Genetics of Spinal Muscular Atrophy and Splicing of Smn Gene
Received: 01-Jan-2022 / Manuscript No. nctj-22-51515 / Editor assigned: 03-Jan-2022 / PreQC No. nctj-22-51515 (PQ), / Reviewed: 17-Jan-2022 / QC No. nctj-22- 51515 / Revised: 22-Jan-2022 / Manuscript No. nctj-22-51515 (R) / Accepted Date: 18-Jan-2022 / Published Date: 31-Jan-2022 DOI: 10.4172/nctj.1000110
Abstract
Spinal muscular atrophy is characterized by loss of motor neurons and muscle atrophy, largely in childhood. It is a devastating neuromuscular disorder. In humans, nearly two identical inverted SMN genes (SMN1, SMN2) are present on chromosome 5q13. Homologous deletion of SMN1 results in SMA.SMA is initiated by low levels of the survival motor neuron protein (SMN) because of inactivating mutations in the encoding gene SMN1. Another functional protein for survival is produced by second duplicate gene SMN2.It produces a shortened, unstable SMN messenger RNA. From alternative splicing it produces a small length fully functional SMN messenger RNA. For SMA clinical severity, SMN2 gene copy number is a good prognostic biomarker. Many therapeutical strategies for spinal muscular atrophy are in clinical trials. Recently, Antisense oligonucleotide (ASO) therapy has been licensed. Though, several factors recommend that complementary strategies may be desirable for the long-term maintenance of neuromuscular disorder. During the establishment of structural connections of neuromuscular system, SMN protein is required in highest amount. Besides, people receiving SMN-based treatments may be vulnerable to delayed symptoms if rescue of the neuromuscular system is incomplete. Hence, for the treatment of CNS and periphery, a comprehensive whole-lifespan approach to SMA therapy is required. This therapy includes both SMN-dependent and SMN-independent strategies for the enhancement of SMN expressions many current and planned clinical trials are designed.
Keywords: Spinal muscular atrophy,SMN genes,Homologous deletion,clinical trials.
Keywords
SMN gene; Atrophy; Spinal muscle; Motor neuron; Protein; Disorder
Introduction
Guido Werdnig documented spinal muscular atrophy in two newborn brothers in 1891, and Johan Hoffmann characterized it in seven further instances from 1893 to 1900. Although the title Werdnig- Hoffmann illness was subsequently used to the severe infantile form of SMA, their cases were really of intermediate intensity; Sylvestre described severe infantile SMA in 1899 and Beevor in 1903 [1]. Wohlfart, Fez, and Eliasson first described a milder form of SMA in the 1950s, with patients retaining the ability to stand and walk and living longer [1]. Kugelberg and Welander went on to describe it in greater detail. All of these descriptions identified and underlined the primary pathology as anterior horn cell degeneration, as well as the relevant clinical symptoms of symmetrical, proximal predominate extremities weakness affecting axial, intercostal, and bulbar muscles [1]. During the next half-century, the severity variability was further identified and classified, and debate developed about whether the infantile, juvenile, and adult forms of SMA reflected one or many disorders. The numerous phenotypes were subsequently codified into a categorization scheme at a Muscular Dystrophy Association-sponsored International Consortium on Spinal Muscular Atrophy in 1991 [2]. This categorization distinguished three forms of SMA based on the maximum degree of motor function and the age of onset. Subsequent changes separated the type 3 group according to age of onset, introduced a type 4 for adultonset cases, and included a type 0 for patients with prenatal onset and death within weeks. Although there are degrees of severity even within a single type, and up to 25% of individuals evade accurate classification, this approach remains relevant in the genetic era and gives significant clinical and prognostic information.
