Research Article Open Access
Genetic Engineering and Improvement of a Zymomonas mobilis for Arabinose Utilization and Its Performance on Pretreated Corn Stover Hydrolyzate
Yat-Chen Chou*, Jeffrey Linger, Shihui Yang and Min ZhangNational Bioenergy Center, National Renewable Energy Laboratory, Golden, CO 80401, USA
- Corresponding Author:
- Yat-Chen Chou
National Bioenergy Center
National Renewable Energy Laboratory
15013 Denver West Parkway Golden, CO 80401,USA
Tel: 3033847766
E-mail: yat.chen.chou@nrel.gov
Received date:: March 18, 2015; Accepted date:: April 21, 2015; Published date:: April 28, 2015
Citation: Chou YC, Linger J, Yang S, Zhang M (2015) Genetic Engineering and Improvement of a Zymomonas mobilis for Arabinose Utilization and Its Performance on Pretreated Corn Stover Hydrolyzate. J Biotechnol Biomater 5:179. doi:http://dx.doi.org/10.4172/biotechnology-biomaterials.1000179
Copyright: © 2015 Chou YC, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Visit for more related articles at Journal of Biotechnology & Biomaterials
Abstract
A glucose, xylose and arabinose utilizing Zymomonas mobilis strain was constructed by incorporating arabinose catabolic pathway genes, araBAD encoding L-ribulokinase, L-arabinose isomerase and L-ribulose-5-phosphate- 4-epimerase in a glucose, xylose co-fermenting host, 8b, using a transposition integration approach. Further improvement on this arabinose-capable integrant, 33C was achieved by applying a second transposition to create a genomic knockout (KO) mutant library. Using arabinose as a sole carbon source and a selection pressure, the KO library was subjected to a growth-enrichment process involving continuous sub-culturing for over 120 generations. Strain 13-1-17, isolated from such process demonstrated significant improvement in metabolizing arabinose in a dilute acid pretreated, saccharified corn stover slurry. Through Next Generation Sequencing (NGS) analysis, integration sites of the transposons were identified. Furthermore, multiple additional point mutations (SNPs: Single Nucleotide Polymorphisms) were discovered in 13-1-17, affecting genes araB and RpiB in the genome. We speculate that these mutations may have impacted the expression of the enzymes coded by these genes, ribulokinase and Ribose 5-P-isomerase, thus attributing to the improvement of the arabinose utilization.
Keywords
Zymomonas mobilis; Arabinose; araBAD; Transposition; Knockout library; Cre-Lox recombination; Next generation sequencing (NGS)
Introduction
Z. mobilis has been recognized as a viable alternative to yeast for ethanol production from biomass for its high conversion yield, high ethanol tolerance, resistance to pretreated hardwood hydrolyzate and fermentation capability at lower pH and in anaerobic conditions [1]. However, the narrow substrate utilization, capable of only using glucose, fructose and sucrose, hinders the application of this organism in the ethanol production from lignocellulosic biomass feedstock. Our laboratory has successfully constructed a glucose/ xylose co-utilizing strain, Z. mobilis 8b, based on ZM4 (ATCC31821), using homologous recombination and transposition integration methods [2]. Moreover, strain 8b demonstrates superior ethanol production performance in a pretreated corn stover hydrolyzate [3].
Within the context of process economics for cellulosic ethanol, it is imperative to maximize sugar utilization. Given that arabinose comprises approximately 5% of the available sugars within corn stover [4], any organism that is unable to utilize this sugar will automatically leave valuable sugars (and potential end-product) behind. Specifically, NREL’s 2011 design report for biochemical conversion of lignocellulosic biomass to ethanol [4] assumes an 85% conversion of arabinose to ethanol in order to meet the aggressive 2012 cost targets of $2.15/gal in year 2007 dollars. Importantly, achieving this 85% conversion reduces the MESP by $0.08/gal. Therefore, we sought to generate an efficient arabinose utilizing strain of Z. mobilis.
Previous genetic engineering work has implemented arabinose utilization capabilities within Z. mobilis strain ATCC39676 [5]. However, its arabinose usage and coordinate ethanol production were limited. We hypothesized that part of this could be due to the strain background used, as ATCC39676 generally seems to be inferior to strains based on ZM4 with regards to ethanol production. For example ZM4-based glucose/xylose co-utilizing, plasmid-bearing strains have proven superior as it relates to ethanol production from oat hull hydrolyzate [6] and resistance to acetic acid inhibition [3,6] as well as elevated temperature [3]. More recently, DuPont has also constructed an arabinose/xylose co-utilizing strain based on the ZM4 host and improved its arabinose utilization by the introduction of an arabinoseproton symporter [7].
We report here the generation of arabinose-utilizing strains of Z. mobilis with superior performance in lab pure sugar media as well as a corn stover derived biomass. These strains were generated through the use of rational genetic engineering, transposon based mutagenesis, and selection/enrichment in arabinose medium. We show these strains are superior not only in the usage of arabinose in defined rich media, but also in process relevant conditions using saccharified pre-treated corn stover as the nutrients source when compared with their parental strain, 8b introduced with an arabinose pathway genes (namely, Strain 33C). We further define the underlying genetic alterations that have occurred within these strains and hypothesize about their roles in increasing arabinose utilization for future metabolic engineering to further enhance arabinose utilization.
Materials and Methods
Bacterial strains and growth conditions
All bacteria and plasmids used in this study are listed in Table 1. Escherichia coli DH5α and SCS110 were cultured in LB medium at 37ºC with shaking (200 rpm) and Z. mobilis strains were cultured microaerophilically (either statically or with gentle shaking) in rich media (“RM”: 10 g/L yeast extract, 2 g/L KH2PO4) supplemented with glucose or arabinose (“RMG” or “RMA,” respectively). For regeneration and selection of Z. mobilis transformants or integrants, mating medium (“MMG”; 10 g/L yeast extract, 5 g/L tryptone, 2.5 g/L (NH4)2SO4, 0.2 g/L K2HPO4 and 50 g/L glucose) was used. All agar media were made with 15 g/L agar. Appropriate antibiotics were added to the media when needed: Spectinomycin (“Sp”, 200 μg/mL for Z. mobilis and E. coli), ampicillin (“Amp”, 100 μg/mL) and kanamycin (“Kan”, 50 μg/mL).
