Opinion Article Open Access
Genetically-encoded Biosensor for Kinases: To See is to Believe?
Pauline Vandame1, Davel Trinel2, Corentin Spriet2* and Jean-François Bodart1
1Université de Lille, Sciences et Technologies, Régulation des Signaux de Division Team, UMR 8576 CNRS, Villeneuve d´Ascq, Site de Recherche Intégrée en Cancérologie(SIRIC ONCOLILLE), France
2Université de Lille, Sciences et Technologies, TISBio, UMR 8576 CNRS, Villeneuve d´Ascq, France
- Corresponding Author:
- Corentin Spriet
Université de Lille, France
E-mail: corentin.spriet@iri.univ-lille1.fr
Received Date:December 22, 2014; Accepted Date: January 13, 2015; Published Date:January 16, 2015
Citation: Vandame P, Trinel D, Spriet C, Bodart JF (2015) Genetically-encoded Biosensor for Kinases: To See is to Believe? Biosens J 4:113. doi:10.4172/2090-4967.1000113
Copyright: © 2015 Vandame P, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Biosensors Journal
Keywords
Genetically-encoded biosensor; KAR; FRET; PKA; ERK; Artifacts
Introduction
This is a quite a truism to assert that scientific community has been for centuries in an hunger for observing life in any of its details, beginning with an anthropomorphized conception of life : Hartsoecker was seeking for germ of life like homunculi in germinal cells, nourishing the preformationist theories in developmental biology. For decades, animalcule and manikins were sought with the frenzy and curiosity of Brobdingnag inhabitants willing to scrutinize Lilliputians' activities. Frustrations came along with development of microscopy and the restraining of "life" into smaller and smaller entities, though microscopes had been bartered to scalpel in the search of life's sparkles. DNA was the support of genetic data, inert as a modular set of information can be, around which were dancing and swinging enzymes, assuming transcription and translation of genes into proteins, which in turn were involved in all the aspects of cell life (migration, proliferation, differentiation, death). Nevertheless, those enzymatic activities remained mainly addressed in vitro, and characterized in context where spatiotemporal patterns of activities were lost : fixed cells enabling snapshots, lysates & fragmented cells. A supplementary level of complexity arose when post-translational modifications of proteins were found to regulate those enzymatic activities. Still, cellular life in motion was challenging science to get properly tracked and seen. Single molecules tracking seemed far out from hand and life was getting far from being observed at a glance, discerned by a naked eye.
Genetically-encoded FRET Reporters for Kinases
Together with other types of sensors, genetically-encoded FRET biosensors for enzymes raised hope to see several of these activities in real time, in living cells and even in living and developing organisms [1]. Those sensors rely on FRET (Förster resonance Energy Transfer). FRET is a non radiative process involving radiation less energy transfer from a donor fluorophore to an appropriately chosen and positioned acceptor fluorophore [2]. For most genetically encoded donor/acceptor pair, this transfer can only occur if they are separated by a distance less than ~10 nm. To build a FRET based sensor, an adapted bioreceptor is tagged on both ends with appropriate fluorophores. The bioreceptor will alter its conformation or will be cleaved upon either analyte/second messenger presence or protein activity and will result in a measurable change in FRET efficiency [3,4].
Among post-translational modifications, a beloved one by scientists is phosphorylation. The latter is brought by proteins kinases, which are included in a large and diverse family of evolutionarily related proteins, whose deregulations have been involved in many pathologies. Genetically-encoded reporters for protein kinases activities are tools for a constructed perception of protein kinase activities, with their intrinsic properties, advantages and limits. Namely Kinase Activity Reporters (KAR), these tools are structurally composed of two fluorescent proteins, chosen for their amenability for FRET experiments, flanking a specific substrate for a specific kinase and a phospho-amino-acid binding domain (PAABD). This PAABD recognizes and binds the phosphorylated substrate, allowing a conformational modification that brings the fluorophores close to each other in order to lead to a measurable FRET signal [2,5-7]. As a limit, one might argue that the FRET signal does not only reflect the activity of the protein kinase, but also the one of protein phosphates(s) acting to remove the phosphates. Thus, KARs are rather reporters for phosphorylation/dephosphorylation balance regarding specific protein-kinases. Specificity of the phosphorylation/dephosphorylation site is assessed usually through the use of pharmalogical compounds, whose specificities are always subjected to caution.