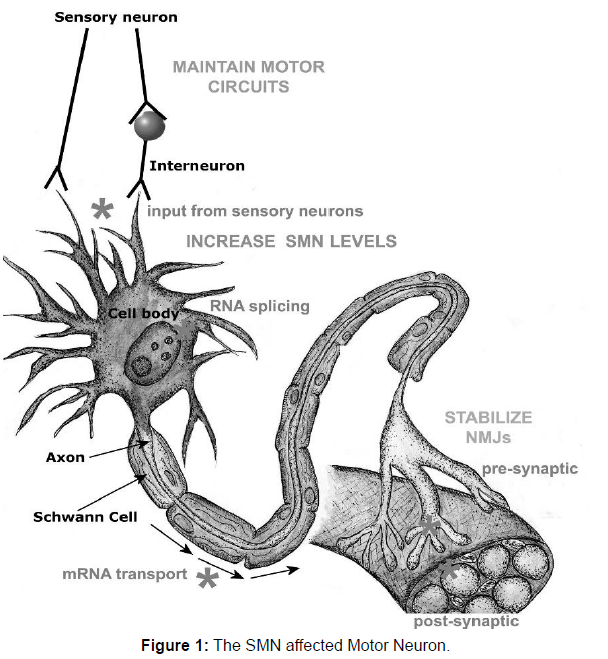
Spinal muscular atrophies (SMA) are fatal inherited disorder characterized by the loss of spinal motor neurons. It is caused by the degeneration of motor neurons of the spinal cord anterior horns, leading to progressive atrophy of proximal muscles, paralysis, respiratory failure, and infant death [2]. SMN is majorly caused when a motor neuron is a deficit of the ubiquitously expressed housekeeping protein ‘‘survival motor neuron’’ (SMN). Many types of spinal muscular atrophy are caused by changes in the same genes. Childhood SMA is severe, generalized muscle weakness at birth or within the next 3 months. Death of the infants occurs usually within the first two months of birth [3]. In Pakistan there are almost 68% of the patients mostly children have SMN gene deletion. It is more common in the countries like Pakistan, Iran, Saudi Arabia as these countries have high rate of consanguinity and high birth rate that’s why 68 percent population of children were affected with this neuromuscular disorder [4]. A full study of the pathophysiology of SMA is beyond the scope of this article; nonetheless, a few remarks are in order. The SMN protein is distributed throughout the cytoplasm and nucleus, where it works as a component of the SMN complex, a multiprotein complex that is required for spliceosomal small nuclear ribonuclear protein synthesis and premRNA splicing. Small nuclear ribonuclear protein synthesis is changed in SMA-affected mouse cells. The SMN protein has also been detected in the axons of motor neurons. One frugal explanation may be that the downstream consequences of altered RNA processing that result from insufficient expression of SMN are not favorable for motor neuron development, survival, or both. In this sense, because the motor neuron transcriptome is unique, a global alteration in splicing, for example, could have a unique effect on the transcriptome of motor neurons. This concept on the pathogenic role of RNA processing abnormalities in motor neuron illnesses is getting popular. Fortunately, based on human genotype phenotype research and preclinical investigations in SMA animal models, a thorough knowledge of the disease’s molecular pathophysiology may not be an absolute requirement for the creation of sensible therapy methods. Nonetheless, knowing the molecular pathogenesis of SMA may give a footing and lead to an understanding of related motor neuron illnesses such as non-SMN spinal muscular atrophies and amyotrophic lateral sclerosis. Many pathogenic mechanisms in SMA are concerned with a large variety of genes. RNA splicing, metabolism and protein synthesis is implicated by SMN, SETX, DCNT1, GARS, RARS2, and LASIL. HSPB1, HSPB8, BSCL2, UBE1, AR, VAPB, DCNT1, MAPT are involved in Protein-folding, aggregation and degradation pathways. Genes DCNT1, DYNC1H1, PLEKHG5, HSPB1, SMN, BICD2, FBX034 are responsible for Axonal guidance and transport (Figure 1).
SMN protein is required in all cells, including motor neurons, to assemble snRNPs in the cytoplasm and transport them into the nucleus for RNA splicing. It is also needed in sensory neurons, to maintain motor circuit activity and MN activity in motor neuron axons to transport actin mRNA to the neuromuscular junction (NMJ) and both pre- and post-synaptically at NMJs for their normal function and stability. The main molecular approach to treatment has been to try to increase SMN levels by increasing SMN production or stability. This may be required systemically, rather than in motor neurons alone and may be complemented by measures to stabilize NMJ stability or maintain sensory inputs to motor neurons. Sites of action of SMN protein are shown in the figure at lower case grayscale, and potential sites for therapeutic intervention in upper case grayscale.