| Strain or plasmid | Description | Reference | |
|---|---|---|---|
| Strain | |||
| Escherichia coli | DH5α | F– Φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rK–, mK+) phoAsupE44 λ– thi-1 gyrA96 relA1 |
Invitrogen |
| SCS110 | rpsL (Str-r) thrleuendA thi-1 lacYgalKgalTara tonAtsx dam dcm supE44 Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15] |
Agilent Technologies |
|
| Zymomonasmobilis | 8b | ZM4 integrated with Tc-PgapxylAB/Cm-Penotaltkt; glucose/xylose utilizing |
our lab; [2] |
| Strain 33 | 8b integrated with loxP-aadA-loxP-Pgap-araBAD; glucose/xylose/arabinose utilizing |
this study | |
| 33C | aadAfree derivative of Strain 33; glucose/xylose/arabinose utilizing | this study | |
| 13-1-17 | isolate from 33C genomic KO library; glucose/xylose/arabinose utilizing | this study | |
| 15-1-34 | isolate from 33C genomic KO library; glucose/xylose/arabinose utilizing | this study | |
| Plasmid | pMOD2Sp | Ampr,Spr; integrative plasmid containing ME-loxP-aadA-loxP-ME for transposition and KO library construction; non-replicative in Z. mobilis |
this study |
| pMOD2Sploxara | Ampr,Spr; integrative plasmid containing ME-loxP-aadA-loxP-PgaparaBAD- ME for transposition; non-replicative in Z. mobilis |
this study | |
Table 1: Bacterial strains and plasmids.
Recombinant DNA techniques
Plasmid DNA isolation, restriction endonuclease digestion, ligation and transformation, agarose electrophoresis and other standard recombinant DNA techniques were carried out according to the manufacturers’ recommendations. Plasmid DNA extraction was carried out using Qiaprep Spin Miniprep kits and gel extraction with Qiaquick kits (Qiagen, CA).
Construction of plasmids and linear DNA for transposition integration
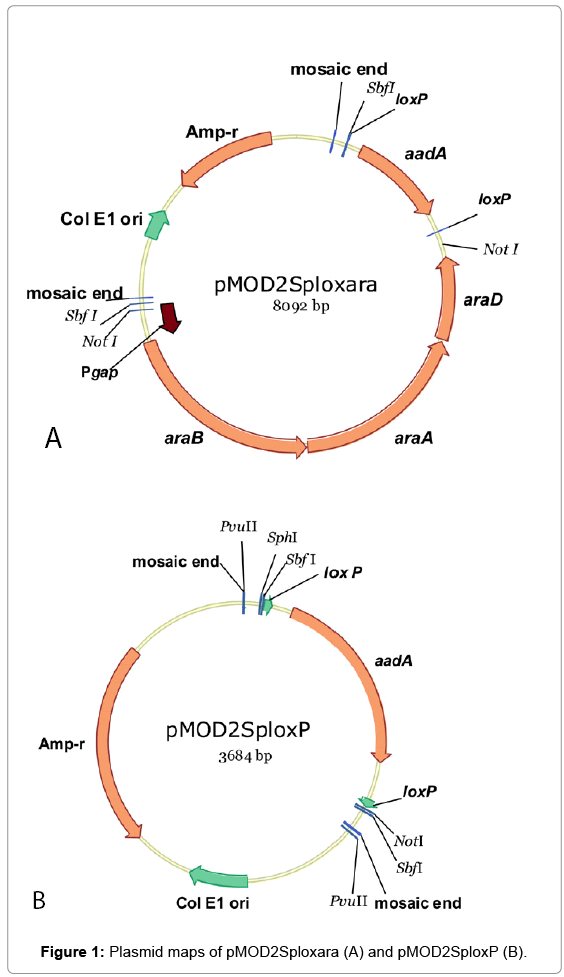
An EZ-Tn5TM transposon construction vector (non-replicative in Zymomonas), pMOD2 (Epicentre, WI) was used to construct plasmids for the introduction of arabinose catabolic pathway genes or transposition mutagenesis. Mosaic ends (ME) flanking a multiple cloning sites in pMOD2 are designed for the cloning and production of transposons. Plasmid pMOD2Sploxara (Figure 1A) was assembled by cloning in SbfI site of pMOD2 a DNA fragment containing two elements: 1) Pgap-araBAD operon [5], and 2) aadA gene (conferring spectinomycin resistance; GI:125380577/DQ975275.1) [8] flanked by loxP sites (loxP-aadA-loxP). The loxP sites were used in conjunction with a plasmid-borne copy of Cre recombinase for the removal of aadA from the genomes of integrants when required. Plasmid pMOD2Splox (Figure 1B) was constructed by NotI digestion of pMOD2Sploxara followed by self-ligation to remove the Pgap-araBAD.
Transposition integration of loxP-aadA-loxP-Pgap-araBAD or ME-loxP-aadA-loxP-ME in Z. mobilis 8b and removal of aadA in the integrants
A commercially available system, EZ::TNTM TransposomeTM Kit (Epicentre, WI) [9] was used to generate transposomes (a DNAtransposase complex or specifically “transposase bound ME - loxP - aad AloxP - Pgap-ara BAD - ME or ME - lox P - aadA - loxP - ME”) for initiating the transposition in Z. mobilis. Integrative plasmid, pMOD2S ploxara, or PvuII - digested ME-loxP-aadA-loxP-ME (from pMOD2Splox) was treated with transposase at room temperature for one hour followed by dialysis against 5% glycerol and 5 mM Tris - HCl (pH7.8) before electroporation/transformation in Z. mobilis 8b [2]. A cre-loxP sitespecific recombination [10] was used to remove the aadA from the integration site in the genome by expressing a Cre recombinase gene (cre) on a plasmid in the integrants.
Transformation of Z. mobilis
Z. mobilis or E.coli cells were transformed with plasmids by electroporation (Bio-Rad Gene Pulser, 0.1-cm gap cuvettes, 1.6 kV, 200 ohms, 25 μFD). Electrocompetent Z. mobilis or E. coli cells were prepared by collecting cells at an optical density (OD600) = 0.4 to 0.6, washed once in ice-cold sterile water followed by 10% glycerol and concentrated approximately 1000-fold. Competent cells were stored at –80ºC as small aliquots for later use. Transformants of E. coli or Z. mobilis were selected on LB or MMG, agar plates, respectively, containing antibiotics. Due to the presence of a restriction/modification system, all plasmids were built in and isolated from a methylationdeficient E. coli strain, SCS110, for successful transformations in Z. mobilis ZM4 derived hosts, 8b or 33C.
