Genetic Diversity and Population Structure of Tef [Eragrostis tef (Zucc.) Trotter] as Revealed by SSR Markers
Received: 21-Jan-2020 / Accepted Date: 05-Feb-2020 / Published Date: 12-Feb-2020 DOI: 10.4172/2329-8863.1000438
Abstract
Tef [Eragrostis tef (Zucc.) Trotter] is an indigenous cereal crop widely cultivated and utilized in Ethiopia. Its grain provides healthy and nutritious human diet while the straw is used as livestock feed. This study was designed to analyze the genetic diversity and population structure of 189 tef genotypes including improved varieties, breeding lines and pure lines derived from germplasm collections using 10 SSR primer pairs. All studied primer pairs were polymorphic and generated a total of 168 alleles with the number of alleles; polymorphic information content and gene diversity per locus ranging from 2 to 26, 0.30 to 0.92 and 0.38 to 0.90 respectively. North Shewa and West Shewa populations had the highest gene diversity unlike breeding lines which had lowest values of all genetic parameters. Analysis of molecular variance revealed 55%, 42% and 3% of the total variation due to variation within individual, among individuals and among populations, respectively. Cluster analysis grouped both populations and individual genotypes into five major clusters. Structure bar-plot also inferred five gene pools, but with high level of admixtures. This study generally revealed substantial variations among the studied genotypes to be used in future tef improvement.
Keywords: Eragrostis tef ; Genetic diversity; SSR markers; Population structure
Introduction
Tef [Eragrostis tef (Zucc.) Trotter] is an indigenous staple cereal crop for about 70 million people in Ethiopia. It is a crop adapting to a wide range of climatic and soil conditions. The grain of tef is excellent source of human food while its straw is used as livestock feed [1,2]. Tef is therefore a crop well-integrated into the socio-economic and cultural values of Ethiopian people and is the most preferred cereal by both its growers and consumers. Besides, its potential use as global and popular life-style diet is also increasing due to its gluten free and healthy nature [3]. However, the productivity of tef is very low compared to other cereal crops due to wider uses of low yielding cultivars, drought and other stresses as well as lodging or falling of the stalk by heavy rain and wind [4].
Ethiopia has rich diversity of tef germplasm resources due to its diverse geographic, climatic and soil conditions. Such diversity in indigenous germplasm resources is a major source of genetic variability for the improvement of tef crop. Ethiopian Biodiversity Institute (EBI) has so far collected and deposited in its gene bank over 5,000 accessions from various tef growing regions of the country to conserve the existing variabilities in our germplasm resources [5].
Various markers systems including phenotypic or morphological as well as molecular markers have been used to assess the extent of genetic diversity in tef germplasm accessions. Earlier tef germplasm characterization based on morphological markers showed existence of huge diversity in terms of morphology and other phenotypic traits [4,6-9]. Molecular markers, on the other hand, are the most efficient tools compared to phenotypic markers in assessing genetic diversity and relationships, classifying germplasm resources. Hence, these markers are very effective to construct genetic linkage maps, employ marker-assisted selection and link phenotypic and genotypic variations [10,11]. Thus, restriction fragment length polymorphism (RFLP), amplified fragment length polymorphism (AFLP), random amplified polymorphic DNA (RAPD) and QTL mapping based on enhanced SSR markers have been so far employed in tef [12-17].
Simple sequence repeats (SSRs) or microsatellites have been used to identify polymorphism among diverse germplasm and are the most recently utilized marker unlike those reported previously in tef. However, studies conducted in tef using SSR markers had some limitations in terms of using representative germplasm resources from various sources. For instance, study conducted by Abraha et al. [18] utilized tef germplasm collected from northern part of Ethiopia alone while Desta et al. [19] focused on germplasm related to acid soil problem alone. Similarly, study by Fikre et al. [20] had also dealt with improved varieties and limited number of landraces. This study was, therefore, performed to assess the extent and patterns of genetic diversity and population structure among tef genotypes from 14 diverse populations using SSR markers.
Materials and Methods
Plant materials
A total of 189 tef genotypes representing 14 populations including 144 pure lines, 35 released varieties and 10 breeding lines were studied (Table 1). Thus, the 144 pure lines were derived from germplasm collection of 12 administrative zones in North and Central parts of Ethiopia by the Ethiopian Biodiversity Institute (EBI) between 1979 and 2011. For this study, 12 pure lines were sampled from each of the 12 Administrative Zones of Tigray, Amhara and Oromia Regional States. Districts or Woredas in Ethiopia from where tef accessions were collected originally are shown in Figure 1. The 35 released varieties, on the other hand, were obtained from seven agricultural research centers in Ethiopia while, eight of the breeding lines were derived from crosses of Kinde × Kaye Murri and Quncho × Kinde. The other two breeding lines were the parents of those crosses: Kaye Murri and Kinde. Kaye Murri is a cultivar having very white seed and compact and purple panicle while Kinde is a mutant line with shorter plant height and better lodging tolerance. Quncho, on the other hand, is a popular released tef variety having wider market demand due to its extra white seed color. The seeds of all breeding lines and their parents were obtained from Debre Zeit Agricultural Research Center (DZARC).
