Genetic Diversity of Plantain (Musa sp.) collection in Cote d'Ivoire revealed by Single Nucleotide Polymorphism (SNP) markers
Received: 17-Jan-2022 / Manuscript No. acst-22-51773 / Editor assigned: 20-Jan-1970 / PreQC No. acst-22-51773(PQ) / Reviewed: 28-Jan-2022 / QC No. acst-22-51773 / Revised: 04-Feb-2022 / Manuscript No. acst-22-51773(R) / Accepted Date: 07-Feb-2022 / Published Date: 10-Feb-2022 DOI: 10.4172/2329-8863.1000495
Abstract
The Plantain (Musa sp.) is a giant perennial rhizomatous herb native to South East Asia and Western Pacific. Precise identification of plantain germplasms is essential for efficient management, exchange and use of plantain genetic resources. Thirty-two accessions of plantain from the National Agricultural Research Center collection and six diploid cultivars of Musa were analyzed using sixty-three single nucleotide polymorphism (SNP) markers. The genetic diversity observed was relatively high among plantain accessions. The mean values of observed heterozygosity were higher than the expected mean heterozygosity values, which confirm Plantain’s highly heterozygous nature. Analysis of Molecular Variance revealed that the genetic diversity mainly resides within the population (which is based on inflorescence characteristics) with 96 % of the total variation. Both, Bayesian clustering analysis and Principal Coordinate Analysis separated the accessions into two and three clusters, respectively. Two cultivars of false horn were clear separated from the bulk of other plantains, suggesting that they are a potential source of useful or rare genes for widening the genetic base of breeding populations derived from the plantains. This Plantain SNP panel is demonstrated here to be proficient tools to assist plantain germplasm management, propagation of planting material, and plantain cultivar authentication.
Keywords
Plantain; Germplasm; genetic diversity; Single Nucleotide Polymorphism; Accessions
Introduction
In Banana (Musa spp.) is a giant perennial herb belonging to the family Musaceae [1]. The edible bananas originated from intraspecific hybridizations between two wild diploid species, Musa accuminata Colla (origin of A genome) and Musa balbisiana (origin of B genome) [2]. The most economically important cultivated types used by farmers worldwide are triploids (2n = 3x = 33 chromosomes) and divided into three major groups: AAA (Cavendish or dessert bananas), ABB (cooking bananas) and AAB (plantain). Among these groups, the plantain subgroup (AAB) is mostly produced for local consumption and is of considerable importance to the agriculture of tropical humid forest regions in Africa, Central and South America, and Asia [3].
Plantain is a staple food for many people in Africa. It is indigenous to South East Asia but are so predominant in the humid lowlands of west and central Africa that this region is now considered as the secondary centre of diversification [2]. These two regions produce more than 70 % of African production (14.5 million tonnes) [4]. Plantain is very appreciated for organoleptic, cooking and agronomic qualities. It is also a major source of income for many people and actors in the supply chain in the rural and urban sectors [5].
In Côte d’Ivoire, plantain has an important role in both the diet of populations and cropping systems, where it serves as shading for the establishment of perennial crops such as cocoa and coffee. Plantain is the third food crop after yam and cassava, with an annual production estimated at 1.8 million tonnes [4]. Despite, the crucial role of plantain in food security in Côte d’Ivoire, production systems have remained extensive with average yields on farm ranging from 5 to 10 tons per hectare [6]. This low production is due to pest and disease constraints among which nematodes, banana weevils, and foliar diseases such as black leaf streak caused by (Mycosphaerella fijiensis) are part of these constraints [7]. Thus, the selection of the best triploids could be used in hybridization programs to develop improved tetraploid hybrids adapted to our specific dishes for increase the productivity of plantain in Côte d’Ivoire and allow producers to get a better income. In this context, the plantain collection of the National Agricultural Research Center has been enriched by several plantain cultivars collected on farm in banana production areas in Côte d’Ivoire. Such collection constitutes the germplasm collection from which valuable genes can be selected. This strategy requires a better knowledge of genetic diversity to improve the choice of plantain parents. A morphological diversity study was conducted by [8] and shows a high morphological diversity of plantain and allowed to set a gene bank of 42 accessions based the morphological descriptors. However, many phenotypic characteristics are highly influenced by the environment [9]. The development and application of technologies based on molecular markers provide the best tools that are able to reveal polymorphism at the DNA sequence level, which are adequate to detect genetic variability between individuals and within the populations [10].
The utilization of DNA molecular markers for plantain germplasm management has been recently reviewed [11-13]. Using SSR and AFLP markers, [3] studied the genetic diversity of 30 plantains accessions constituting a representative sample of the phenotypic diversity from the CARBAP collection. They found a very narrow genetic base of this cultivars group. Out of 9 loci SSR used, only one polymorphic locus was observed. With the AFLP marker, on 633 bands provided by 8 primer pairs, only one methylation insensitive polymorphism level appears (0.2%) and all plantains are grouped into two distinct clusters. This result supports the hypothesis of a unique origin (i.e., a unique meiotic event) of plantain landraces and total absence of sexuality in the evolution of plantains. To date, little is known about the genetic diversity within the plantain subgroup. The most common hypothesis is that this group is composed of very similar genotype, many of which arose as mutant ‘sports’ [3]. However, important morphological variation is observed amongst plantains landraces, particularly in inflorescence characters, plant size and several other characters which are commonly used for classifying germplasm [14].
