Genetic Relationship of Rhododendron ripense Makino to Japanese Evergreen Azalea Cultivars Evaluated by SSR Markers
Received: 09-Nov-2017 / Accepted Date: 25-Nov-2017 / Published Date: 11-Dec-2017
Abstract
Evergreen azalea cultivars as the important ornamental shrubs and pot plants have been developed based on the genetic resources of Japanese wild azaleas. Rhododendron ripense Makino is one of the representative wild azalea species endemic to West Japan and the ancestral parent of evergreen azalea cultivars. In this study, we focused on this large flowered riverside azalea and the genetic contribution to azalea cultivars was evaluated by means of SSR markers. The results of PCoA using SSR data made plot distributions, which would reflect the taxon and genetic relationship between cultivar groups and their ancestral species. All individuals of R. ripense which classified in subsection Scabra makes distribution group apart from R. kaempferi and R. kiusianum group which classified in subsection Tsutsusi. The cultivars of R. ripense, Ryukyu azalea (R. × mucronatum) and ‘Oomurasaki’ (R. × pulchrum) are all included in this R. ripense plot group. These results of this study suggested the important genetic contribution of R. ripense to the development of these azalea cultivars.
Keywords: Ancestral parent; Breeding history; Evergreen azalea; Genetic resources
Introduction
In the genus Rhododendron (Ericaceae), the species in the subgenus Tsutsusi , section Tsutsusi , are important genetic resources for evergreen azaleas used as ornamental shrubs or pot azaleas in the world. Among the native azalea species in Japan, such as R. kaempferi , R. kiusianum, R. indicum, R. eriocarpum, R. macrosepalum and R. ripense have the high ornamental value. By means of mutation and hybrid selection from the natural populations of these wild azaleas, hundreds of azalea cultivars have been developed since the Edo era (1603-1867) [1]. The development of each cultivar groups, such as Edo Kirishima, Ryukyu, Hirado, Satsuki and Kurume are the fruits of Japanese horticulture history [2,3].
Rhododendron ripense Makino belongs to subgenus Tsutsusi ; section Tsutsusi subsection Scabra [4], distributes in rocky riverside areas of the Chugoku and Shikoku areas of West Japan [5]. This Japanese endemic species has been considered to be the important ancestral parent of evergreen azalea cultivars, especially, Ryukyu azalea (R. × mucronatum) and ‘Oomurasaki’ (R. × pulchrum) [6]. And also, this large flowered azalea is one of the best ornamental performance species with high adaptability of environmental conditions [7,8].
In this study, the genetic contributions of R. ripense to the development of Japanese azalea cultivars were evaluated by using SSR (Simple sequence repeat) markers. With the comparison of the genetic distance calculated by SSR genotypes, the genetic relationship of R. ripense to azalea cultivars and also to other species was investigated.
Materials and Methods
Plant materials
Azalea species and cultivars used in this study were collected from wild habitat in Japan and/or the collection of Niigata Prefectural Botanical Garden, and the Plant Breeding Laboratory of Shimane University. They included 73 individuals of R. ripense from 23 wild habitats of 12 rivers in the Chugoku (47) and Shikoku (26) areas (Figure 1). And also 45 individuals of R. kaempferi , 19 individuals of R. kiusianum, 15 individuals of R. eriocarpum and 25 individuals of R. macrosepalum were used. Furthermore, 53 cultivars from the cultivar groups of Ryukyu azalea (R. × mucronatum), Ookirishima (R. × pulchrum), Kurume azalea (R. × obtusum), R. macrosepalum and R. ripense were analyzed.