Genomic Knockout (KO) library construction based on strain 33C using the transposition integration method
An aadA-free, arabinose utilizing strain, 33C, was further used to construct a genomic library with non-specific insertion of the aadA gene by transposition. A transposome (PvuII digested ME-loxP-aadAloxP- ME fragment of pMOD2Splox, bound by transposases) was used to transform strain 33C by electroporation. Colonies that appeared on MMG plates with Sp were collected by scraping and rinsing the plates with a small volume of RM with 5% glucose and Sp. Collected cells were added with 1/3 volume of the same medium and incubated at 33ºC for 1 h before aliquoting and freezing in the presence of 20% glycerol. Multiple transpositions and transformations were conducted in order to obtain sufficient colonies to represent the coverage of a Z. mobilis genome.
Enrichment of 33C knockout (KO) library in medium containing arabinose
Frozen library cells were thawed and revived for 2 h at 35°C in 20-mL modified rich medium containing 10 g/L yeast extract, 2 g/L KH2PO4, 7.5 g/L glucose, 2.5 g/L L-arabinose, 1g/L MgSO4 and Sp200. The revived cultures were used to inoculate 100-mL the same medium containing different sugar composition (5 g/L glucose and 5 g/L L-arabinose) in shake flasks as seeds. Seeds were incubated at 35°C overnight before inoculating RMA5 (50 g/L L-arabinose, initial OD600 = 0.05) for the enrichment process. Growth was monitored by OD measurement. RMA5 grown cultures at mid to late exponential phase were diluted into fresh RMA5 daily (or every 2-3 days, dependent upon the growth rates) to enrich for the fast growers in arabinose. As a nonmutated control, 33C was subjected to the same enrichment process. Periodically, cultures were streaked on RMA5 agar plates for single colony isolation. Large colonies were selected and grown in RMA5 and preserved at -80°C for later evaluation.
Evaluation of arabinose utilizing Z. mobilis in mixed sugar medium in shake flasks
For down selection of the fast arabinose growers from the KO library, frozen stocks of single isolates from the enrichment process were evaluated in shake flasks containing RM supplemented with sugars – 1% glucose; 4% xylose; 1% arabinose (RMG1X4A1). Frozen stocks were inoculated in 3-mL RMG2 (2% glucose) for 6 hours at 33°C, then diluted into 20-mL RMG6 (6% glucose) as seeds and grown at 33°C for 16 hours with shaking. Seed cultures were concentrated by centrifugation before inoculating the mixed sugar medium (RMG1X4A1) in shake flasks (initial OD = 0.05). Fermentation was conducted at 33°C with 100 rpm shaking. The initial pH of the medium was 6.0 but was not controlled or monitored during the fermentation.
Pretreated, saccharified corn stover slurry (“PCS Saccharified Slurry”) production
For slurry A, the corn stover feedstock (with 34% total solids content) used in this study was knife-milled and de-acetylated using 0.4% NaOH (w/w) at 80°C followed by a dilute-acid pretreatment (0.5% H2SO4, 150°C, 20 minutes). Pretreated corn stover (PCS) was neutralized to pH 5.1 using 30% ammonium hydroxide. Enzymatic hydrolysis was performed using Novozymes’ Cellic CTec 2 at a loading of 40 mg protein/g cellulose at 48°C for 96 hours. Slurry B was produced without the de-acetylation step. For fermentation purposes, the slurries were supplemented with RM and diluted with water to an equivalence of 20% (for slurry A) or 15% (for slurry B) total solids (TS) in the original corn stover feedstock before the inoculation of cultures.
Evaluation of arabinose utilizing Z. mobilis in the saccharified slurries (20% and 15% total solids for slurries A and B, respectively)
Two strains selected from the mixed sugars fermentation were evaluated in fermentors (Sixfor) containing PCS saccharified slurry at 20% total solids (TS) for slurry A and 15% TS for slurry B. Frozen cultures were revived in 4-mL RMA5 or RMG5 for 15-16 h followed by inoculation in 20-mL RMG5 for 6 h as preseeds. Seeds were prepared by diluting the preseeds in 100-mL seed medium (RM with 10% glucose and 2% xylose) supplemented with 1 g/L sorbitol and grown for 17 -18 h before inoculating the fermentors. Fermentors were filled with 180 mL of saccharified slurry (containing RM), to which 20-mL seeds were added, bringing the TS concentration to approximately 18% (slurry A) and 13.5% (slurry B) for the fermentations. Temperature was maintained at 33°C and pH 5.8 with KOH. Initial OD was 1.0. Glucose, xylose and arabinose concentrations were measured by high performance liquid chromatography (HPLC) using an Agilent 1100 series HPLC (Santa Clara, CA) with a carbohydrate column (Shodex SP0810) at 85oC and a Bio-rad de-ashing guard p/n 125-0118. Mobile phase used was water and flow rate at 1 mL/min. Ethanol, acids and furfural were measured by the HPLC equipped with an organic acid column (Bio-rad HPX-87H) at 55°C and a Bio-rad H+guard p/n 125- 0129. Sulfuric acid (0.01N) was used as the mobile phase, at a flow rate of 0.6 mL/min. Refractive Index detector (RID) was used in both HPLC analyses.
Genomic resequencing through next-generation sequencing (NGS)
Genomic DNA was extracted using Qiagen DNAEasy Kit and sent to the University of Utah for next-generation sequencing using Illumina Hiseq2000 after passing the quality requirements. The quality of FASTQ genome resequencing data was checked using FastQC program (http://www.bioinformatics. babraham.ac.uk/ projects/fastqc/). Data passing the quality control was imported into CLC Genomics Workbench (version 5.1, CLCBio, Denmark) for data analysis to identity the potential small genetic changes such as SNPs (Single Nucleotide Polymorphisms) and Indels (Insertions/ Deletions). Briefly, sequencing reads were trimmed to remove reads with poor quality and then mapped against the reference genome sequence followed by resequencing analysis using both Probabilistic Variant Detection and Quality-based Variant Detection tools. The variants identified were filtered to remove those with less than 80% confident frequency level [11] before comparing different strains. In addition, the genomic loci of transposon insertion were determined through identifying the reads containing the transposon sequences and subsequently Blasting (BlastN, NCBI) the flanking sequences (of the transposon) against the reference genome, Z. mobilis ZM4 [12].