Single nucleotide polymorphisms (SNPs) are defined as variations in single nucleotide positions in genome sequences. It is one of the latest technologies developed and is considered an ideal co-dominant marker system for assessing genetic diversity [15, 16]. Advantages of SNP markers include their abundance and wide distribution throughout the genome [17]. Compared to SSR markers, SNP analysis can be done without requiring DNA separation by size and can, therefore, be automated in high-throughput assay formats [18]. The diallelic nature of SNPs offers much lower error rate in allele calling and raises the level of consistency between laboratories. These advantages have resulted in the increasing use of SNPs as the markers of choice for accurate genotype identification and diversity analysis of several crops, such as: Cacao (Theobroma cacao) [15], oil palm (Elaeis guineensis) [19], pineapple (Ananas comosus) [20], Coffee (Coffea sp.) [16]. Nonetheless, this most powerful tool for germplasm management has not been directly applied to plantain germplasm identification in Africa.
In 2010, the National Center for Agricultural Research (CNRA) was nominated as Centre of excellence for the West Africa Agriculture Productivity Program (WAAPP) for plantain. The objective is to select promising varieties for farmers’ in West Africa and optimize the husbandry practices. The program started with a farm survey carried out on harvest season during which plantains accessions were collected. Results are presented here on the level of genetic diversity, evaluated with SNP markers.
Materials and Methods
Plant material
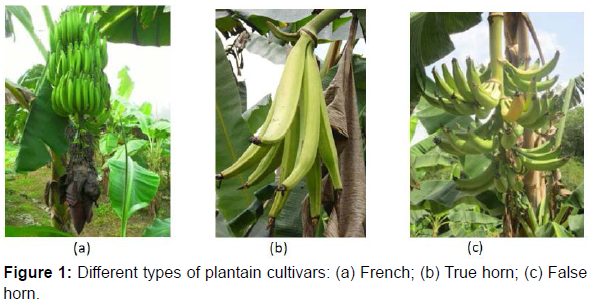
A total of 32 of the 42 plantain landraces maintained at the germplasm collection of the National Agricultural Research Center in Azaguié, Côte d’Ivoire, and six diploid accessions (used as reference) was used in this study (Table 1). The accessions are composed of three inflorescence types. The first one, ‘True Horn’ types is characterized by the absence of male flowers at maturity and the presence of very large horn-like fruits, the second cultivar, ‘French’ Types with persistence of male flowers at maturity and smaller fruit size and the third cultivar, ‘False Horn’ characterized by a restricted number of neutral flowers but without any male bud at maturity (Figure 1).
| Accession name | Bunch types | Source of introduction | Genome type |
|---|---|---|---|
| Agnirin | French | Gagnoa, Côte d'Ivoire | AAB |
| Banablé_(Corne_4) | False horn | Abengourou, Côte d'Ivoire | AAB |
| Eniaba_(N'Zélouka) | French | Abengourou, Côte d'Ivoire | AAB |
| Foua | French | Blouaflé, Côte d'Ivoire | AAB |
| French_clair | French | Akoupé, Côte d'Ivoire | AAB |
| Ghana_Agnirin | French | Abengourou, Côte d'Ivoire | AAB |
| Laknao | French | Côte d'Ivoire | AAB |
| Ngletia_1 | French | Côte d'Ivoire | AAB |
| Ninglinnin_1 | French | Blouaflé, Côte d'Ivoire | AAB |
| Zakoi | French | Côte d'Ivoire | AAB |
| French_sombre | French | Cameroun | AAB |
| Bindi | French | Côte d'Ivoire | AAB |
| Monthan | French | Vietnam | ABB |
| Bandiè_2 | False horn | Côte d'Ivoire | AAB |
| Corne_1 | False horn | Akoupé, Côte d'Ivoire | AAB |
| Corne_4 | False horn | Côte d'Ivoire | AAB |
| Corne_5 | False horn | Côte d'Ivoire | AAB |
| Corne_bout_rond | False horn | Côte d'Ivoire | AAB |
| Corne_tacheté | False horn | Abengourou, Côte d'Ivoire | AAB |
| Ehô | False horn | Côte d'Ivoire | AAB |
| Lorougnon | False horn | Abidjan, Côte d'Ivoire | AAB |
| Lorougnon_2 | False horn | Aboisso, Côte d'Ivoire | AAB |
| Big_Ebanga | False horn | Cameroun | AAB |
| Orishele | False horn | Nigeria | AAB |
| Red_Ebanga | False horn | Nigéria | AAB |
| N'zelou-ka | French | Congo | AAB |
| Ndoria | True horn | Abidjan, Côte d'Ivoire | AAB |
| Oleakotoi | True horn | Akoupé, Côte d'Ivoire | AAB |
| M'bomo | True horn | Congo | AAB |
| Iba | True horn | Nigéria | AAB |
| Kokoida | False horn | Aboisso, Côte d'Ivoire | AAB |
| kpatrè_gnon | True horn | Gagnoa, Côte d'Ivoire | AAB |
| Pisang_Jari_Buaya_A | Acuminata | Solomon Islands | AA |
| KirKinan | Acuminata | AA | |
| Kuspaka_A | Acuminata | AA | |
| Tani_B | Balbisiana | BB | |
| Fenjiao | Balbisiana | BB | |
| Pisang_glutuk_wulung | Balbisiana | BB | |
| Accuminata and Balbisiana are not a bunch types | |||
Table 1: List of plantain germplasms and diploid accessions used in SNP genotyping.