SSR marker analysis
Total genomic DNA was extracted from -80°C freeze fresh leaves of each plant by modified CTAB methods [9]. Each DNA samples were genotyped by using 3 pairs of SSR primers (AZA002, AZA003 and AZA008) [10,11]. PCR amplification was carried out with a thermal cycler (ASTEC, PC320). The size of fragments was determined by ABIPRISM310 genetic analyzer and GeneScan™ programs. To evaluate the characteristics of each wild azalea species, the mean number of alleles per locus (NA), observed heterozygosity (HO), expected heterozygosity (HE), and total number of private alleles (PA) were calculated using GenAlEx 6.503 [12]. Genetic relationships between all individuals of the wild species and cultivars were evaluated by a principal co-ordinate analysis (PCoA) of codominant genotypic distance [13] using GenAlEx 6.503 to visualize the relationship among the accessions in a scatter-plot.
Results and Discussion
In SSR analysis using 3 SSR primer sets, totally 78 alleles including polymorphic patterns were obtained in all samples. With the primer set of AZA002, 34 alleles including 5 common alleles between species were detected in the range of 139-295bp. And the primer sets of AZA003 and AZA008 detected 17 alleles in the range of 144-184bp, 23 alleles in the range of 152-239bp, respectively. The genetic diversity parameters are shown in Table 1. The genetic diversity observed in the five wild species differed among species, as indicated by the expected heterozygosity (HE) which ranged from the Chugoku area of R. ripense (0.539) to R. macrosepalum (0.898). Among five wild species, R. ripense , R. macrosepalum and R. kaempferi harbored from 3 to 14 private alleles.
| N | NA | HO | HE | PA | ||
| R. ripense | Chugoku area | 47 | 9.33 | 0.376 | 0.539 | 3* |
| Shikoku area | 26 | 8.00 | 0.705 | 0.589 | 3* | |
| R. macrosepalum | 25 | 16.33 | 0.800 | 0.898 | 14 | |
| R. kaempferi | 45 | 16.33 | 0.733 | 0.890 | 7 | |
| R. kiusianum | 19 | 10.33 | 0.702 | 0.747 | 0 | |
| R. eriocarpum | 15 | 5.67 | 0.511 | 0.627 | 0 |
Table 1: Population genetic parameters estimated from 3 SSR markers for 5 azalea species. N total number of analysed samples, NA means number of alleles per locus, HO observed heterozygosity, HE expected heterozygosity, PA total number of private alleles.
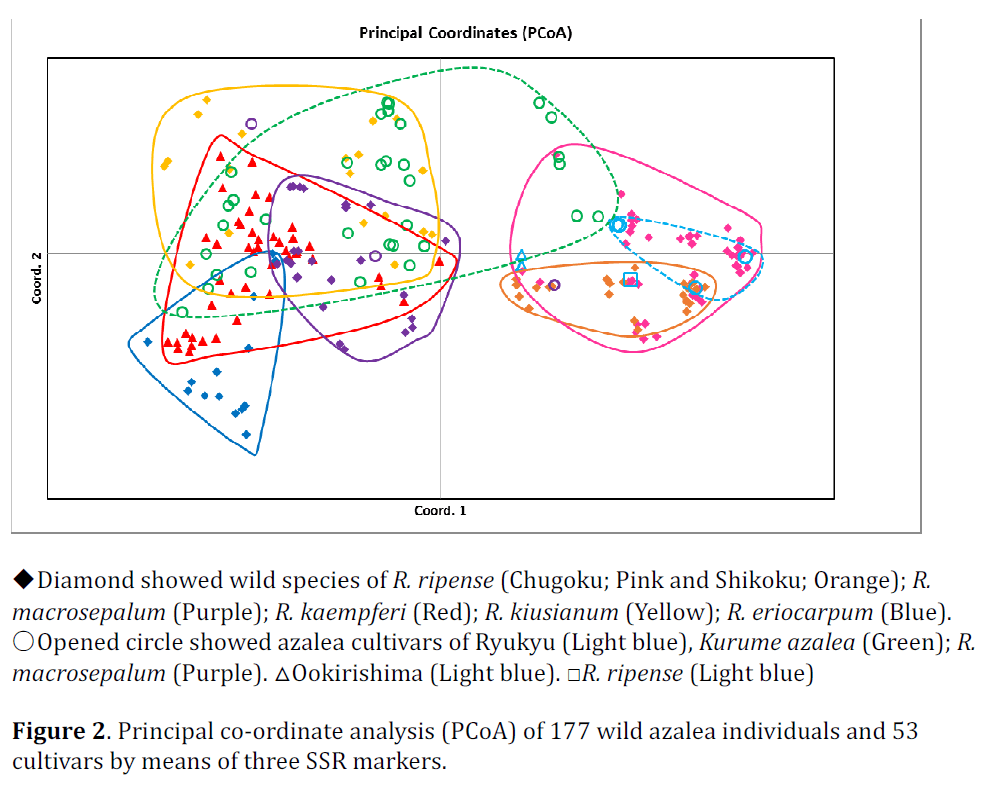
PCoA using SSR data of wild species and cultivars made the plot distributions which would reflect the taxon and genetic relationship between cultivar groups and their ancestral species (Figure 2). Individuals of R. kaempferi and R. kiusianum which classified in subsection Tsutsusi series Kaempferia made overlap distribution, and most of Kurume azalea cultivars were also overlapped in this group. The breeding history of this small flower cultivar group which had developed from these two species was corresponding to these distribution patterns. R. eriocarpum classified in subsection Tsutsusi and R. macrosepalum classified in subsection Scabra also located in the vicinity of these groups.