| Population | Number | Name | Altitude (m a. s. l) |
|---|---|---|---|
| Central Tigray | 12 (1-12) | Acc. nos. 19132-2, 19132-3, 19166-1,19253-1,19253-2, 234407-1, 234407-2, 237184-1, 237205-2, 243513-1, 243513-3 & 243520-2 | 1350-2640 |
| East Tigray | 12 (13-24) | Acc. nos. 15297-1, 15297-2,15299-1,15299-3,19201-1,19201-3, 19202-2, 234460-1, 234460-2, 234460-3, 242540-1 and 242540-2 | 1979-2632 |
| West Tigray | 12 (25-36) | Acc. nos. 9419-1, 9444-2, 9444-3, 19241-1, 19241-2, 234435-2, 237236-3, 237236-4, 237239-3, 243526-2, 243526-4 and 243526-5 | 1260-2054 |
| East Gojam | 12 (37-48) | Acc. nos. 9545-1, 9556-1, 19516-1, 19516-3, 55221-1, 55221-2, 212698-2, 229768-1, 229768-3, 229768-4, 55046-2 and 55046-3 | 1470-2650 |
| East Gojam | 12 (49-60) | Acc. nos. 19394-1, 19443-3, 19452-4, 19506-2, 19506-4, 242140-3, 242144-1, 242144-3, 242155-1, 242155-3, 55029-2 and 55029-3 | 1890-2735 |
| North Gonder | 12 (61-72) | Acc.9448-1, 9448-2, 9451-2, 9469-2, 9472-2, 9472-4,19343-2, 242186-3, 242186-4, 243540-1, 243540-3 and 243540-4 | 1840-2208 |
| South Gonder | 12 (73-84) | Acc. nos. 19341-2, 19341-3, 19367-2, 19374-1, 55293-2, 212717-0, 212720-1, 225919-2, 225919-3, 225919-4, 225919-7 and 242160-1 | 1804-2950 |
| North Wello | 12 (85-96) | Acc. nos. 55104-3, 215196-1, 215200-1, 215200-2, 215200-3, 234356-4, 234985-2, 234993-1, 234993-3, 237148-1, 237148-5 and 243501-2 | 1520-2950 |
| South Wello | 12 (97-108) | Acc. nos. 212607-2, 212612-3, 212614-1, 212614-2, 225898-1, 242214-1, 242214-2, 243491-2, 2433497-2, 243504-1, 243504-2 and 243504-3 | 1550-3090 |
| East Shewa | 12 (109-120) | Acc. nos. 15361-3, 17335-1, 18460-0, 18466-2, 18466-3, 236963-1, 236963-2, 236965-1,236965-3,236967-1, 236967-2 and 236972-0 | 1657-2303 |
| North Shewa | 12 (121-132) | Acc. nos. 9559-1, 9559-2, 15309-2, 15309-3, 15322-1, 15322-2, 18385-2, 212482-1, 236745-2, 236746-0, 236748-2 and 236957-1 | 1260-2670 |
| West Shewa | 12 (133-144) | Acc. nos. 17365-1, 17371-3, 18410-1, 18410-2, 18410-3,18414-2, 18414-4, 18423-3, 236757-2, 236760-1, 236760-4 and 236760-6 | 1640-2674 |
| Improved varieties | 35 (145-179) | Enatite (DZARC), Asgori (DZARC), Magna (DZARC), Wellenkomi (DZARC), Menagesha (DZARC), Melko (DZARC), Tsedey (DZARC), Gibe (DZARC), Ziquala (DZARC), Dukem (DZARC), Holetta Key (HARC), Ambo Toke (HARC), Gerado (DZARC), Koye (DZARC), Key Tena (DZARC), Gola (SARC), Ajora (ArARC), Genete (SARC), Zobel (SARC), Dima (AARC), Yilmana (AARC), Dega Tef (DZARC), Gimbichu (DZARC), Amarach (DZARC), Quncho (DZARC), Gudurru (BARC), Gemechis (MARC), Mechare (SARC), Kena (BARC), Etsub (AARC), Laketch (SARC), Simada (DZARC), Boset (DZARC), Kora (DZARC) and Were-Kiyu (SARC) | - |
| Breeding lines | 10 (180-189) | Kaye Murri (cultivar, parent), Kinde (Mutant line, parent), Quncho × Kinde (RIL-85), Quncho × Kinde (RIL-91), Quncho × Kinde (RIL-96), Kinde × Kaye Murri (RIL-11), Kinde × Kaye Murri (RIL-302), Kinde × Kaye Murri (RIL-44), Kinde × Kaye Murri (RIL-69) and Kinde × Kaye Murri (RIL-81) | - |
Note: DZARC, HARC, SARC, ArARC, AARC, BARC and MARC refer to the Debre Zeit, Holetta, Sirinka, Areka, Adet, Bako and Melkassa Agricultural Research Centers, respectively.