Genetically-encoded kinase sensors offer also the opportunity to drive their expression towards a specific compartment. Though most sensors are diffusible through the cytoplasm and nucleus, one might be tempted to add specific sequence to address the sensor either in the cytoplasm or the nucleus, as it was done for Mitogen-Activated Protein Kinase (MAPK) Erk activity reporters [8], or to direct sensors towards more local structures such as microtubules [9,10]. Noteworthy, one cannot discard that forcing location of sensors or addition of an addressing sequence could alter the sensor FRET intrinsic properties, and remodel the endogenous distribution and availability of the kinases docked to the sensor.
Choosing the Adequate KAR
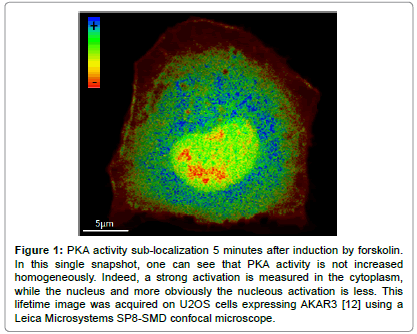
Nevertheless, the observation of a balance of kinase/phosphatase activity by microscopy in living cells can brought accurate insights into cellular processes, without holding an exhaustive approach of a pathway [11]. As an example, Protein Kinase A (PKA) is a pleiotropic protein kinase involved in a variety of cellular processes. Activation is provided by the binding of four molecules of cAMP to the regulatory sub-units, driving a conformational change releasing an active dimer of catalytic sub-units [11]. An holistic approach of the PKA pathway would lead to consider the pathway's regulation in each cell type, to measure the flux and compartmentalization of cAMP within cells and consider activities of several PKA targets. As a first step to intend to unravel the regulation of PKA activity in space and time during cell cycle and cellular reorganization at the onset of mitosis, we measured PKA activity using the AKAR variants (Protein Kinase A Kinase Activity Reporters). An example of sublocalization is presented in Figure 1. We highlighted in HeLa cells a substantial increase of PKA activity during mitosis, which ends with telophase. Spatially, hot spots of high levels of PKA activity were revealed for the first time near to the chromosomal plate during metaphase and anaphase. This observation was correlated to increased rates of chromosomes misalignment at the spindle during metaphase that could result in aneuploidy, when PKA was impaired [8]. Another approach could have been to monitor cAMP level using sensors such as tEPACvv [12,13]. However, one can expect that while cAMP has different targets and can be localized in microdomains by phospho-diesterase while PKA is sub localized by anchoring proteins (AKAP) the response could have been different. Therefore, while belonging to the same pathway each element of the cascade can be involved in several cell functions and can thus be regulated differently in space and time.
Figure 1: PKA activity sub-localization 5 minutes after induction by forskolin. In this single snapshot, one can see that PKA activity is not increased homogeneously. Indeed, a strong activation is measured in the cytoplasm, while the nucleus and more obviously the nucleous activation is less. This lifetime image was acquired on U2OS cells expressing AKAR3 [12] using a Leica Microsystems SP8-SMD confocal microscope.
Thus, one should keep in mind that sensors are focused on a specific element and interpretation has to be made accordingly. One has also to consider the complexity of pathways where PKA plays as a node of the network. Monitoring the phosphorylation of one particular sequence within the sensors will not necessarily reflect its involvement in all functions of the considered kinase. Also, the peptide substrate has to be chosen carefully, according to the specificity of the kinase, if known.