Methodology
Protein families related to spinal muscular atrophy
On the basis of mode of inheritance spinal muscular atrophy is classified into two distinct forms i.e. Distal spinal muscular atrophy and the other one is proximal spinal muscular atrophy [5]. Distal spinal muscular atrophy is a slowly progressive disease with a rare bulbar involvement. It may be dominant, named as distal hereditary motor neuropathy (dHMN) and recessive, distal spinal muscular atrophy (DSMA), Mutation in the chaperones like HSPB1, HSPB3, HSPB8 resulting in the dysfunction of protein folding. Mutation in GARS gene alters the transfer RNA sequence which is responsible for amino acylation [6].
Heat shock proteins (HSP) is the most abundant protein family in organisms. In Human genome there are almost ten different types of HSPB named as HSPB1-HSPB10. Some of them are expressed ubiquitously like (HSPB1, HSPB5, HSPB6, HSPB8) while some of the family members are tissue specific in nature like HSPB2, HSPB3, HSPB4, HSPB7, HSPB9, HSPB10 [7]. There are three major domains of HspB family that i.e. C-terminal domain, N-terminal domain and highly conserved α-Crystallin domain [8]. The genes responsible for the distal hereditary motor neuropathy have diverse function such as (HSPB1, HSPB8, BSCL2) in the Protein folding, (IGHMBP2, GARS) in the RNA metabolism, (HSPB1, DYNC1H1, DCTN1) axonal transport and (ATP7A and TRPV4) in cation-channel dysfunction [9] (Table 1).
| Gene | Location | Mutation | Inheritance |
|---|---|---|---|
| Hspb1 | 7q11.23 | Transition404c-t | Ad |
| Hspb8 | 12q24.23 | 423g→c, (transversion) (belgian, czech) 421a→g, (transition) (bulgarian and english) |
Ad |
| Hspb3 | 5q11.2 | Transversion21g-t | Ad |
| Fbxo38 | 5q32 | Transition616t>c | Ad |
| Smar | 11q13 | Het 1178g>a Het 1284+5g>a |
Ar |
| Gars1 | 7p14.3 | 815t>g | Ad |
| Bscl2 | 11q12.3 | 263a→g transition (austrian, italian, english) 269c→t missense (brazilian family) |
Ad |
| Reep1 | 2p11.2 | 303+1 7gtaatat>ac |
Ad |
| Ighmbp2 | 11q13.3 | Nonsense 5’mutation (c.138t>a) 3frame shifts C.2911_2912 Delag: P. Arg971glufs |
Ar |
| Slc5a7 | 2q12.3 | 1497delg | Ad |
| Dctn1 | 2p13.1 | 175g>a | Ad |
| Dnajb2 | 2q35 | Transition (352+1g>a) |
Ar |
| Wars | 14q32.2 | Heterozygous mutation c.770a>g | Ad |
| Atp7a | Xq21.1 | 4156c>t (family a) Transition 2981c>t (family b) Transition |
Xlr |
| Mtatp6 | ----------- | 9185t>c | Mitochondria |
| Mtatp8 | ----------- | (m.8403t>c) | Mitochondria |
| Ar: Autosomal recessive; xlr: x-linked recessive; ad: autosomal dominant; mtatp: mitochondrial adenosine triphosphate. | |||
Table 1: Variants responsible for DSMA in populations. AR: Autosomal Recessive; XLR: X-linked Recessive; AD: Autosomal Dominant; mtATP: Mitochondrial Adenosine Triphosphate.
Types of Spinal muscular atrophy
Proximal spinal muscular Atrophy: Proximal spinal muscular atrophy is the autosomal recessive disorder causes the degeneration of the α-spinal motor neurons in brain stem and spinal cord. It is categorized into three types [4].
Werdnig-Hoffmann (SMA type 1): It is reported that about 50% patient were diagnosed with this severe type of SMA. Infants with SMA type-1don’t have the ability to sit unsupported and, usually do not survive further than the first 2 years. These patients have intense hypotonia, flaccid paralysis, and no head control. Natural motility is generally poor and movements of limbs are typically not observed. In the most severe forms, decreased inside the womb movements suggest prenatal onset of the disease with severe weakness and joint contractures [10].
Dubowitz syndrome (SMA type2): Dubowitz syndrome is a rare autosomal recessive disorder and was first identified in children. Its manifest growth retardation, microcephaly, short stature, facial features, skin eruptions, and mild to severe mental retardation. Patients are able to sit unsupported and some of them are able to attain standing position, but they do not have the ability to walk independently. Joint contractures and kyphoscoliosis are very common severe type II patients [10].