Table 1: Tef populations used in the study.
DNA extraction and PCR amplification
Leaf tissues were harvested from 3-4 weeks old seedlings of 189 tef genotypes shown in a 40 mm diameter pot using sterile soil in a long day growth room at the Institute of Plant Sciences, University of Bern, Switzerland. The harvested leaf tissues were pressed onto FTA cards (Sigma, Germany) until the card was soaked with a leaf extract. Samples were sent to Sci-Corp (the former INCOTEC) laboratory, South Africa for DNA extraction and SSR study. The DNA was isolated using Whatman FTA Card extraction protocol following the manufacturer protocol. Ten polymorphic SSR markers were selected for PCR amplification based on previous study report [21] (Table 2). The PCR amplicons were fluorescently labeled and separated via capillary electrophoresis on an ABI 3130 automatic sequencer (Applied Bio systems, Pietermaritzburg, 3201, South Africa).
| No | Primer | Forward primer | Reverse primer (5’…3’) | Repeat type and size | Expected size (bp) | Source |
|---|---|---|---|---|---|---|
| 1 | CNLTs 538 | CCATCTTAGCTTTGGCGAGA | ACAAGAGGCAACAAGCCAGA | AG18AGA20 | 176 | (Zeid, Belay, Mulkey, Poland, and Sorrells, 2011) |
| 2 | CNLTs 42 | ATGCATGGATGGATGGCTA | TTACCCAATTGCCCTAGCTG | TC27 | 179 | |
| 3 | CNLTs 136 | TGAGAAGGTAATAACTGGTGAAGC | CAAGGTTTACACACCGTGACTT | CT18 | 246 | |
| 4 | CNLTs 150 | AACACGTCCTTGCCGTATTC | CGGGGTAGCCATAGCCTAAT | AG5&AG16 | 225 | |
| 5 | CNLTs 157 | GGATCCGACATGACGTGTAGT | CACAGAATGAGATTGGGGAGA | CT18 | 168 | |
| 6 | CNLTs 315 | ATAGCTGCTCCGTTTTGCAT | GGTCCACTTGGCATTCTGTT | AG8 TAG6 | 233 | |
| 7 | CNLTs 416 | AACAGATACAGTTGGAGACAGAAATG | CTCTGAGTGCGTCGCAAG | AG19 | 151 | |
| 8 | CNLTs 438 | CTAACCGGCGGCGAGAGA | CTGCCACATGCGTCGTTAGA | GA14 | 153 | |
| 9 | CNLTs 484 | GAGATCCTACCACGGCGATA | CGCTTTCCCCTCCTTTTGTA | GA18 | 157 | |
| 10 | SSR3.3 | GGGAAGAGGAGTGTACAGA | CCCTGGCAACTGCTTTAAGA | (AAG)6 | 226 | (Cannarozzi et al., 2014) |
Table 2: Description of SSR markers used in the current study.
Data analysis
Data from all entries were summarized and converted into input files suitable for various analyses using convert software version 1.31 [22]. Analysis of molecular variance (AMOVA), determination of the number of alleles (Na), observed heterozygosity (Ho) and gene diversity (UHe) were determined using GenAlEx software version 6.5 [23,24]. Power Marker software v.3.25 was also employed to compute polymorphic information content, major allele frequency and Nei’s unbiased genetic distance [25]. Cluster analysis was carried out based on population and individual genotypes using Pop tree2 [26] and DARwin software version 6.0.13 respectively [27]. The dendrogram for population was viewed and edited in tree view while that of individual genotypes was edited for visualization using Fig-Tree software version 1.4.3 [28].
Bayesian model-based clustering method was employed using STRUCTURE software version 2.3.4 assuming population admixture through inferred ancestry to determine the population structure [29,30]. Twenty independent simulations were performed for each number of assumed sub populations (K) ranging from 1 to 14. This was made to determine the most likely number of populations (K) by using a burn-in period of 100,000 and a Markov Chain Monte Carlo (MCMC) run length of 150,000. Zipped results from structure software was submitted to STRUCTUR HARVESTER online software version 0.6.92 to determine the appropriate number of subpopulations (delta K) that explains the structure of the studied genotypes as suggested by Earl and Vonholdt, Evanno et al. [31,32]. Besides, the same zipped results from structure software and names of 14 populations written on notepad were submitted to CLUMPAK (Cluster Markov Packager Across K) beta version to identify an optimal alignment of inferred clusters across different values of K [33].
