 227: 263-272.
- DiNieri JA, Wang X, Szutorisz H, Spano SM, Kaur J, et al. (2011) Maternal cannabis use alters ventral striatal dopamine D2 gene regulation in the offspring. Biol Psychiatry 70: 763-769.
- Belsky J, Beaver KM (2011) Cumulative-genetic plasticity, parenting and adolescent self-regulation. J Child Psychol Psychiatry 52: 619-626.
- Arranz MJ, de Leon J (2007) Pharmacogenetics and pharmacogenomics of schizophrenia: a review of last decade of research. Mol Psychiatry 12: 707-747.
- Liu JC, Baker RE, Chow W, Sun CK, Elsholtz HP (2005) Epigenetic mechanisms in the dopamine D2 receptor-dependent inhibition of the prolactin gene. MolEndocrinol 19: 1904-1917.
- Li J, Guo Y, Schroeder FA, Youngs RM, Schmidt TW, et al. (2004) Dopamine D2-like antagonists induce chromatin remodeling in striatal neurons through cyclic AMP-protein kinase A and NMDA receptor signaling. J Neurochem 90:1117-1131.
- Febo M, Akbarian S, Schroeder FA, Ferris CF (2009) Cocaine-induced metabolic activation in cortico-limbic circuitry is increased after exposure to the histone deacetylase inhibitor, sodium butyrate. NeurosciLett 465:267-271.
- Noble EP (2003) D2 dopamine receptor gene in psychiatric and neurologic disorders and its phenotypes. Am J Med Genet B Neuropsychiatr Genet 116B: 103-125.
- Archer T, Oscar-Berman M, Blum K, Gold M (2012) Neurogenetics and Epigenetics in Impulsive Behaviour: Impact on Reward Circuitry. J Genet Syndr Gene Ther 3: 1000115.
Citation: Blum K, Braverman ER, Kreuk F, Dushaj K, Li M, et al. (2014) Genome Wide Sequencing Compared to Candidate Gene Association Studies for Predisposition to Substance Abuse a Subset of Reward Deficiency Syndrome (RDS): Are we throwing the Baby Out with the Bathwater?. Epidemiol 4:158. DOI: 10.4172/2161-1165.1000158
Copyright: © 2014 Blum K, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 16015
- [From(publication date): 6-2014 - Nov 23, 2024]
- Breakdown by view type
- HTML page views: 11537
- PDF downloads: 4478