Kugelberg-Welander disease: SMA type3 (Kugelberg-Welander disease) causes muscle weakness in infancy. People with this condition will stand and walk independently; there are some genes responsible for the proximal spinal muscular atrophy. Scoliosis and some medical problems like poor mobility, obesity and osteoporosis arise in those patients who lose ambulation. Respiratory system requires most care as it became weakened it will never recover again and make breathing difficulties (Table 2).
| Gene /locus | Position | Mutation | Disease |
|---|---|---|---|
| Trpv4 | 12q24.11 | 805c>t 806g>a Messene 946c-t Transition |
Scapuloperoneal sma |
| Dync1h1 | 14q32.31 | Heterozygous 1750a-c | Lower extremity predominant sma type 1 |
| Bicd2 | 9q22.31 | 320c>t (dutch family) C.2108c>t (canada) 563a>c (netherlands) |
Lower extremity predominant sma type 2 |
| Vapb | 20q13.32 | Heterozygous 166c-t (transition) |
Finkel-type late onset sma |
| Lmna | 1q22 | Exon 3 Codon208, Delag |
Adult-onset proximal sma, Followed by cardiac involvement |
| TFG | 3q12.2 | 854C>T | Hereditary motor and sensory neuropathy, Okinawa type |
| AR | Xq12 | CAG repeat (Exon 1 AR gene) |
Spinal and bulbar muscular atrophy |
Table 2: Genes responsible for the occurrence of the proximal spinal muscular atrophy.
Function of SMN protein
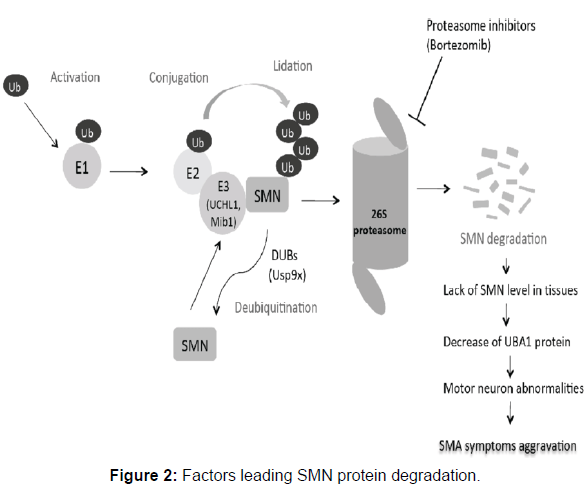
SMN is a 294 amino acid long chain weighs of about 38 kilo Daltons. It is found in a large amount within the growth cones of motor neurons. SMN involved in ribonucleoprotein biogenesis. It plays an important role in splicing machinery. Gemins, spliceosomal U-snRNPs, SMA proteins and profilins makes the SMN complex. Gemin-8 is portion of the SMN complex. In gemin, spliceosomal snRNP are assembled in the cytoplasm and splicing of pre mRNA occurs in the nucleus. When electron microscopy demonstrated SMN localization in the dendrites and axons of motor neurons in the developing rat spinal cord, it was the first hint that SMN had a role other than its typical actions in the spliceosome. It has been proposed that SMN protein localizations go from mostly nuclear during development to more cytoplasmic and axonal localizations in adult neurons. SMN was also discovered in the growth cones of cultured motor neurons, and live cell imaging revealed puncta positive for SMN actively transporting bi-directionally along axons. SMN co-localizes with some SMN complex parts in the axon, such as Gemin, although Sm proteins are extremely rare in distal neurites, and most axonally localized SMN granules lack Sm proteins. The neuron-specific protein neurochondrin is essential for SMN’s normal cytoplasmic localization, and neurochondrin did not co-localize with snRNPs, indicating that SMN is engaged in processes other than splicing. According to recent research, SMN can bind to the -COP component of the COPI vesicle. The COPI system, a Golgi-derived vesicular transport system, is involved in intracellular trafficking in neurites, which is required for neuronal cell process maturation. -COP deficiency was discovered to alter SMN localization inside growth cones, resulting in its buildup within the trans-Golgi network. -COP deficiency inhibited neurite production in NSC-34 cells and primary cortical neurons, resulting in shorter map2-positive dendrites and tau-positive axons, and both -COP and SMN are necessary for proper neurite formation. This suggests that SMN has a function in trafficking for neuronal expansion and the development of the axonal and synaptic cytoskeleton. Following these initial findings, fluorescent in situ hybridization tests against the polyA tails of mRNA demonstrated a more than 50% drop in mRNA transcript localization along the axon of primary motor neurons after SMN suppression (Figure 2).