Results and Discussion
SSR markers and their level of polymorphism
All the 10 studied loci were polymorphic and generated a total of 168 alleles with a mean of 16.8 alleles per locus (Table 3). The observed number of alleles and Shannon diversity index (I) per locus ranged from 2 and 0.55 for SSR 3.3 to 26 and 2.15 for CNLT 538, respectively. The observed total number of alleles in this study is higher than the 148 alleles reported by Desta et al. (2015) using 16 SSR markers while it is closer to the 164 alleles reported by Abraha et al. (2016) using 10 SSR markers. Such variations in number of alleles could be due to differences in the employed number and types of genotypes and markers [34-36]. On the other hand, the range of alleles observed per locus in this study is closer to the earlier reports of 2 to 27 alleles and 8 to 23 alleles while it is far larger than the 5-7 alleles per locus [18-21].
| Locus | MAF | Na | I | Ho | He | uHe | Nm | PIC |
|---|---|---|---|---|---|---|---|---|
| CNLT 538 | 0.15 | 24 | 1.66 | 0.33 | 0.74 | 0.79 | 3.03 | 0.91 |
| CNLT 42 | 0.15 | 22 | 2.15 | 0.61 | 0.86 | 0.9 | 3.12 | 0.92 |
| CNLT 150 | 0.24 | 14 | 1.73 | 0.82 | 0.79 | 0.82 | 2.03 | 0.83 |
| CNLT 157 | 0.35 | 10 | 1.43 | 0.01 | 0.71 | 0.74 | 3.88 | 0.77 |
| SSR 3.3 | 0.76 | 2 | 0.54 | 0.47 | 0.36 | 0.38 | 1.57 | 0.3 |
| CNLT 136 | 0.27 | 17 | 1.56 | 0.05 | 0.74 | 0.78 | 8.74 | 0.85 |
| CNLT 315 | 0.52 | 12 | 1.23 | 0.95 | 0.64 | 0.67 | 3.04 | 0.62 |
| CNLT 416 | 0.24 | 25 | 1.85 | 0.61 | 0.8 | 0.84 | 1.79 | 0.86 |
| CNLT 438 | 0.15 | 18 | 1.83 | 0.26 | 0.8 | 0.85 | 4.09 | 0.9 |
| CNLT 484 | 0.21 | 24 | 2.02 | 0.97 | 0.83 | 0.87 | 1.54 | 0.87 |
| Mean | 0.28 | 16.8 | 1.6 | 0.5 | 0.73 | 0.76 | 3.28 | 0.78 |
Note: MAF: Major Allele Frequency; Na: Observed number of alleles per locus; I: Shannon’s Information Index; Ho: observed heterozygosity; uHe: Un-biased heterozygosity (gene diversity); Nm: Gene flow; PIC: polymorphic information content.
Table 3: Informativeness and values of different diversity indices for 10 loci.
The polymorphic information content (PIC) is generally used to measure the informativeness of a genetic marker. In this study, the PIC values ranged from 0.30 for SSR 3.3 to 0.90 for CNLT 42 with a mean of 0.76 (Table 3). This is a bit different from the previous findings of 0.05 to 0.86, 0.02 to 0.95 and 0.64 to 0.94 being reported based on 39, 16 and 10 SSR markers, respectively [18,19,21]. About 90% of the markers we have employed here were selected from Zeid et al. (2012). As a result, the PIC values in this study are closer to the earlier investigation by Zeid et al. (2012). The fact that about 75% of our studied loci had PIC values greater than the mean (0.78) reveals excellent discriminatory power of the studied markers.
Marker CNLT 42 exhibited the largest gene diversity (0.97), Shannon diversity index (2.15) and lowest major allele frequency (MAF=0.15) while he highest MAF (0.75) and the smallest Shannon diversity (0.52) and PIC (0.30) values were recorded for SSR 3.3. Besides, CNLT 42, CNLT 484, CNLT416 and CNLT150 had also the highest gene diversity values. In general, markers having the largest number of alleles had the lowest MAF unlike those with lowest number of alleles. The lowest PIC and gene diversity values of SSR 3.3 suggest that the sequence for its development is highly conserved in the studied tef genotypes. In this study, CNLT-42 had the highest values for most studied parameters and this is in line with earlier studies by Desta et al. Abraha et al. and Zeid et al. [18,19,21]. The positive and significant correlation observed between allele number and gene diversity (r=0.85, p=0.01) and that of number of alleles and PIC (r=0.87, p=0.01) reveals that loci with higher number of alleles have high PIC and gene diversity. Such kind of loci is therefore useful in assessing genetic diversity in crop germplasm including tef.