◆Diamond showed wild species of R. ripense (Chugoku; Pink and Shikoku; Orange); R. macrosepalum (Purple); R. kaempferi (Red); R. kiusianum (Yellow); R. eriocarpum (Blue). ○Opened circle showed azalea cultivars of Ryukyu (Light blue), Kurume azalea (Green); R. macrosepalum (Purple). △Ookirishima (Light blue). □R. ripense (Light blue)
Figure 2: Principal co-ordinate analysis (PCoA) of 177 wild azalea individuals and 53 cultivars by means of three SSR markers.
All individuals of R. ripense which classified in subsection Scabra makes distribution group apart from other species, and the population of the Shikoku area was gathered inside of the population of Chugoku area. R. ripense ‘Wakasagi’, 6 cultivars of Ryukyu azalea, 4 cultivars of Ookirishima, and 2 presumed cultivars of R. macrosepalum are all included in this large flowered R. ripense group. These results back up the important genetic contribution of R. ripense to the development of Ryukyu azalea (R. × mucronatum) and ‘Oomurasaki’ (R. × pulchrum) cultivars. And the results of AFLP marker analysis also supported these genetic relationship [14,15]. Six cultivars of Kurume azalea groups plotted inside or the vicinity of R. ripense distribution area. These distributions also support the genetic relationship between R. ripense and some of Kurume azalea cultivars because of the similarity leaf surface morphology [16].
As a result of this study, all cultivars of Ryukyu azalea and Ookirishima groups and a part of Kurume azalea cultivars were included in the R. ripense plot distribution of PCoA using SSR data. The genetic contribution of R. ripense to the development of some of Japanese azalea cultivars was suggested by SSR marker analysis. The detail of the genetic relationship between species and cultivars will be explicated by the future investigation with using other SSR markers reported by De Keyser [17]. And the investigation of functional gene related to the important ornamental trait; flower color and flower shape [18] will be informative for the study of development history of cultivars from wild azaleas.
References
- Ito I, Creech JL (1984) A Brocade Pillow: Azaleas of Old Japan. Weatherhill, New York and Tokyo.
- Kobayashi N, Handa T, Miyajima I, Arisumi K, Takayanagi K (2007) Introgressive hybridization betweenRhododendron kiusianum and R. kaempferi (Ericaceae) in Kyushu, Japan based on chloroplast DNA markers. Edinburgh Journal of Botany 64: 283-293.