SNP data mining was performed using sequence data of Musa sp genotypes which were deposited in the NCBI Sequence Read Archive (SRA) database. These SRA reads were downloaded from the database and mapped on the Musa sp reference genome [21] using BWA program. The Genome Analysis Toolkit (GATK) package v 3.520 was upstream or 80 bp downstream were eliminated. From the discovered putative SNPs, a subset of 288 putative SNPs was selected for validation test using the nanofluidic array genotyping system (Fluidigm Corporation, South San Francisco, California, United States). The primers of the selected 288 SNPs were designed by Fluidigm and applied on the selected Musa cultivars for validation. Based on the validation result, the top 200 SNPs with high repeatability were used for further analysis of genetic diversity.
DNA isolation and SNP genotyping
Genomic DNA was extracted from fresh leave using the modified Mixed Alkyl Trimethyl Ammonium Bromide (MATAB) method. The DNA extracts obtained were quantified using a Nano drop UV Vis 2000 spectrophotometer (Thermo Scientific, USA). SNP genotyping was performed at USDA-ARS, Sustainable Perennial Crops Lab, Beltsville, MD, USA, using the Fluidigm 96.96 Dynamic Array TM (Fluidigm Corporation, South San Francisco, California, United States). Each 96.96 Dynamic Array can run 96 samples against 96 SNP assays generating a total of 9216 data points in a single experiment. One key feature of this protocol is the inclusion of a specific targeted amplification (STA) reaction [22], which allows the enrichment of template molecules for each individual Integrated Fluidic Circuit® (IFC) reaction that facilitates the multiplexing during genotyping. An advantage to STA is that it allows the use of limited or low-quality DNA samples and reduces bias that may occur when samples are loaded to the 96 sample wells of the IFC. The STA reaction was performed as described in the Fluidigm SNP Genotyping User Guide, PN 68000098 Rev I1 [23]. The STA master mix consisted of 2.5 μL of TaqMan® Taq polymerase (Life Technologies, Carlsbad, CA), PreAmp Master Mix (2X), 1.25 μL of Pooled assay mix (0.2X), and 1.25 μL of genomic DNA for a total reaction volume of 5.0 μL.
PCR was performed with an initial denaturation step of 95°C for 10 min, followed by 14 cycles of a 2-step amplification profile consisting of 15 sec at 95°C and 4 min at 60°C. The resulting amplified DNA was then diluted 1:5 in TE buffer in order to reduce the concentration of any remaining PCR by-products. Samples were then genotyped using the nanofluidic 96.96 Dynamic ArrayTM IFC (Integrated Fluidic Circuit; Fluidigm Corp.). The 96.96 Dynamic Array IFC for SNP genotyping was described [22]. End-point fluorescent images of the 96.96 IFC were acquired on an EP1TM imager (Fluidigm Corp, South San Francisco, CA, USA). The data was recorded with Fluidigm Genotyping Analysis Software.
Genetic structure and diversity parameters
The following parameters were calculated using GenAlEx 6.5 software (Peakall and Smouse, 2012) to analyze the diversity among and within genetic groups of plantain trees: allelic frequency (Ne), observed heterozygosity (Ho), the expected heterozygosity (He), the Shannon diversity index (I) and the fixation index (Fis). Mann-Whitney U test was used to compare observed and expected heterozygosity values. Molecular Analysis of Variance (AMOVA) was performed to determine the level of genetic differentiation between and within populations.
Differences between populations were measured using PhiPT (analogue of binding index Fst), as implemented in the GenAlEx 6.5 program [24]. The significance of AMOVA was tested using a nonparametric permutation approach with 999 permutations. The pairwise genetic distances defined by and calculated using the DISTANCE procedure implemented in GenAlEx 6.5 were used to perform the Principal Coordinate Analysis (PCoA) [24].
Then, the Polymorphism Information Content (PIC) which assesses the discriminatory capacity of a marker in a population was calculated using Power Marker software (version 3.25) [25].
The dissimilarity matrix was used to make unrooted tree using Neighbor-joining (NJ) method using Darwin v 6.5 software [26]. The reliability of the dendrogram was tested by a bootstrap of 1000.
Finally, the Bayesian model-based classification algorithm implemented in STRUCTURE v2.3.4 software [27] was applied to refine the genetic structure of the collection. STRUCTURE analysis was performed with five iterations of K values (assumed number of subpopulations). The value of K varied from 1 to 10, with 100,000 repetitions of Markov Chain Monte Carlo (MCMC) and a break-in period of 50,000, using the mixing model. The optimal number of groups (optimal K) for the set of data was determined using the Evanno method [28] implemented in the STRUCTURE HARVESTER software [29].
Results
SNP genotyping and identification of duplicate genotype Among the 200 SNPs panel chosen to study genetic diversity of plantain, 63 were successful for genotyping. For the 63 markers generated consistent results, four were monomorphic across the 32 plantain samples. A total of 59 polymorphic SNPs were retained for further analysis of this sample set. Individual genotype matching (pairwise comparisons) based on the 59 SNPs identified no duplicates or synonymous. Each plantain accession has unique SNP profiles.