Results
Introduction of arabinose catabolic pathway in Z. mobilis 8b
To create a glucose/xylose/arabinose utilizing Z. mobilis strain, arabinose catabolic pathway genes from E. coli (araBAD, a threegene operon encoding L-ribulokinase, L-arabinose isomerase and L-ribulose-5-phosphate-4-epimerase) were introduced in the genome of a glucose/xylose utilizing host 8b [2] adopting a transposition method with a non-replicative plasmid pMOD2Sploxara (Figure 1A) and transposase. A native promoter of glyceraldehyde-3-phosphate dehydrogenase gene (ZMO0177;“Pgap”), isolated from Z. mobilis ZM4 was used to drive the expression of araBAD [5]. Antibiotic marker gene, aadA was included in the integration fragment for the selection of transformants/integrants. Multiple Spr integrants were isolated and subjected to screening processes in RMA medium (2% arabinose) in test tubes. In this initial screening/selection process, integrants exhibiting incremental improvement in growth rates when serially cultured in the same medium (Supplementary, Figure S1) were further used for single colony isolation on RMA plates (5% arabinose). One colony, namely “Strain 33” demonstrated the highest arabinose utilization in a preliminary shake flask evaluation in RMG0.8X4.5A0.6 (0.8%glucose, 4.5%xylose and 0.6% arabinose) (Supplementary, Figure S2). While Strain displayed the ability to utilize arabinose, the efficiency was lower than desired. Thus, we chose to use this strain as the base “chassis” for further genetic manipulation to improve the arabinose conversion efficiency. Due to the very limited antibiotic markers that are available to Zymomonas in general, we first created an aadA free derivative of Strain 33 taking advantage of the Cre-Lox recombination system. Two copies of short loxP sequence (flanking aadA) were built in the integration cassette for this very purpose. Marker gene, aadA was removed from the integration cassette, aadA- Pgap-araBAD, leaving only the Pgap-araBAD in the genome of Strain 33. The resultant aadA-free, arabinose-utilizing derivative was named 33C. A mixed sugar (glucose, xylose and arabinose) fermentation in shake flasks was conducted to compare the performance of strains 33 and 33C. Results confirmed the latter strain maintained the same fermentation capability as its parent pertaining to sugar utilization and ethanol yield (Supplementary, Figure S3).
Knockout library construction based on 33C and the selection/enrichment in arabinose medium
It has been reported that adaptation improved sugar utilization in Zymomonas [2,13] and Pseudomonas [14] after rational metabolic engineering. Therefore, we additionally reasoned that following implementation of arabinose metabolic genes, other un-specified genetic changes might be required to achieve optimal arabinose metabolism. The simple incorporation of heterologous genes might render native metabolic networks out of balance. Accordingly, we proposed using transposon based mutagenic method, which maximizes the diversity of genomic loci for the insertion (of an antibiotic marker gene) to potentially mutagenize any genes or regulatory elements in the genome. This library of mutants are then collectively subjected to continuous growth in the presence of arabinose to encourage the selection/enrichment of superior arabinose utilizing strains of Z. mobilis as compared with previous generations.
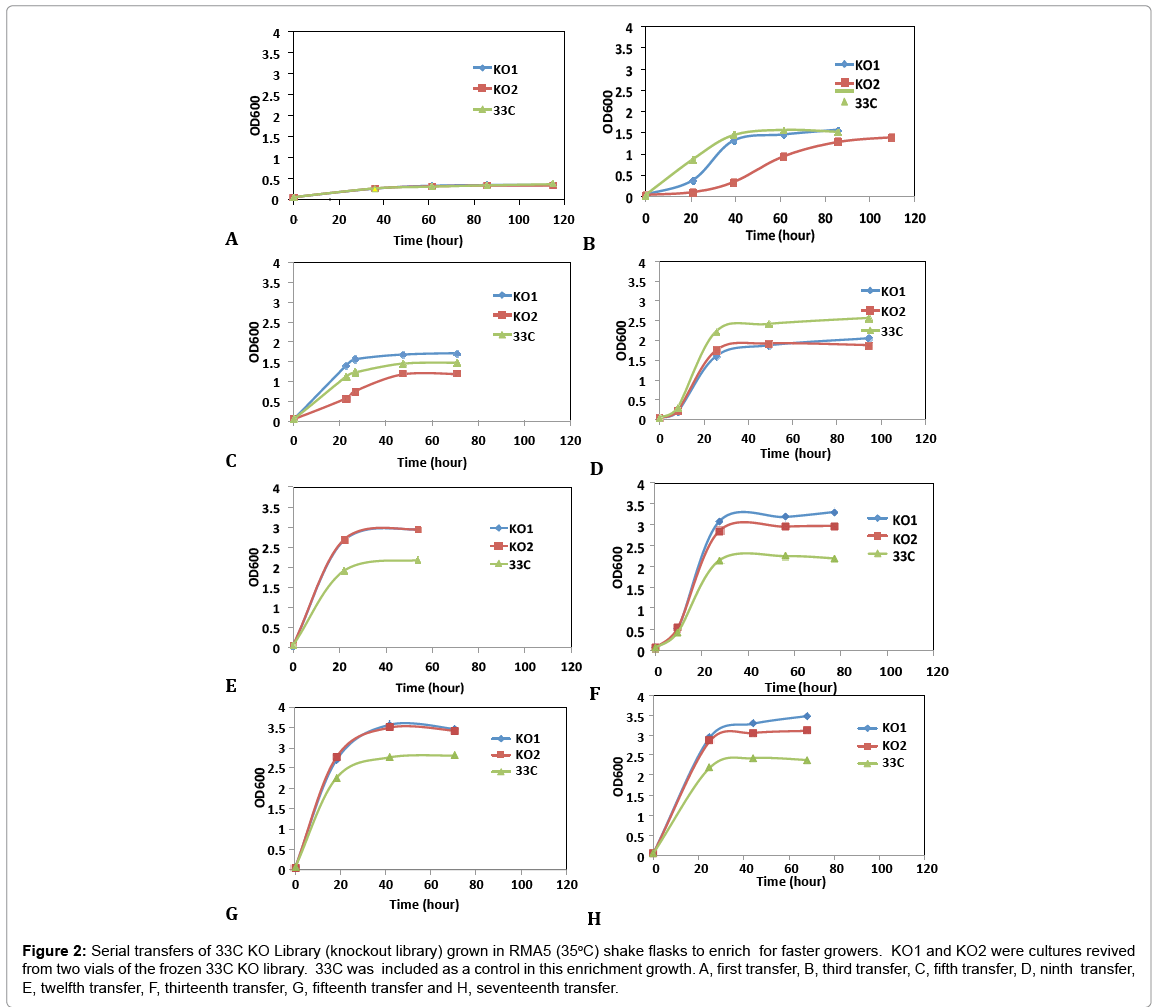
A second transposition process was deployed to the aadA-free, arabinose utilizing strain, 33C for the construction of a genomic knockout (KO) library to further improve on arabinose growth. This mutant library was generated by non-specific insertions of a transposome cassette, PvuII digested fragments (ME-loxP-aadA-loxPME) treated with transposases, in the genome of 33C by electroporation. Nearly 20,000 Spr colonies, representing over eight-fold coverage of Z. mobilis genome, were collected and preserved at -80°C as the library. To select and enrich for the fast growers in arabinose medium, library cells were revived from frozen stocks (KO1 and KO2, two individual vials serving as replicates) and inoculated in rich medium with arabinose as the sole carbon source (RMA5) at 35°C. Cultures at exponential growth phase were transferred into fresh RMA5 for 17 consecutive transfers. Figure 2 shows representative examples of the growth curves from these transfers. 33C was grown and transferred alongside the library cultures as a control. This was done to understand the rate of adaptation in homogeneous parent culture, as compared to our heterogeneous transposon disruption library.