- Kobayashi N (2013) Evaluation and application of evergreen azalea resources of Japan. ActaHorticulturae 990: 213–219.
- Yamazaki T (1996) A revision of the genus Rhododendron in Japan, Taiwan, Korea, and Sakhalin. Tokyo, Tsumura Laboratory.
- Kobayashi N, Oji M, Ureshino K, Nakatsuka A, Hosoki T (2008) Evaluation of genetic resources of Rhododendronripense Makino in San-in area, Japan, based on morphological characters and chloroplast DNA polymorphism. Horticultural Research (Japan) 7: 181-187.
- Inobe T (1971) A study of the autogenesis of RhododendronhortenseNakai. The Journal of Japanese botany. 46: 311-315.
- Kobayashi N, Morita T, Miyazaki M, Adachi F, Ban T (2010) Root characteristics of evergreen azaleas—root development of transplanted plants—. Horticultural Research (Japan) 9: 1-5.
- Scariot V, Caser M, Kobayashi N (2013) Evergreen azaleas tolerant to neutral and basic soils: breeding potential of wild genetic resources. Acta Horticulturae 990: 287–291.
- Kobayashi N, Horikoshi T, Katsuyama H, Handa T, Takayanagi K (1998) A simple and efficient DNA extraction method from the plants, especially from woody plants. Plant Tissue Culture and Biotechnology 4: 76-80.
- Dendauw J, De Riek J, Arens P, Van Bockstaele E, Vosman B, et al. (2001) Development of sequenced tagged microsatellite site (STMS) markers in azalea. Acta Horticulturae 546: 193-197.
- Scariot V, De Keyser E, Handa T, De Riek J (2007a) Comparative study of the discriminating capacity and effectiveness of AFLP, STMS and EST markers in assessing genetic relationships among evergreen azaleas. Plant Breeding 126: 207-212.
- Peakall R, Smouse PE (2012) GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28: 2537-2539.
- Smouse PE, Peakall R (1999) Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity 82: 561-573.
- Ueno M, Kita K, Kurashige Y, Yukawa T, Handa T (2006) Estimation of the origin of Ryukyu azalea ‘Shiroryukyu’, Ookirishima azalea ‘Oomurasaki’ and ‘Akebono’ by AFLP markers. Jour Japan Soc Hort Sci 75 (Suppl.1): 179.
- Scariot V, Handa T, De Riek J (2007b)A contribution to the classification of evergreen azaleacultivars located in the Lake Maggiore area (Italy)by means of AFLP markers. Euphytica 158: 47–66.
- Okamoto A, Nonaka M, Suto K (2000) Characteristics of dorsal leaf surface, implicating the relationship between Ryukyu azalea [Rhododendron mucronatum (Blume) G. Don] and Kurume azalea (R. obtusum (Lindley) Planch). Jour Japan Soc Hort Sci 69: 103-108.
- De Keyser E, ScariotV, Kobayashi N, Handa T, De Riec J(2010) Azalea phylogeny reconstructed by means of molecular techniques. Protocols for In Vitro Propagation of Ornamental Plants: 349-364. Series: Methods in Molecular Biology, Vol. 589.
- Nakatsuka A, Mizuta D, Kii Y, Miyajima I, Kobayashi N (2008) Isolation and expression analysis of flavonoid biosynthesis genes in evergreen azalea. Scientia Horticulturae 118: 314-320.
Citation: Kobayashi N, Sugai K, Tsuji T, Nakatsuka A (2017) Genetic Relationship of Rhododendron Ripense Makino to Japanese Evergreen Azalea Cultivars Evaluated by SSR Markers. J Plant Genet Breed 1: 101
Copyright: ©2017 Kobayashi N, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Share This Article
Open Access Journals
Article Usage
- Total views: 4571
- [From(publication date): 0-2017 - Dec 20, 2024]
- Breakdown by view type
- HTML page views: 3836
- PDF downloads: 735