Discarding Artifacts: Chemical Inhibitors and Dead Reporters
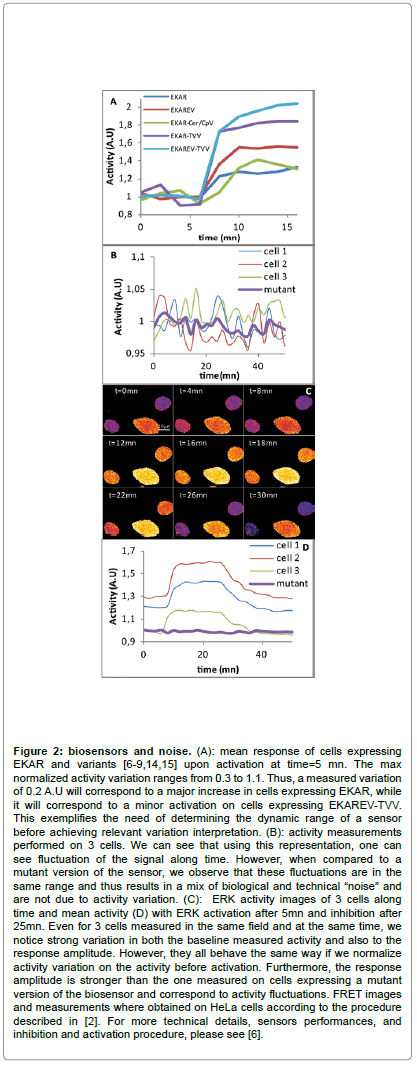
Fanciness of genetically-engineered kinase(s) reporters toolbox shall not occult the fact that insights are gained and only validated through the use of adequate controls. An unhandy idea is that sensors response must be carefully analyzed. Indeed, what you see is not to be believed, because what you see is not what you get as a response at the end of the analysis [14,15]. Many parameters shall be taken in account regarding the sensor itself such as level of expression, kinetics and dynamic range. Furthermore, cells are noisy-crowded environments, from which it remains delicate to extract the specifically attended signal from the biosensor. “Cellular noise” arise randomly from variable biophysical characteristics of cells and variable quantities resulting i.e. from differences in expression levels, ion concentration, auto-fluorescence or pH to just mention a few of them. To get rid of artifact in an adequate manner, one might first use, if available, drugs that can activate and impair protein activity in order to determine the dynamic range of a sensor and thus to discriminate a real activity from this biological “noise” (Figure 2). Being mandatory, a dead-reporter must be used for any genetically-encoded kinase reporter: the dead reporter is a construct where a point mutation is made up to replace the phosphorylable residue by a non phosphorylable one, thereby suppressing any possible change in FRET signal. In case of PKA sensor, we built a dead or inactive form, AKAREV T>A, by replacing the phosphorylable Threonine by an Alanine. In such conditions, no conformational changes into the sensor structure were attended to occur, and no aberrant and/or irregular variations were expected, providing a control "baseline" [6].
Figure 2: biosensors and noise. (A): mean response of cells expressing EKAR and variants [6-9,14,15] upon activation at time=5 mn. The max normalized activity variation ranges from 0.3 to 1.1. Thus, a measured variation of 0.2 A.U will correspond to a major increase in cells expressing EKAR, while it will correspond to a minor activation on cells expressing EKAREV-TVV. This exemplifies the need of determining the dynamic range of a sensor before achieving relevant variation interpretation. (B): activity measurements performed on 3 cells. We can see that using this representation, one can see fluctuation of the signal along time. However, when compared to a mutant version of the sensor, we observe that these fluctuations are in the same range and thus results in a mix of biological and technical “noise” and are not due to activity variation. (C): ERK activity images of 3 cells along time and mean activity (D) with ERK activation after 5mn and inhibition after 25mn. Even for 3 cells measured in the same field and at the same time, we notice strong variation in both the baseline measured activity and also to the response amplitude. However, they all behave the same way if we normalize activity variation on the activity before activation. Furthermore, the response amplitude is stronger than the one measured on cells expressing a mutant version of the biosensor and correspond to activity fluctuations. FRET images and measurements where obtained on HeLa cells according to the procedure described in [2]. For more technical details, sensors performances, and inhibition and activation procedure, please see [6].
Prospective
In conclusion, patience, caution and humility are still requested to interpret signals changes in cells populations where inter-cellular variability exists. Cell by cell analysis enabled by genetically encoded kinases reporters provides pertinent tools to unravel the kinasesignatures associated to cellular decision. Thus, cells behaviors can be addressed in a more subtle manner. Data observed might either exclude a kinase activity from truly being associated together with a cellular mechanism or decision, and/or open the question that the studied phenomenon may rely on noise-utilizing information process, or either characterize a specific protein-kinase activity signature with a cell fate, in a noise-robust context.