Figure 2: Serial transfers of 33C KO Library (knockout library) grown in RMA5 (35oC) shake flasks to enrich for faster growers. KO1 and KO2 were cultures revived from two vials of the frozen 33C KO library. 33C was included as a control in this enrichment growth. A, first transfer, B, third transfer, C, fifth transfer, D, ninth transfer, E, twelfth transfer, F, thirteenth transfer, G, fifteenth transfer and H, seventeenth transfer.
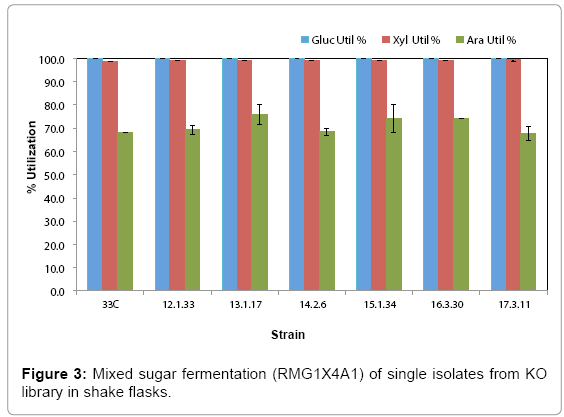
Library cultures showed higher final cell density and faster growth rate than the control starting from 12th transfer, suggesting an improved strain(s) had populated and become dominant in the culture. Further transfers showed stabilized growth patterns between library cultures and control 33C. Single colonies were isolated from the 5th to 17th transfers on RMA5 plates. Representative large colonies from each transfer were evaluated in a pure sugar medium (RMG1X4A1) without pH control for the purpose of down-selection. Based on arabinose utilization, two colonies (13-1-17 and 15-1-34, from 13th and 15th transfers respectively) exhibiting higher arabinose utilization than 33C and the rest of the isolates (Figure 3), were chosen for evaluation in a PCS saccharified slurry. Strain 16-3-30 though relatively high arabinose utilization, was not included in the hydrolyzate evaluation because of the lower ethanol yield (data not shown)
Fermentation performance of select library cultures in corn stover hydrolyzates
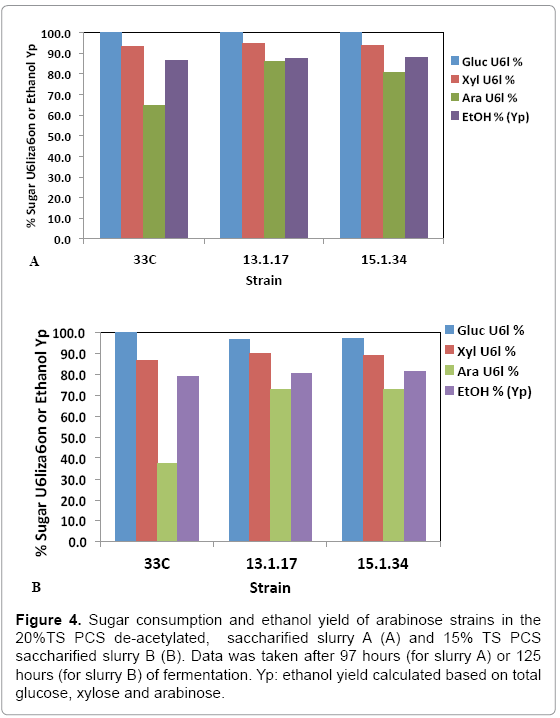
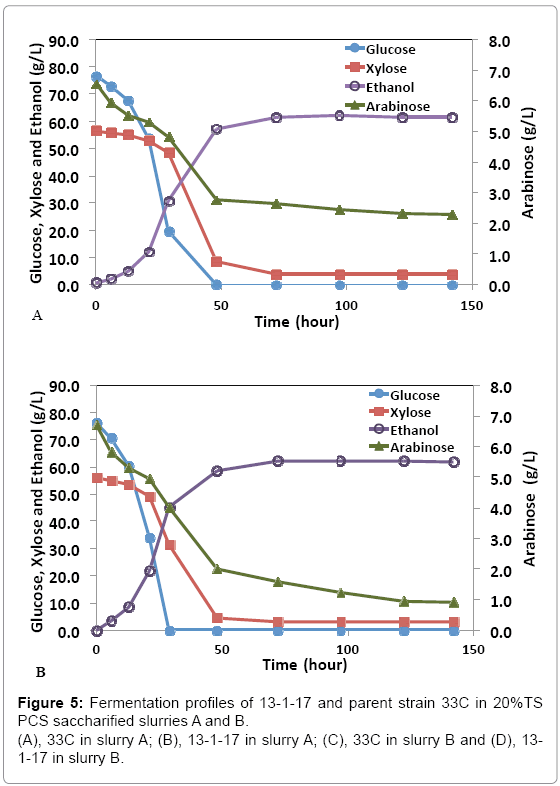
While pure sugar fermentation indicated promising results by 13-1- 17 and 15-1-34, it is crucial that the strains can perform in a less favorable environment (yet closer to the industrial operation application), i.e. hydrolyzates produced through dilute-acid pretreatment, which is known to generate compounds that are inhibitory to microorganisms [15]. Strains 13-1-17 and 15-1-34, demonstrating improvement of arabinose utilization in a pure sugar medium, were evaluated in a pHcontrolled fermentation with PCS saccharified slurries (Slurry A: 20% or Slurry B: 15% total solids, pretreated and enzymatically hydrolyzed). Due to the limited quantity of each slurry, we were not able to incorporate duplicates of each strain using the same slurry. Rather, we used two different slurries as substrates in the two independent fermentation experiments to show the consistent improvement of strains 13-1-17 (and 15-1-34) over the parent strain 33C. Figure 4 summarizes sugar consumption and ethanol yield. Both 13-1-17 and 15-1-34 showed a dramatic increase in arabinose utilization compared with 33C, verifying the superior performance of the arabinose strains. For example, strain 13-1-17 was able to consume 86% and 73% arabinose while the parent strain, 33C only 64% and 37% in slurries A and B, respectively. Fermentation profiles of 13-1-17 in slurries A and B are shown in Figure 5. Co-utilization of sugars is observed though a preference of the sugars in the order of glucose, xylose and arabinose also appears to be present, indicated by the rate of consumption. It is recognized that the ethanol titers were similar between 13-1-17 (and 15-1-34) and 33C despite increased arabinose consumption. We are currently unsure of the metabolic fate of the arabinose, but ultimately for the production of cellulosic ethanol it will be important to drive this flux towards ethanol production. We speculate that perhaps this phenomenon may be a result of the low energetic state of the cultures at the end of the fermentation, in which relatively high concentration of ethanol was already present from the conversion of glucose and xylose which are consumed more rapidly than arabinose (Figure 5). However, future flux experiments will be required to ascertain the fate of the arabinose metabolized during late-fermentation.