SNP-Based polymorphism and genetic diversity
The summary of genetic diversity statistics is presented in Table 2. The minor allele frequencies of these SNPs ranged from 0.013 (Musa114) to 0.734 (Musa032) with an average of 0.158. The mean information index was 0.447, ranging from 0.054 (Musa114) to 0.794 (Musa199). The observed heterozygosity values ranged from 0.000 (Musa290) to 1.000 (Musa022) with an average of 0.337, whereas the average expected heterozygosity was 0.290 ranging from 0.025 (Musa114) to 0.535 (Musa19). The mean Polymorphic Information Content (PIC) value was 0.375 with a maximum of 0.619 for the Musa199 and a minimum of 0.059 observed with Musa303 locus (Table 2). Thirteen SNP marker obtained a PIC value greater than 0.5.
| SNP ID | Minor allele frequency | Information index | Observation heterozygosity | Expected heterozygosity | Inbreeding coefficient | PIC |
|---|---|---|---|---|---|---|
| Musa022 | 0.5 | 0.693 | 1 | 0.5 | -1 | 0.457 |
| Musa032 | 0.734 | 0.555 | 0.532 | 0.371 | -0.389 | 0.378 |
| Musa037 | 0.459 | 0.688 | 0.918 | 0.495 | -0.856 | 0.454 |
| Musa041 | 0.079 | 0.239 | 0 | 0.138 | 1 | 0.21 |
| Musa049 | 0.472 | 0.688 | 0.944 | 0.495 | -0.905 | 0.488 |
| Musa054 | 0.381 | 0.648 | 0.628 | 0.456 | -0.383 | 0.517 |
| Musa060 | 0.258 | 0.558 | 0.455 | 0.373 | -0.206 | 0.416 |
| Musa106 | 0.183 | 0.475 | 0.367 | 0.298 | -0.225 | 0.465 |
| Musa111 | 0.319 | 0.605 | 0.472 | 0.416 | -0.169 | 0.533 |
| Musa114 | 0.013 | 0.054 | 0.026 | 0.025 | -0.04 | 0.139 |
| Musa130 | 0.068 | 0.238 | 0.137 | 0.124 | -0.075 | 0.234 |
| Musa132 | 0.167 | 0.42 | 0.267 | 0.26 | -0.058 | 0.511 |
| Musa148 | 0.028 | 0.616 | 0.511 | 0.389 | -0.281 | 0.551 |
| Musa152 | 0.03 | 0.102 | 0.061 | 0.055 | -0.1 | 0.281 |
| Musa163 | 0.028 | 0.52 | 0.278 | 0.306 | 0.008 | 0.375 |
| Musa168 | 0.026 | 0.179 | 0.103 | 0.091 | -0.13 | 0.299 |
| Musa170 | 0.411 | 0.668 | 0.822 | 0.475 | -0.722 | 0.53 |
| Musa173 | 0.444 | 0.67 | 0.543 | 0.477 | -0.139 | 0.578 |
| Musa174 | 0.03 | 0.668 | 0.576 | 0.428 | -0.331 | 0.494 |
| Musa178 | 0.015 | 0.704 | 0.795 | 0.476 | -0.662 | 0.502 |
| Musa191 | 0.226 | 0.408 | 0.124 | 0.282 | 0.511 | 0.5 |
| Musa199 | 0.048 | 0.794 | 0.248 | 0.535 | 0.538 | 0.619 |
| Musa208 | 0.233 | 0.512 | 0.4 | 0.337 | -0.181 | 0.478 |
| Musa222 | 0.197 | 0.493 | 0.393 | 0.314 | -0.246 | 0.271 |
| Musa229 | 0.033 | 0.108 | 0.067 | 0.06 | -0.111 | 0.21 |
| Musa236 | 0.014 | 0.205 | 0.083 | 0.115 | 0.273 | 0.194 |
| SNP ID | Minor allele frequency | Information index | Observation heterozygosity | Expected heterozygosity | Inbreeding coefficient | PIC |
| Musa247 | 0.306 | 0.586 | 0.418 | 0.401 | -0.065 | 0.398 |
| Musa254 | 0.03 | 0.483 | 0.343 | 0.281 | -0.194 | 0.366 |
| Musa255 | 0.03 | 0.253 | 0.121 | 0.143 | 0.154 | 0.267 |
| Musa256 | 0.043 | 0.156 | 0.086 | 0.08 | -0.07 | 0.234 |
| Musa257 | 0.03 | 0.492 | 0.338 | 0.284 | -0.184 | 0.401 |
| Musa259 | 0.026 | 0.09 | 0.051 | 0.047 | -0.083 | 0.059 |
| Musa261 | 0.487 | 0.692 | 0.974 | 0.499 | -0.952 | 0.375 |
| Musa263 | 0.384 | 0.622 | 0.545 | 0.433 | -0.26 | 0.511 |
| Musa267 | 0.03 | 0.51 | 0.112 | 0.297 | 0.498 | 0.383 |