Patterns of tef genetic diversity
The results of all studied population genetic diversity indices are summarized in Table 4 showing that improved varieties had the highest number of different alleles (8.40) compared to breeding lines (4.0). This shows that larger populations are expected to have wider genetic diversity compared to smaller and newly establish ones [37]. North Shewa (1.77) followed by North Gonder (1.76) and West Shewa (1.74) had the highest Shannon’ s information index while, North Shewa (0.82) followed by West Shewa (0.81) and East Gojam (0.81) had the highest gene diversity (Table 4). These administrative zones which had high gene diversity and Shannon information index could be considered as genetic diversity hot spots and potential in situ conservation areas for tef crop. Hence, they are useful for future germplasm collections and conservation. In this study, most genotypes from germplasm accessions had high gene diversity due to more chances of coevolving in nature than those under artificial selection. The breeding lines, however, had the lowest values of all genetic parameters may due to artificial selection towards homogenous populations which drastically reduces the diversity. In the present study, analysis of Percentage Polymorphic Loci (PPL) has showed that the entire loci were polymorphic in the 14 studied populations.
| Population | Na | I | Ho | He | uHe | F | PPL |
|---|---|---|---|---|---|---|---|
| C. Tigray | 7.1 | 1.66 | 0.47 | 0.74 | 0.78 | 0.33 | 100 |
| E. Tigray | 6.7 | 1.59 | 0.52 | 0.73 | 0.76 | 0.26 | 100 |
| W. Tigray | 5.6 | 1.43 | 0.46 | 0.69 | 0.72 | 0.31 | 100 |
| E. Gojam | 7.1 | 1.7 | 0.52 | 0.77 | 0.81 | 0.25 | 100 |
| W. Gojam | 7.6 | 1.68 | 0.46 | 0.74 | 0.77 | 0.34 | 100 |
| N. Gonder | 7.9 | 1.76 | 0.47 | 0.76 | 0.79 | 0.34 | 100 |
| S. Gonder | 7.1 | 1.63 | 0.38 | 0.73 | 0.77 | 0.43 | 100 |
| N. Wello | 5.7 | 1.44 | 0.48 | 0.69 | 0.74 | 0.27 | 100 |
| S. Wello | 7.3 | 1.69 | 0.52 | 0.76 | 0.79 | 0.27 | 100 |
| E. Shewa | 6.6 | 1.52 | 0.48 | 0.71 | 0.74 | 0.3 | 100 |
| N. Shewa | 7.5 | 1.77 | 0.53 | 0.78 | 0.82 | 0.27 | 100 |
| W. Shewa | 7.2 | 1.74 | 0.53 | 0.77 | 0.81 | 0.26 | 100 |
| Improved Varieties | 8.4 | 1.68 | 0.46 | 0.73 | 0.75 | 0.33 | 100 |
| Breeding Lines | 4 | 1.12 | 0.5 | 0.6 | 0.64 | 0.24 | 100 |
| Mean | 6.84 | 1.6 | 0.48 | 0.73 | 0.76 | 0.3 | 100 |
Note: Na: No. of different Alleles; I: Shannon's Information Index; Ho: Observed Heterozygosity; He: expected Heterozygosity; uH: Unbiased Expected Heterozygosity; F: fixation index; PPL: Percentage of polymorphic loci.
Table 4: Summary of population diversity parameters for 10 SSR loci.
Population genetic differentiation
Analysis of Molecular Variance (AMOVA) showed existence of highly significant (p<0.01) variations among populations, genotypes within populations and within genotypes from various sources (Table 5). Thus, the largest genetic variation (55%) was attributed to variation with in individuals followed by the variation among individuals with in populations (42%) and variation among populations (3%). Such low level of genetic differentiation among our studied tef populations is in line with the previous reports [18,21] and contrary to the high genetic differentiation among genotypes within population [19]. The overall Fstatistics also exhibited significant genetic differentiation among populations (FST=0.033), among individuals within population (FIS=0.436) and within individuals (FIT=0.455) (Table 5). This estimated FST value is considered to be low to moderate may be due to high variability resulting from gene flow among and within individuals (Table 4) [38]. The observed little differentiation among tef populations could be due to extensive migration and exchange of genes through hybridization during varietal development [18,39].
| Source | df | SS | MS | Variance estimated | % | F Statistic | p values |
|---|---|---|---|---|---|---|---|
| Among populations | 13 | 121.22 | 9.33 | 0.14 | 3 | FST=0.033 | 0.001 |
| Among individual | 175 | 994.71 | 5.68 | 1.73 | 42 | FIS=0.436 | 0.001 |
| Within individual | 189 | 422 | 2.23 | 2.23 | 55 | FIT=0.455 | 0.001 |
| Total | 377 | 1537.93 | - | 4.1 | 100 | - | - |
Table 5: AMOVA showing variation among populations, among individuals within populations and within individual tef genotypes from different sources.