SMN2 gene as main modifier
The SMN2 gene can generate some full-length SMN protein; it is the primary phenotypic modifier in SMA patients. Homozygous SMN2 absence is observed in 5% of the healthy population and has no phenotypic consequence [11]. Patients with spinal muscular atrophy who have had both copies of the SMN1 gene deleted or disrupted, on the other hand, have at least one copy of the SMN2 gene. Prenatal death is caused by the complete lack of SMN genes [12]. The number of SMN2 copies normally ranges between 1 and 4, with only a few cases reaching 8 copies [13]. The increased copy number of SMN2 is induced by duplication or gene conversion of SMN1 into SMN2 and has been linked to a milder form of the illness [14]. Several observations of asymptomatic patients with homozygous deletion of the SMN1 gene and elevated SMN2 copy number [15,16] support the role of the SMN2 gene as a primary disease modulator. In addition, an inverse relationship has been discovered between SMN2 copy quantity and survival time.
Despite several evidences of the influence of SMN2 copy number on SMA phenotype, certain cases of families with significant differences in disease severity among siblings with the same number of SMN2 copies [16-18] have been described. In some situations, asymptomatic carriers of a homozygous deletion of the SMN1 gene have less than five SMN2 copies, but in others, six SMN2 copies are inadequate to avoid SMA symptoms. Some SMA type I patients have three copies of the SMN2 gene, whereas SMA type III patients have only two copies [19]. Variability in expression between SMN2 copies, either by SMN2 gene mutations or the action of splicing factors, might explain such differences. Factors that interact with the SMN protein, modulate its stability, and are involved in numerous pathways affecting motor neuron survival are potentially interesting study areas as potential modifiers of SMA severity. The SMN2 gene copy number is complete (Figure 3).
Conclusion
SMA is an autosomal recessive neuronal disorder. SMN1 (survival motor neuron) is a faulty gene in SMA. SMN1 is present on chromosome 5. Mainly, it is important for nerve cells called lower motor neurons. SMN2 gene is most commonly found in humans. SMN2 provide directions for making SMN protein like SMN1. Additional SMN2 genes offered full size SMN protein. A mutation in the VAPB gene on chromosome 20 is responsible for Finkel type SMA. This gene produce VAPB protein, related with the membrane that surrounds the endoplasmic reticulum, where newly-made proteins attain three-dimensional structure and can be prepared to transport within the cell or to the surface of cell. Proline is replaced by the amino acid serine at position 56 due to mutation, which leads to SMA. Unfolded protein response cannot be activated by the mutated protein. As a result, protein aggregation occurs. In this condition, motor neurons are susceptible to cell death. SMA treatment has mostly consisted of supportive and palliative care for more than 100 years, since its original description. Over the last decade, there has been a significant advance in doctors’ capacity to treat the various respiratory, dietary, orthopedic, rehabilitative, mental, and social difficulties that most of these patients suffer. There were few clinical studies in SMA prior to the 1990s since there was no apparent molecular target. Studies were mainly conducted with repurposed pharmacologic drugs that had demonstrated promising outcomes in other diseases characterized by muscle weakness, such as amyotrophic lateral sclerosis or muscular dystrophy. However, the clarification of the genetic and molecular underpinnings of SMA has proposed numerous viable treatment options based on the basic premise of raising the production of the SMN protein. Pharmacologic or gene-based therapies to increase SMN2 expression (resulting in more full-length SMN mRNA), antisense oligonucleotide–based therapies to promote exon 7 incorporation into SMN2-derived mRNA transcripts, and virus-mediated therapies to replace the entire SMN1 gene are among these strategies. Human clinical studies employing RNA-based and gene therapy techniques are currently being conducted at a quick pace, indicating that these approaches are being developed at a fast rate.
References
- Dubowitz V (2009) Ramblings in the history of spinal muscular atrophy Neuromuscul Disord 19: 69-73.
- Munsat TL (1991) Workshop report. International SMA collaboration Neuromuscul Disord 1: 81.