| Musa271 | 0.028 | 0.529 | 0.394 | 0.314 | -0.225 | 0.399 |
| Musa275 | 0.178 | 0.428 | 0.028 | 0.268 | 0.873 | 0.24 |
| Musa278 | 0.017 | 0.736 | 0.908 | 0.504 | -0.804 | 0.536 |
| Musa286 | 0.289 | 0.548 | 0.411 | 0.366 | -0.15 | 0.539 |
| Musa287 | 0.026 | 0.199 | 0.118 | 0.107 | -0.097 | 0.144 |
| Musa290 | 0.456 | 0.631 | 0 | 0.441 | 1 | 0.56 |
| Musa293 | 0.015 | 0.062 | 0.03 | 0.029 | -0.048 | 0.231 |
| Musa301 | 0.193 | 0.483 | 0.326 | 0.307 | -0.071 | 0.47 |
| Musa303 | 0.026 | 0.09 | 0.051 | 0.047 | -0.083 | 0.059 |
| Musa304 | 0.113 | 0.308 | 0.115 | 0.181 | 0.154 | 0.279 |
| Musa305 | 0.417 | 0.615 | 0.5 | 0.424 | -0.172 | 0.438 |
| Musa306 | 0.015 | 0.71 | 0.352 | 0.48 | 0.29 | 0.595 |
| Musa310 | 0.028 | 0.752 | 0.822 | 0.507 | -0.622 | 0.522 |
| Musa312 | 0.312 | 0.619 | 0.624 | 0.427 | -0.456 | 0.379 |
| Musa314 | 0.033 | 0.617 | 0.197 | 0.387 | 0.367 | 0.577 |
| Musa317 | 0.03 | 0.428 | 0.209 | 0.233 | 0.027 | 0.348 |
| Musa319 | 0.028 | 0.212 | 0.056 | 0.117 | 0.455 | 0.359 |
| Musa320 | 0.026 | 0.48 | 0.323 | 0.273 | -0.171 | 0.316 |
| Musa321 | 0.056 | 0.15 | 0.111 | 0.093 | -0.2 | 0.256 |
| Musa324 | 0.026 | 0.09 | 0.051 | 0.047 | -0.083 | 0.166 |
| SNP ID | Minor allele frequency | Information index | Observation heterozygosity | Expected heterozygosity | Inbreeding coefficient | PIC |
| Musa327 | 0.027 | 0.112 | 0.053 | 0.051 | -0.042 | 0.116 |
| Musa335 | 0.028 | 0.096 | 0.056 | 0.051 | -0.091 | 0.166 |
| Musa336 | 0.026 | 0.687 | 0.103 | 0.452 | 0.766 | 0.406 |
| Musa340 | 0.154 | 0.425 | 0.308 | 0.258 | -0.184 | 0.371 |
| Mean | 0.158 | 0.447 | 0.337 | 0.29 | -0.095 | 0.375 |
| PIC: Polymorphic Information Content | ||||||
Table 2: Minor allele frequency, information index, heterozygosity, inbreeding coefficient, and PIC of the 59 SNP loci scored on 32 plantain accessions.
AMOVA showed that both the within-population and the between-population variations were highly significant (FST= 0.036; P < 0.05). However, the estimated molecular variance in the set of 32 plantain accessions is low 12.854. Only 4% (0.464) of the total molecular variance was due to difference between the three germplasm groups, whereas 96% (12.390) was due to individual differences within subpopulations (Table 3).
Cluster and Assignment Analysis
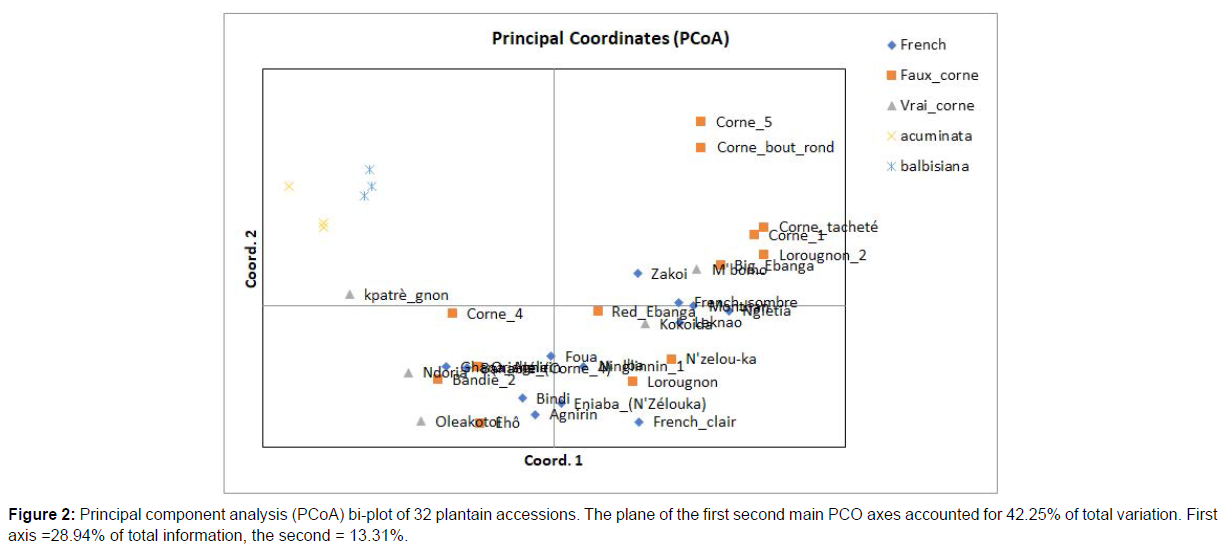
The genetic relationships among the 32 plantain accessions and 6 diploid accessions (3 M. acuminata and 3 M. balbisiana) are presented in the principal coordinates analysis plot (Figure 2). PCoA based on SNP allele frequencies revealed a clear differentiation between plantain genotypes and diploid accessions. The first and second axes explained 28.94% and 13.31% of the total variance, respectively. Three main clusters were identified, cluster 1 only contained two genotypes of the “False Horn” type (Corne_bout_rond and Corne_5), while cluster 2 contained the diploid cultivars and the third cluster includes all other plantain accessions. Cluster 3 further contained small sub-clusters that not grouping based the inflorescence characteristics. Similarly, no geographical patterns of clustering were observed.