Genetic distance between populations
The pair-wise population Nei’s unbiased genetic distance ranged from 0.16 between populations of West Gojam and improved varieties to 0.52 between populations of North Wello and the breeding lines [39] (Table 6). The next highest genetic distance was observed between populations of breeding lines (BrL) and that of East Shewa (0.48), East Tigray (0.48) and East Gojam (0.47) respectively. The mean genetic distance of each population from the other population, on the other hand, ranged from 0.26 (improved varieties) to 0.41 (breeding lines) showing that the breeding lines are the most distantly related population to the other populations. The pair-wise genetic differentiation (FST values) ranged from 0.03 for West Tigray vs West Shewa to 0.11 between breeding lines and that of East Gojam, East Shewa and West Gojam populations (Table 6). In pair-wise population FST, the values observed between breeding lines and that of the remaining populations other than improved varieties is very large. Such close relationship between breeding lines and improved varieties could be due to utilization of genetically related parents for their development.
Cluster analysis and population genetic structures
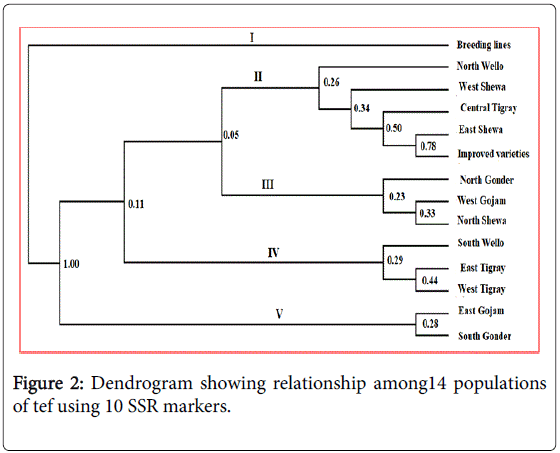
Cluster analysis based on 14 studied populations resulted in the formation of five distinct groups (Figure 2). Cluster-I was solitary and consisted of the population of breeding lines alone while cluster-II consisted of five populations including those from North Wello, West Shewa, Central Tigray, East Shewa and improved varieties. The third cluster consisted of populations from North Gonder, West Gojam and North Shewa while the fourth cluster included populations from West Tigray, East Tigray and South Wello. The fifth and the last cluster consisted of populations from South Gonder and East Gojam. The patterns of grouping of populations across all clusters clearly showed existence of high gene flow along geographical borders due to proximity and exchange of germplasm resources. Thus, improved varieties were clustered with accessions from East Shewa, North Shewa, West Shewa and North Wello may be due to physical appearance of the releasing centers in these zones to facilitate extension and diffusion of improved varieties into their surroundings.
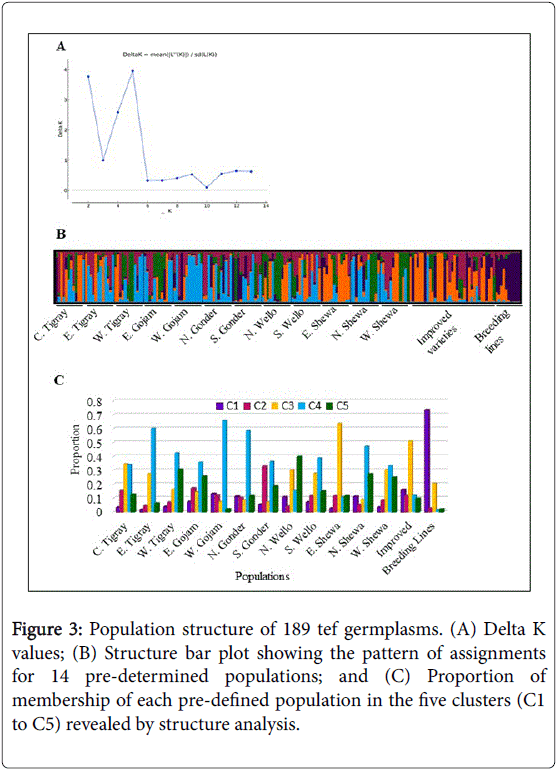
The Bayesian approach-based assignment of 189 individual tef genotypes to different populations and determination of their population structure using STRUCTURE outputs predicted K=5 to be the most likely number of clusters (Figure 3). In such clustering, structure bar plot revealed five gene pools with wide genetic admixtures and the proportion of membership of 14 pre-defined population in each of the five cluster is also summarized (Figure 3). The breeding lines (73.3%) and improved varieties (16.0%) constituted the first cluster with little admixture from other populations. On the other hand, populations from South Gonder (32.7%), East Gojam (17.1%), Central Tigray (15.5%) and West Gojam (12.1%) constituted the second cluster. The third cluster was composed of mostly populations from East Shewa (63.7%), improved varieties (50.9%), Central Tigray (34.6%), West Shewa (30.4%) and North Shewa (30.1%). Similarly, the fourth cluster consisted of populations from West Gojam (65.6%), East Tigray (60.3%), North Gonder (58.5%), North Shewa (47.2%) and West Tigray (42.5%). The fifth cluster consisted of a significant level of admixture of populations from North Wello (39.6%), West Tigray (30.5%), North Shewa (27.2%) and West Shewa (25.4%). In general, this analysis of population structure revealed weak sub-division among the genotypes from the 14 predetermined populations grouped into five gene pools with potential admixtures. Hence, it is only the breeding lines which tended to form a distinct group with over 70% representation while, the remaining 13 populations showed high level of genetic admixtures with the various gene pools.