Figure 4: Sugar consumption and ethanol yield of arabinose strains in the 20%TS PCS de-acetylated, saccharified slurry A (A) and 15% TS PCS saccharified slurry B (B). Data was taken after 97 hours (for slurry A) or 125 hours (for slurry B) of fermentation. Yp: ethanol yield calculated based on total glucose, xylose and arabinose.
Genetic characterization 13-1-17 and 33C through Next Generation Sequencing
In addition to generating a superior arabinose utilizing strain, it would be very informative if we could understand the causative underlying genetic alterations leading to this phenotypic enhancement. The results would help identify the genes or regions in the genome, which are disrupted by the insertions, giving us insights into the genetic basis for improved growth. To verify the genomic insertion loci of transposon and identify other potential genetic changes besides transposon insertion occurring during arabinose selection/enrichment process, we further employed Next Generation Sequencing (NGS) technology to resequence the entire genomes of top performing arabinose-utilizing strains.
Through NGS-based genome resequencing, we located the integration loci of the transposons in the genomes of 33C and 13-1- 17. Transposon loxP-aadA-loxP-Pgap-araBAD was inserted in a native plasmid of 33C, disrupting a hypothetical protein gene. The second transposon containing only aadA (created during the KO library construction) was inserted within ZMO1650 in the chromosome of 13- 1-17. This latter insertion site was surprising in that we cannot envision a scenario where the disruption of this gene, encoding a beta-lactamase would lead to improved arabinose utilization. However, NGS also showed that additional mutations had occurred in these strains whose role in enhanced arabinose utilization can be rationally understood. Accordingly, we believe that transposon insertion events have little to do with the enhanced phenotype, and rather the SNPs that occurred by chance are primarily responsible for the phenotype of enhanced arabinose utilization in this work.
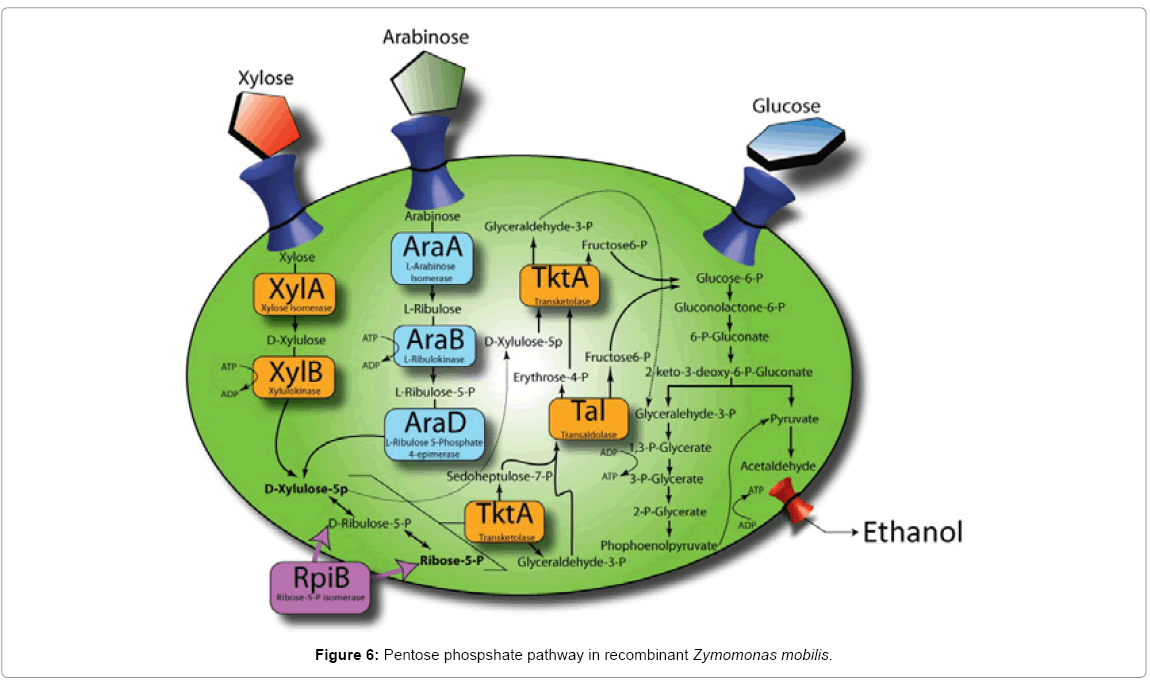
SNPs in strains 33C and 13-1-17 relative to their parents were revealed (Table 2). Specifically, mutations, which we speculated to be relevant to arabinose utilization, were identified within the araB itself in strains 33C and 13-1-17, suggesting the possibility that direct arabinose pathway modulation had occurred. When we compared the sequence in 33C to the published sequence of E. coli, [16] from which the araBAD was used to construct the transposon loxP-aadA-loxP-Pgap-araBAD, we observed that 33C had acquired 3 independent mutations in the araB gene (R127E, S128A, A490G), encoding ribulokinase. Furthermore, in 13-1-17 the A490G mutation was further altered to A490V (a G to V amino acid mutation from 33C to 13-1-17). This strongly implicates altered ribulokinase activity (either through altered protein levels or kinetic properties) as being central to the enhanced phenotype from the initial integrants (which did not grow on arabinose readily), to 33C then to 13-1-17. Additionally, a mutation in the putative Ribosome Binding Site (RBS) of the gene rpiB (ZMO1200, encoding ribose-5-Pisomerase, RpiB) was identified in strain 13-1-17. RpiB promotes the isomerization from D-ribulose-5-P to Ribose-5-P and lies right at the junction of the pentose phosphate pathway (Figure 6).