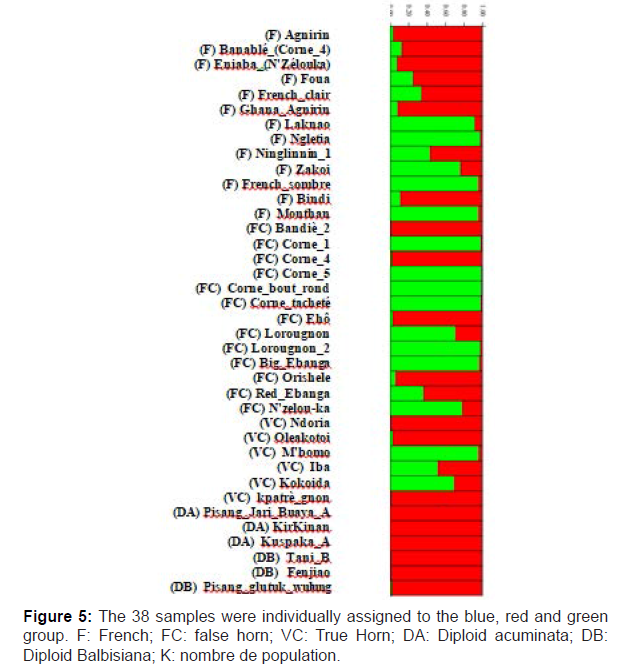
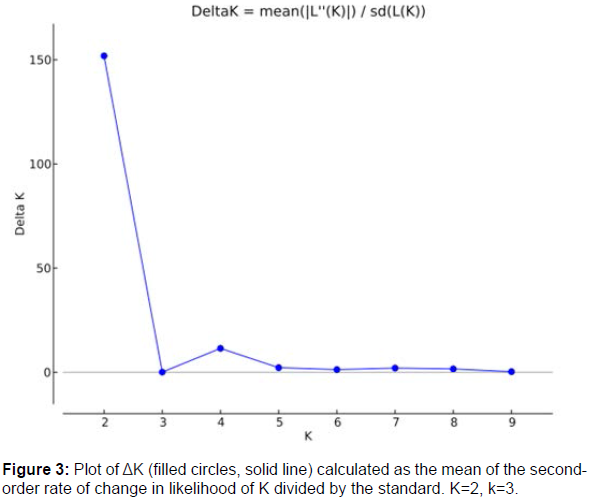
The Bayesian analysis using the software STRUCTURE was applied in the population structure analysis. The highest value of ΔK was found at K = 2, suggesting that the samples consisted of two main genetic groups (Figure 4 and 5). All the diploid germplasms were assigned to one Bayesian cluster, whereas a high rate of admixture observed in the plantain germplasms. “Kpatrè_gnon”, “Ndoria”, “Corne_4, Bandiè_2”, “Agnirin” and “Orishele” accessions presented a highly proportions of the diploid cultivar genomes. “Ninglinnin”, “Red_Ebanga”, “Iba”, “French clair” and “Foua” cultivars appeared as a hybrid genotype between the plantain and diploid bananas groups. To further illuminate the diversity within the plantain germplasm, the clustering result at K = 3 is also presented in Figure 5. The diploid germplasm remained as a single cluster at K = 3, but the plantain cultivars were split into three sub-clusters. The three populations (True Horn, False Horn and French) did not show a strong subpopulation structure. However, two false horn cultivars (Corne_5 and Corne_bout_rond) differed from all other genotypes. This partitioning was fully compatible with the principal coordinate analysis (Figure 3).
Discussion
Assessment of genetic diversity represents essential tools for germplasm management and plant breeding efforts. Although the use of molecular markers to analyse genetic diversity among germplasm of plantains is well described [3, 11-13], very little plantain genetic diversity studies were performed using SNP markers. In this study, we used 63 SNPs for the assessment of genetic identity of 32 plantain landraces maintained at the germplasm collection of the National Agricultural Research Center in Azaguié, Côte d’Ivoire and six diploid cultivars of Musa.
Accurate and precise genotype identification is especially important in vegetative propagated plant species such as plantains. In the present study, it has been shown that a set of 59 SNP markers is highly accurate in genotype identification and genetic structure. Results from multiple cultivars of the same inflorescence types showed 100 % concordance, demonstrating that the nanofluidic system is a reliable platform for generating plantain DNA fingerprints with high accuracy. This finding suggests that these SNP markers are able to identify polymorphism of plantain germplasm, compared to SSR markers which not can make distinguished of 30 plantain landraces from the CARBAP [3]. Thus, these 59 SNPs constitute a cost-effective marker resource suitable for plantain germplasm characterization.
PIC values revealed by SNP markers in this study were 0.375. Compared to the previous studies on plantain, this level of genetic diversity is high [12]. Reported genetic diversity of 25 West African plantains, and obtained PIC values of 0.24 and 0.15 for AFLP and RAPD, respectively. The PIC value can be classified into three classes: slightly informative (PIC < 0.25), reasonably informative (0.5 > PIC > 0.25), and highly informative (PIC > 0.5) [30]. Based on this classification, thirteen SNP markers were classified as highly informative. These results suggest that SNP can be used as an effective type of molecular markers for identification and estimation of genetic diversity in plantain.
Observed heterozygosity (HO) values (0.337) recorded in this analysis was lower than the mean value reported for pineapple (Ananas comosus) 0.465 [20], longan (Dimocarpus longan) 0.406 [18] and maize (Zea mays) 0.43 [31], all revealed by SNP markers. Furthermore, the heterozygosity value of plantains attained from the present work was lower than that reported on the same germplasms using simple sequence repeat (SSR) markers (0.897) [32]. The higher Observed heterozygosity detected by SSR markers is likely due to the multi-allelic nature of the SSRs. This result supports previous studies on maize which showed a higher HO values for SSR (0.801) as compared to SNPs (0.319) [33]. Although varying across the loci, the mean values of observed heterozygosity (Ho=0.337) were higher than the expected mean heterozygosity values (He=0.290), which confirms Plantain’s highly heterozygous nature on the target SNPs. Indeed, plantain (AAB) originated from intraspecific hybridizations between two wild diploid species, Musa accuminata Colla (donor of A genome) and Musa balbisiana (donor of B genome) [2].