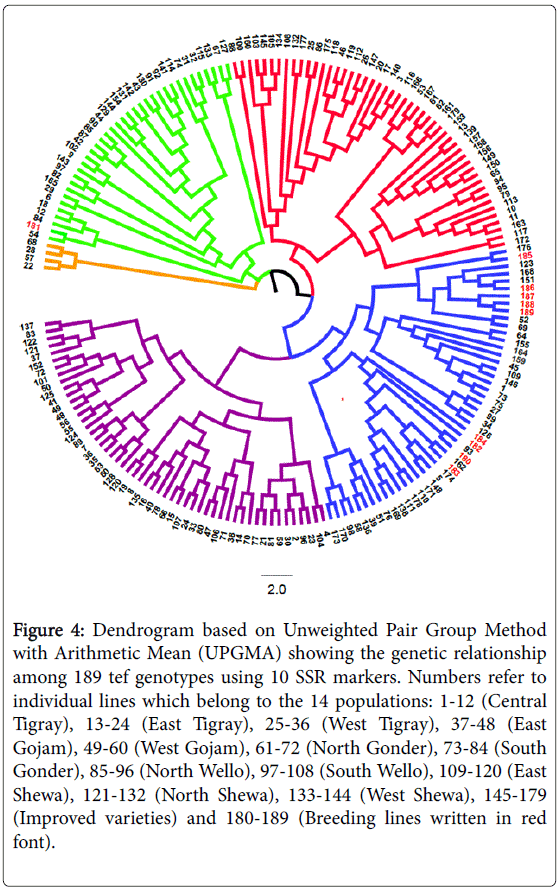
The UPGMA clustering grouped the 189 individual tef genotypes into five groups consisting of four to 52 genotypes from various populations (Figure 4). Cluster-I consisted of four genotypes from four different populations while Cluster-II consisted of 36 genotypes including one breeding line, two improved varieties and 33 genotypes from all administrative zones of collections. Cluster-III consisted of 49 genotypes composed of 17 improved varieties and 32 genotypes from all zones of collections. Cluster-IV consisted of 48 genotypes including 15 improved varieties, nine breeding lines and 24 genotypes from 10 administrative zones of collections. Furthermore, Cluster-V consisted of 52 genotypes composed of one improved variety and 51 genotypes from all zones of collections. The observed number of clusters for individual genotypes is larger than the previously reported three clusters [18,19]. In this clustering of individual genotypes, the 144 tef pure lines derived from germplasm collections were grouped into all the five clusters with various proportion. The 35 improved varieties, however, were distributed throughout four of the five clusters (Cluster- II, III, IV and V). Surprisingly, all breeding lines other than the mutant parental line (Kinde) were grouped under two sub clusters of cluster- IV. Thus, the first sub cluster of this cluster consisted of five breeding lines (genotypes listed from 185 to 189 in Table 1) while the second sub cluster consisted of four breeding lines (genotypes number 180, 182, 183 and 184). This indicates that the SSR markers we have employed are very powerful to show the genetic relationship among the studied tef genotypes.
Figure 4: Dendrogram based on Unweighted Pair Group Method with Arithmetic Mean (UPGMA) showing the genetic relationship among 189 tef genotypes using 10 SSR markers. Numbers refer to individual lines which belong to the 14 populations: 1-12 (Central Tigray), 13-24 (East Tigray), 25-36 (West Tigray), 37-48 (East Gojam), 49-60 (West Gojam), 61-72 (North Gonder), 73-84 (South Gonder), 85-96 (North Wello), 97-108 (South Wello), 109-120 (East Shewa), 121-132 (North Shewa), 133-144 (West Shewa), 145-179 (Improved varieties) and 180-189 (Breeding lines written in red font).
Conclusion
In the present study, both cluster analysis and structure bar plot revealed five groups of genotypes. However, the latter analysis showed existence of high level of admixtures among the various gene pools. In such groupings, the improved varieties were clustered with the pure lines derived from Central Ethiopia while distantly related to those from northern and northwestern Ethiopia. The SSR markers identified to have high PIC values are useful in future characterization and conservation of tef germplasm. North Shewa, West Shewa, East Gojam and North Gonder which had high values of gene diversity and Shannon diversity index could be important hot spot for in-situ conservation of tef germplasm. Though the present study revealed sufficient level of variations among the studied genotypes, using a greater number of genotypes and markers is essential to capture enough diversity in the tef germplasm resources.