| 8b | 33C | 13-1-17 | |
|---|---|---|---|
| araBADin native plasmid (ref. 8b) |
no | yes | yes |
| aadAin ZMO1650 (ref. 8b) | no | no | yes |
| araBmutation (ref. E .coli) | no | Yes: R127E,S128A, A490G | Yes: R127E, S128A, A490V |
| ZMO0353 mutation (ref. 8b) | no | no | Yes: A376V |
| ZMO1200 mutation (ref. 8b) | no | no | yes |
ref: referenced genome sequence
ZMO1650: β--lactamase gene
ZMO0353: 4--diphosphocytidyl--2C--methyl--D--erythritol synthase gene
ZMO1200: ribose--5--phosphate isomerase Gene (rpiB)
Table 2: Transposon insertions and mutations identified through Next Generation Sequencing.
One final SNP that was identified in 13-1-17 was within ZMO0353, encoding a 4-diphosphocytidyl-2C-methyl- D-erythritol synthase (A376V), acting as an intermediate step within the methylerythritol phosphate pathway.
Discussion
We set out to utilize numerous techniques to improve arabinose utilization in our current highest performing ethanologen, Z. mobilis strain 8b. In order to generate this strain we attempted strategies including independent and combinatorial overexpression of the individual arabinose pathway genes, Global Transcription Machinery Engineering (GTME) [17], gene overexpression libraries (data not shown), and as described here transposon6 based genomic knockout libraries. The advantage of using transposons for mutagenesis is that a large and diverse collection of mutations can be generated in random locations throughout the genome, rendering greater chances of beneficial effect to the organisms from the mutations. Ironically, while this method yielded the highest performing strain as it relates to arabinose usage, we believe that this phenotype was not derived from the transposon integration event, but rather unrelated spontaneous mutations that occurred within the genome of this strain, or the synergetic effect of transposon integration and spontaneous mutations. However, this is speculative and has not yet been confirmed experimentally. Specifically, we identified mutations affecting three independent proteins via NGS-based genome resequencing. The relationship between the gene mutations and the desired phenotype (e.g. improved arabinose utilization) can then be tested by re-creating the insertion mutation(s) in the parental strain (e.g. 33C) and then examining the phenotype.
The first protein that was found altered in strain 13-1-17 compared to 33C was 4-diphosphocytidyl-2C-methyl-D-erythritol synthase (A376V) encoded by ZMO0353. This protein plays a key step in the methylerythritol phosphate pathway (MEP), but it is difficult to theoretically tie this mutation with the observed phenotype. However, a common metabolite link between the MEP pathway and the pentose phosphate pathway is D-glyeraldehyde-3-phosphate. It is possible that this mutation is related to metabolic flux alterations between these two pathways. However, recapitulation of this mutation, and further analysis, including flux balance analysis, will be required to understand its effect on arabinose usage.
The second subset of these mutations was located within the araB gene encoding ribulokinase that we introduced from E. coli leading to these translational changes: R127E, S128A, and A490G in strain 33C. While 33C is greatly inferior to its derivative strain 13-1-17 for arabinose utilization, it is markedly improved over the very first araBAD transformant/integrant, selected on Sp (Supplementary Figure S1). This integrant was cultured in arabinose containing medium, which likely provided the selective pressure on this strain to isolate the mutations found within 33C. Interestingly, compared to 33C, strain 13- 1-17 has an additional mutation to araB, which again targeted amino acid residue 490 (G490V). It will be interesting to assay these various mutant forms of araB to understand their effect on the ribulokinase activity, and pathway flux as a whole.
The third mutation we identified in 13-1-17 (compared to 33C) was located within the predicted RBS of the ribose 5-P-isomerase (RpiB) encoded by ZMO1200. As this gene functions within the pathway of pentose conversion to ethanol, we reasoned that this gene influenced flux through the pathway. RpiB plays a role in the isomerization of ribulose-5-P to ribose 5-P. We hypothesize that insufficient levels of ribose-5-P represent a bottleneck within this pathway in strain 33C, which was then upregulated by the rpiB mutation in strain 13-1-17. This is supported by work reported in Caimi et al. [18], which suggests that rpiB overexpression leads to improved xylose usage. Given that arabinose usage and xylose usage converge at a branch-point further up in the pathway than where RpiB acts (Figure 6), it is logical that rpiB overexpression would also lead to improved arabinose usage. To ascertain whether this mutation leads to an increased or decreased expression of rpiB, a detailed activity assay to compare strains13-1-17 and its parent, 33C, for this enzyme will be called for.