Analysis of molecular variance (AMOVA; Table 3) revealed that only 4 % of the variation was allocated among populations and 96% was due to individual differences within populations. The low genetic differentiation exhibited by between populations can be explained by the highly narrow genetic base of plantains. Indeed, many past studies on diversity of plantains shown that the highly narrow genetic base [3, 11-13]. The results of present study were also found comparable to several study in the species to asexual reproduction. AMOVA analysis in Brazilian cassava sweet varieties revealed that genetic diversity (97%) is mainly contained within the populations [34].
| Source | ddl | Sum of squares | Mean squares | Estimate variation | Percentage | Statistics | Value | P value | |
|---|---|---|---|---|---|---|---|---|---|
| Among populations | 2 | 43.63 | 21.815 | 0.464 | 4% | FST | 0.036 | 0.004 | |
| Intra-population | 61 | 755.808 | 12,390 | 12.39 | 96% | ||||
| Total | 63 | 799.438 | 12.854 | 100% | |||||
| FST: Fixation index ddl: Degrees of freedom |
|||||||||
Table 3: Analysis of Molecular Variance (AMOVA) of three populations based on 59 SNP markers.
Similarly, Random Amplified Polymorphic DNA analysis of 43 wild bananas (Musa acuminata) reveals that 81% of the genetic variation is attributable to within-population differences [35]. Sweetpotato (Ipomoea batatas) populations from different agro-climatic zones in China and other countries also revealed 83.53 % of molecular variance occurred within populations [36]. These results suggest that the level of genetic variation in plants are directly associated with breeding system. According to [37] the reproductive biology is an important factor in determining the genetic structure. This information can be applied in selecting a sampling strategy for genetic conservation. The AMOVA results obtained here suggest that future germplasm expeditions should collect more individuals within a limited number of populations. PCoA based on SNP allele frequencies revealed three clusters, whereas the Bayesian analysis separated the 32 plantain germplasms and 6 diploid cultivars in two genomic groups. The SNP markers used in this study were not able to clearly differentiate the plantains into their distinct morph groups based on inflorescence characteristics (Figure 2). This result is in agreement with the earlier plantain studies based on RAPD marker [11, 13], which showed that this marker was unable to cluster the plantain cultivars in a meaningful morphological order. These observations confirm the hypothesis that morphological diversity in the plantains apparently arose from somatic mutations in an hypervariable region of the genome from a very limited number of botanically different clonal sources [38]. However, two genotypes (“Corne_bout_rond” and “Corne_5”) separated from the bulk of other plantains (Figure 2 and 4). This may suggest that these cultivars are a potential source of useful or rare genes for widening the genetic base of breeding populations derived from the plantains. The clustering analysis also did not show any geographical distribution pattern. Similar observations were obtained with the earlier studies of genetic diversity on plantain germplasms [3, 11-13]. This observation could be the result of frequent seed exchanges (rejection) among different regions. Indeed, farmers traditionally use plantain to create the shade necessary for the proper development of young cocoa and coffee trees. The spatio-temporal dynamics of plantain production are related to those of the coffee / cocoa pair. Thus, the communities attached to coffee and cocoa cultivation have contributed to the dissemination of plantain genetic resources across different production areas.
Conclusion
This study showed that SNP markers have proven to be useful tools for assaying genetic polymorphisms, genetic relationships and identification of plantain cultivars of the collection from Côte d’Ivoire. The relatively high level of the genetic diversity within population suggested these accessions can be regarded as potential sources of genetic tank for in vivo conservation and are a good representation of the genetic diversity in these plants. The clear separation of two cultivars of false horn from the other plantains suggests that they are a potential source of useful or rare genes. These plants should be exploited for improvement of the plantains.
Acknowledgement
We wish to thank Dr Dapeng Zhang, Sustainable Perennial Crops Laboratory, Beltsville Agricultural Research Center, USDA/ARS, for her help in molecular analysis of plantain cultivars with the SNP markers.
References
- Pattanaik AK, Sharma K, Anandan S, Blümmel M (2010) Food-feed crops research: a synthesis. Ani Nutri Feed Techno10: 1-10.
- Blümmel M, Duncan AJ, Lenné JM (2020) Recent Advances in Dual Purpose Rice and Wheat Research: a synthesis. Field Crops Res 253: 107823.
- Addis A (2014) Plant Variety Release, Protection and Seed Quality Control Directorate. Field crops Res 223: 1-5
- AOAC (1990) Official Methods of Analysis: Changes in Official Methods of Analysis Made at the Annual Meeting. J Assoc Off Anal Chem 22: 5-6
- Van Soest PJ, Robertson JB (1985) Analysis of forages and fibrous foods. Cornell University.
- NRC (2001) Nutrient requirements of dairy cattle. NRC: 381.
- Menke KH, Steingass H (1988) Estimation of the energetic feed value obtained from chemical analysis and in vitro gas production using rumen fluid. Anim Res Dev 28: 7-55.
- Padmore JM (1990) Protein (crude) in animal feed. Official Methods of Analysis: 70-74.
- Jifar H (2018) Analyses of Phenotypic and Molecular Diversity, Genotype by Environment Interaction and Food-feed Traits in Tef [Eragrostistef (Zucc.) Trotter]. Doctoral dissertation.