Acknowledgements
Pauline Vandame was granted by the University of Lille 1 and the Region Nord- Pas-de-Calais. We thank the personal of the BICeL-Lille1-HB Facility for access to the microscopy systems and technical advices. We also thank, Jin Zhang, Karel Svoboda, Jun-ichi Miyazaki, and Kees Jalink (The Nederlands Kanker Instituut) as sources for material cited in this article.
References
- Kamioka Y, Sumiyama K, Mizuno R, Sakai Y, Hirata E, Kiyokawa E, Matsuda M (2012) Live imaging of protein kinase activities in transgenic mice expressing FRET biosensors. Cell StructFunct 37:65-73
- Vandame P, Sipieter F, Spriet C, Leray A, Vincent P, Trinel D, Bodart JF, Riquet FB, Heliot L (2013) From FRET imaging to practical methodology for kinase activity sensing in living cells. Prog Mol Biol Transl Sci 113:145-216
- Li IT, Pham E, Truong K (2006) Protein biosensors based on the principle of fluorescence resonance energy transfer for monitoring cellular dynamics. BiotechnolLett 28:1971-82.
- Newman RH, Zhang J (2014) The Design and Application of Genetically-encodable Biosensors based on Fluorescent Proteins. Method Mol. Biol1071:1-16.
- Zhang J, Allen MD (2007) FRET-based biosensors for protein kinases: illuminating the kinome. Mol Biosyst 3:759-65.
- Vandame P, Spriet C, Riquet F, Trinel D, Cailliau-Maggio K, Bodart JF (2013) Optimization of ERK activity biosensors for both ratiometric and lifetime FRET measurements. Sensors (Basel) 14:1140-54
- Komatsu N, Aoki K, Yamada M, Yukinaga H, Fujita Y, et al. (2011) Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol Biol Cell 22:4647-56
- Vandame P, Spriet C, Trinel D, Gelaude A, Caillau K,et al. (2014) Thespatio-temporal dynamics of PKA activity profile during mitosis and its correlation to chromosome segregation. Cell Cycle 13:3232-40.
- Tan L, Kapoor TM (2011) Examining the dynamics of chromosomal passenger complex (CPC)-dependent phosphorylation during cell division. ProcNatlAcadSci108:16675-80.
- Harvey CD, Ehrhardt AG, Cellurale C, Zhong H, Yasuda R, et al. (2008) A genetically encoded fluorescent sensor of ERK activity. Proc. Nat. Acad. Sci105:19264–19269.
- Pearce LR, Komander D, Alessi DR (2010) The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol 11:9-22.
- Allen MD, Zhang J (2006) Subcellular dynamics of protein kinase A activity visualized by FRET-based reporters. Biochem Biophys Res Commun 348:716-721.
- Klarenbeek JB, Goedhart J, Hink MA, Gadella TW, Jalink K (2011) A mTurquoise-based cAMP sensor for both FLIM and ratiometric read-out has improved dynamic range PLoS One 6: e19170.
- Pouvreau S (in press) Spatiotemporal mapping of PKA activity using biosensors. Cell Cycle.
- Niwa H, Yamamura K ,Miyazaki J (1991) Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108: 193-200.
Relevant Topics
- Amperometric Biosensors
- Biomedical Sensor
- Bioreceptors
- Biosensors Application
- Biosensors Companies and Market Analysis
- Biotransducer
- Chemical Sensors
- Colorimetric Biosensors
- DNA Biosensors
- Electrochemical Biosensors
- Glucose Biosensors
- Graphene Biosensors
- Imaging Sensors
- Microbial Biosensors
- Nucleic Acid Interactions
- Optical Biosensor
- Piezo Electric Sensor
- Potentiometric Biosensors
- Surface Attachment of the Biological Elements
- Surface Plasmon Resonance
- Transducers
Recommended Journals
Article Tools
Article Usage
- Total views: 14682
- [From(publication date):
June-2015 - Apr 04, 2025] - Breakdown by view type
- HTML page views : 10071
- PDF downloads : 4611