Acetate has been shown to be inhibitory to the growth and fermentation of Z. mobilis [15]. Our results confirmed this observation in that all strains performed worse in the non de-acetylated hydrolyzate (Figure 4B) compared with in the de-acetylated hydrolyzate (Figure 4A). It is noteworthy that the improvement on arabinose utilization yield was greater for strains 13-1-17 and 15-1-34 compared with 33C when tested in the non de9 acetylated hydrolyzate. Though there is no identified mutation(s) that can explain the acetate toxicity in these strains, we suspect that acetate played a role in the performance difference between the improved and the parental strains. Further investigation in this area (e.g. examining the by-product formation profile) will be important to understand the relationship between arabinose utilization efficiency and acetate toxicity.
Regardless of the genetic mechanisms underlying the phenotypic enhancements of these strains, the functional outcome is that through the methods we employed, we have generated a strain of Z. mobilis that is excellent in its ability to utilize glucose, xylose and arabinose, while simultaneously producing high titers and yields of ethanol. Furthermore, these traits were shown to be relevant when using an industrial relevant feedstock, saccharified slurry of dilute acid pretreated corn stover. The ability of strain 13-1-17 to utilize 100% of the glucose, 95% of the xylose and over 85% of the arabinose in a 20% TS hydrolyzate represents a significant step forward in the realization of economically viable cellulosic ethanol.
Conclusions
A glucose, xylose and arabinose utilizing Zymomonas mobilis strain was constructed by incorporating arabinose catabolic pathway genes, araBAD in a glucose, xylose co-fermenting host, 8b, using a transposition integration approach. Through a second transposition to generate a mutagenic knockout library and the selection/enrichment culturing process, strain 13-1-17 was isolated. Subsequent fermentation evaluation indicated significant improvement in metabolizing arabinose in dilute acid pretreated, saccharified corn stover slurry. Next Generation Sequencing of the genomic DNA from strain 13-1-17 suggested several mutations, which may have impacted the enhanced fermentation performance.
Acknowledgements
This work was funded by Bioenergy Technologies Office of U.S. Department of Energy. The authors would like to thank Ali Mohagheghi, Andrew Lowell, and Robert Nelson for preparing the hydrolyzates, and Philip T. Pienkos for critical reading of this manuscript.
References
- Lawford HG (1988) A new approach to improving the performance of Zymomonas in continuous ethanol fermentations. ApplBiochemBiotechnol 17: 203-219.
- Zhang M, Chou YC, Howe W, Eddy C, Evans K, et al. (2007) Zymomonas pentose-sugarfermenting strains and uses thereof. Patent 7: 223-575.
- Mohagheghi A, Dowe N, Schell D, Chou YC, Eddy C, et al. (2004) Performance of a newly developed integrant of Zymomonasmobilis for ethanol production on corn stoverhydrolysate. BiotechnolLett 26: 321-325.
- Humbird D, Davis R, Tao L, Kinchin C, Hsu D, et al. (2011) Process Design and Economics for Biochemical Conversion of Lignocellulosic Biomass to Ethanol: Dilute-Acid Pretreatment and Enzymatic Hydrolysis of Corn Stover. NREL/TP-5100-47764. Golden, CO: National Renewable Energy Laboratory.
- Deanda K, Zhang M, Eddy C, Picataggio S (1996) Development of an arabinose-fermenting Zymomonasmobilis strain by metabolic pathway engineering. Appl Environ Microbiol 62: 4465-4470.
- Lawford HG, Rousseau JD, Tolan JS (2001) Comparative ethanol productivities of different Zymomonas recombinants fermenting oat hull hydrolyzate. Applied Biochemistry and Biotechnology 91-93: 133-146.
- Yang JJ (2011) Zymomonas with improved arabinose utilization. US20110143408 A1.
- Dubois V, Parizano MP, Arpin C, Coulange L, Bezian MC, et al. (2007) High genetic stability of integrons in clinical isolates of Shigella spp. of worldwide origin. Antimicrob Agents Chemother 51: 1333-1340.
- Hoffman LM, Jendrisak JJ, Meis RJ, Goryshin IY, Reznikof SW (2000) Transposomeinsertional mutagenesis and direct sequencing of microbial genomes. Genetica 108: 19-24.
- Sternberg N, Hamilton D (1981) Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J MolBiol 150: 467-486.
- Yang S, Land ML, Klingeman DM, Pelletier DA, Lu TY, et al. (2010) Paradigm for industrial strain improvement identifies sodium acetate tolerance loci in Zymomonasmobilis and Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 107: 10395-10400.
- Yang S, Pappas KM, Hauser LJ, Land ML, Chen GL, et al. (2009) Improved genome annotation for Zymomonasmobilis. Nat Biotechnol 27: 893-894.
- Agrawal M, Mao Z, Chen RR (2011) Adaptation yields a highly efficient xylose-fermenting Zymomonasmobilis strain. BiotechnolBioeng 108: 777-785.
- Meijnen JP, de Winde JH, Ruijssenaars HJ (2008) Engineering Pseudomonas putida S12 for efficient utilization of D-xylose and L-arabinose. Appl Environ Microbiol 74: 5031-5037.
- Franden MA, Pilath HM, Mohagheghi A, Pienkos PT, Zhang M (2013) Inhibition of growth of Zymomonasmobilis by model compounds found in lignocellulosic hydrolysates. Biotechnol Biofuels 6: 99.
- Lee N, Gielow W, Martin R, Hamilton E, Fowler A (1986) The organization of the araBAD operon of Escherichia coli. Gene 47: 231-244.
- Alper H1, Stephanopoulos G (2007) Global transcription machinery engineering: a new approach for improving cellular phenotype. MetabEng 9: 258-267.
- Caimi PG, McCole L, Tao L, Tomb JF, Viitanen PV (2012) Xylose 1 utilization in recombinant Zymomonas. US 2012/0156746 A1.
Relevant Topics
- Agricultural biotechnology
- Animal biotechnology
- Applied Biotechnology
- Biocatalysis
- Biofabrication
- Biomaterial implants
- Biomaterial-Based Drug Delivery Systems
- Bioprinting of Tissue Constructs
- Biotechnology applications
- Cardiovascular biomaterials
- CRISPR-Cas9 in Biotechnology
- Nano biotechnology
- Smart Biomaterials
- White/industrial biotechnology
Recommended Journals
Article Tools
Article Usage
- Total views: 15797
- [From(publication date):
August-2015 - Mar 31, 2025] - Breakdown by view type
- HTML page views : 11140
- PDF downloads : 4657