- SAS (2002) System Analysis Software. Version 9.0. SAS Institute Inc. Cary, North Carolina.
- Minitab (2007) Minitab Statistical Software, Release 15 for Windows. Pennsylvania State College, Pennsylvania, USA.
- Raivo K (2019) Pheatmap: pretty heatmaps. R package version 1(8).
- Kassambara A, Mundt F (2020) factoextra: extract and visualize the results of multivariate data analyses. R package version 1(5), 337-354.
- Wickham H (2016) ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag .
- Jifar H, Assefa K, Tadele Z (2015) Grain yield variation and association of major traits in brown-seeded genotypes of tef [Eragrostis tef (Zucc.) Trotter]. Agric Food Secur4 (7): 1-9.
- Assefa K, Tefera H, Merker A, Kefyalew T, Hundera F (2001) Quantitative trait diversity in tef [Eragrostis tef (Zucc.) Trotter] germplasm from Central and Northern Ethiopia. Genet. Resour Crop Evol48: 53-61.
- Kassier SB (2002) Comparative responses of fodder and grain teff [Eragrostis tef (zucc.) Trotter] cultivars to spatial, Temporal and nutritional management. RSA.
- DZARC (1989) Debre Zeit Agricultural Research Center Annual Research Report for 1988/89,. Debre Zeit, Ethioia: Debre Zeit Research Center.
- Feyissa F, Kebede G, Assefa G (2015) Dynamics in nutritional qualities of tef and wheat straws as affected by storage method and storage duration in the central highlands of Ethiopia. Afr J Agric Res 10(38): 3718-3725.
- Fekadu D, Walelegn M, Terfe G (2017) Indexing Ethiopian Feed Stuffs Using Relative Feed Value: Dry Forages and Roughages, Energy Supplements, and Protein Supplements. J Biol Agric and Healthcare. 7:57-60.
- Fekadu D, Bediye S, Silesh Z (2010) Characterizing and predicting chemical composition and in vitro digestibility of crop residue using near infrared reflectance spectroscopy (NIRS). Livest Res Rural Dev 22:5- 2
- Bediye S, Sileshi Z (1989) The composition of Ethiopian feedstuffs. Livest Res Rural Dev
- Miller D (2009) Tef Grass: A new alternative. Livest Res Rural Dev 44: 345-346.
- Ketema S (1993) Tef (Eragrostis tef): Breeding, Genetic Resources, Agronomy, Utilization and Role in Ethiopian Agriculture. Addis Ababa: Agri Rese.
- Ashby JA (2009) The impact of participatory plant breeding. Plant breeding and farmer participation, 649-671.
- Bellon MR (1991) The ethno-ecology of maize variety management: a case study from Mexico. Human Ecology 19:389-418.
- Qazi HA, Rao PS, Kashikar A, Suprasanna P, Bhargava S (2014) Alterations in stem sugar content and metabolism in sorghum genotypes subjected to drought stress. Funct Plant Biol 41:954-962.
- Biggs S (2008) The lost 1990s? Personal reflections on a history of participatory technology development. Development in Practice 18:489-505.
- Ceccarelli S, Grando S (2019) From participatory to evolutionary plant breeding. In Farmers and Plant Breeding 231-244.
- Ceccarelli S (2012) Landraces: importance and use in breeding and environmentally friendly agronomic systems. Agrobiodiversity conservation: securing the diversity of crop wild relatives and landraces. CAB International 103-117.
- Ceccarelli S, Grando S, Tutwiler R, Baha J, Martini AM, et al. (2000) A methodological study on participatory barley breeding I. Selection phase. Euphytica 111:91-104.
- Ceccarelli S, Guimarães EP, Weltzien E (2009) Plant breeding and farmer participation. Food and Agriculture Organization of the United Nations, Rome, Italy.
- Chiffoleau Y, Desclaux D (2006) Participatory plant breeding: the best way to breed for sustainable agriculture? International journal of agricultural sustainability 4:119-130.
- Cleveland DA, Daniela S, Smith SE (2000) A biological framework for understanding farmers’ plant breeding. Economic Botany 54:377-394.
- Acquaah G (2012) Principles of plant genetics and breeding. Wiley-Blackwell, Oxford.
- Aly RSH (2013) Relationship between combining ability of grain yield and yield components for some newly yellow maize inbred lines via line x tester analysis. Alex J Agric Res 58: 115-124.
- Comstock RE, Robinson HF (1952) Estimation of average dominance of genes. Heterosis 2:494-516.
- Griffing B (1956) Concept of general and specific combining ability in relation to diallel crossing systems. Aust J Biol Sci 9: 463-493.
Indexed at, Cross ref, Google Scholar
Indexed at, Cross ref, Google Scholar
Indexed at, Google Scholar , Cross ref
Google Scholar, Crossref , Indexed at
Google Scholar , Cross Ref, Indexed at
Google Scholar, Crossref , Indexed at
Google Scholar, Cross Ref, Indexed at
Google Scholar, Cross Ref, Indexed at
Citation: Cyrille KKG, N’da désiré P, Siaka T, Laetitia KD, Olivier AGJ, et al. (2022) Genetic Diversity of Plantain (Musa sp.) collection in Côte d’Ivoire revealed by Single Nucleotide Polymorphism (SNP) markers. Adv Crop Sci Tech 10: 495. DOI: 10.4172/2329-8863.1000495
Copyright: © 2022 Cyrille KKG, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 4445
- [From(publication date): 0-2022 - Dec 06, 2025]
- Breakdown by view type
- HTML page views: 3747
- PDF downloads: 698