- Farrar MA, Kiernan MC (2015) The genetics of spinal muscular atrophy: progress and challenges Neurother 12:290-302.
- Lefebvre S, Bürglen L, Reboullet S, Clermont O,Burlet P et al.(1995) Identification and characterization of a spinal muscular atrophy-determining gene Cell, 80:155-165.
- Karim K, Dileep D, Munim S. (2020). Prenatal diagnosis of rare genetic conditions at a tertiary care hospital in Karachi J Pak Med Assoc 70:724-727
- Rossor AM, Kalmar B, Greensmith L, Reilly MM(2012)The distal hereditary motor neuropathies. J. Neurol 83:6-14.
- Bansagi B, Griffin H, Whittaker RG, Antoniadi T, Evangelista T,et al.(2017) Genetic heterogeneity of motor neuropathies Neurology 88:1226-1234.
- Nefedova VV, Muranova LK, Sudnitsyna MV, Ryzhavskaya AS, & Gusev NB (2015) Small heat shock proteins and distal hereditary neuropathies Biochem. (Mosc.)80: 1734-1747.
- Ackerley S, Kalli A, French S, Davies KE,Talbot K, et al. (2006) A mutation in the small heat-shock protein HSPB1 leading to distal hereditary motor neuronopathy disrupts neurofilament assembly and the axonal transport of specific cellular cargoes Hum. Mol. Genet 15:347-354.
- Penttilä S, Jokela M, Bouquin H, Saukkonen AM, Toivanen J,et al.(2015) Late onset spinal motor neuronopathy is caused by mutation in CHCHD 10 Ann. Neurol. 77: 163-172.
- Hofmann Y, Lorson CL, Stamm S, Androphy EJ, & Wirth B(2000) Htra2-β1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2). Proceedings of the PNAS 97: 9618-9623.
- Simic G (2008) Pathogenesis of proximal autosomal recessive spinal muscular atrophy.Acta Neuropathol 116: 223-234.
- Vitali T, Sossi V, Tiziano F, Zappata S, Giuli A et al.(1999) Detection of the survival motor neuron (SMN) genes by FISH: further evidence for a role for SMN2 in the modulation of disease severity in SMA patients. Hum. Mol 8:2525-2532.
- Vander Steege G, Grootscholten PM, Cobben JM, Zappata S, Scheffer H,et al. (1996) Apparent gene conversions involving the SMN gene in the region of the spinal muscular atrophy locus on chromosome 5. Am J Hum Genet 59 : 834-838
- Jędrzejowska M, Borkowska J, Zimowsk J, Kostera-Pruszczyk A, Milewski M et al.(2008) Unaffected patients with a homozygous absence of the SMN1 gene Eur. J. Hum. Genet 16: 930-934.
- Zheleznyakova GY, Kiselev AV, Vakharlovsky V G, Rask-Andersen M, Chavan R et al. (2011) Genetic and expression studies of SMN2 gene in Russian patients with spinal muscular atrophy type II and III. BMC Med Genet 12:1-9.
- Prior TW, Swoboda, KJ, Scott HD, & Hejmanowski AQ(2004)Homozygous SMN1 deletions in unaffected family members and modification of the phenotype by SMN2. Am J Med. Genet 130 : 307-310.
- Helmken C, Hofmann Y, Schoenen F, Oprea G., Raschke H, et al.(2003) Evidence for a modifying pathway in SMA discordant families: reduced SMN level decreases the amount of its interacting partners and Htra2-beta1 Hum Genet 114: 11-21.
- McAndrew PE, Parsons DW, Simard LR, Rochette C, Ray PN, et al. (1997) Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. The Am J Hum Genet 60: 1411-1422.
Indexed at Google Scholar Cross link
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Indexed at Google Scholar Crossref
Citation: Sohail S, Asif F, Asif MA, Ashraf A, Nisar M, et al. (2022) Genetics of Spinal Muscular Atrophy and Splicing of Smn Gene. Neurol Clin Therapeut J 6: 110. DOI: 10.4172/nctj.1000110
Copyright: © 2022 Sohail S, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Open Access Journals
Article Tools
Article Usage
- Total views: 2974
- [From(publication date): 0-2022 - Apr 07, 2025]
- Breakdown by view type
- HTML page views: 2390
- PDF downloads: 584