Acknowledgements
This research was financially and technically supported by Syngenta Foundation for Sustainable Agriculture and University of Bern through its collaboration with Ethiopian Institute of Agricultural Research. We would also like to thank Ethiopian Biodiversity Institute for germplasm provision and Addis Ababa University for its various supports. Furthermore, Regula Blösch and Annett Weichert should receive special thanks for their unreserved guidance and assistance during the entire research work at the University of Bern.
Conflicts of Interest
Authors declare no conflicts of interest.
References
- Ketema S (1997) Tef, Eragrostis tef (Zucc.) Trotter. In, Promoting the conservation and use of underutilized and neglected crops. Int Pla Gen Res Inst 12: 50.
- Bediye S, Fekadu D (2001) Potential of tef straw as livestock feed. Narrowing the Rift: Tef Research and Development, pp: 245-254.
- Spaenij-Dekking L, Kooy-Winkelaar Y, Koning F (2005) The Ethiopian cereal tef in celiac disease. New Eng J med 353: 1748-1749.
- Assefa K, Cannarozzi G, Girma D, Kamies R, Chanyalew S, et al. (2015) Genetic diversity in tef [Eragrostis tef (Zucc.) Trotter]. Front Plant Sci 6: 117.
- Tesema A (2013) Genetic diversity of tef in Ethiopia, in Achievements and Prospects of Tef Improvement. pp: 15-20.
- Chanyalew S, Assefa K, Metaferia G (2013) Phenotypic and molecular diversity in tef. Achievements and Prospects of Tef Improvement. pp: 21-31.
- Jifar H, Assefa K, Tadele Z (2015) Grain yield variation and association of major traits in brown seeded genotypes of tef [Eragrostis tef (Zucc.) Trotter]. Agri Food Sec 4: 7.
- Kefyalew T, Tefera H, Assefa K, Ayele, M (2000) Phenotypic diversity for qualitative and phenologic characters in germplasm collections of tef (Eragrostis tef). Gen Res Crop Evol 47: 73-80.
- Plaza-Wüthrich S, Cannarozzi G, Tadele Z (2013) Genetic and phenotypic diversity in selected genotypes of tef [Eragrostis tef (Zucc.)] Trotter. Afric J Agric Res 8: 1041-1049.
- Agarwal M, Shrivastava N, Padh H (2008) Advances in molecular marker techniques and their applications in plant sciences. Plan cell repo 27: 617-631.
- Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Tren biotech 23: 48-55.
- Zhang D, Ayele M, Tefera H, Nguyen HT (2001) RFLP linkage map of the Ethiopian cereal tef [Eragrostis tef (Zucc) Trotter]. Theor Appli Gen 102: 957-964.
- Ayele M, Nguyen HT (2000) Evaluation of amplified fragment length polymorphism markers in tef, Eragrostis tef (Zucc.) Trotter, and related species. Plan Breed 119: 403-409.
- Ayele M, Tefera H, Assefa K, Nguyen HT (1999). Genetic characterization of two Eragrostis species using AFLP and morphological traits. Hereditas 130: 33-40.
- Bai G, Tefera H, Ayele M, Nguyen HT (1999) A genetic linkage map of tef [Eragrostis tef (Zucc.) Trotter] based on amplified fragment length polymorphism. Theor Appli Gen 99: 599-604.
- Bai GH, Ayele M, Tefera H, Nguyen HT (2000) Genetic diversity in tef [Eragrostis tef (Zucc) Trotter] and its relatives as revealed by Random Amplified Polymorphic DNAs. Euphytica 112: 15-22.
- Zeid M, Belay G, Mulkey S, Polland J, Sorrells ME (2011) QTL mapping for yield and lodging resistance in an enhanced SSR-based map for tef. Theor Appli Gene 122: 77-93.
- Abraha MT, Shimelis H, Laing M, Assefa K, Amelework B (2016) Assessment of the genetic relationship of tef (Eragrostis tef) genotypes using SSR markers. South Afric J Botany 105: 106-110.
- Desta EA (2015) Pre-breeding of Tef [Eragrostis tef (Zucc.) Trotter] for Tolerance to Aluminum Toxicity.
- Fikre T, Tesfaye K, Assefa K (2018) Genetic diversity of Ethiopian tef [(Eragrostis tef (Zucc.)Trotter] released and selected farmers’ varieties along with two wild relatives as revealed by microsatellite markers. J Crop Sci Biotech 21: 367-374.
- Zeid M, Assefa K, Haddis A, Chanyalew S, Sorrells ME (2012) Genetic diversity in tef(Eragrostis tef) germplasm using SSR markers. Field Crop Res 127: 64-70.
- Glaubitz JC (2004) CONVERT: A user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol Eco Not 4: 309-310.
- Peakall R, Smouse PE (2006) GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol Eco Not 6: 288-295.
- Peakall R, Smouse PE (2012) GENALEX 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28: 2537-2539.
- Liu K, Muse SV (2005) PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 21: 2128-2129.
- Takezaki N, Nei M, Tamura K (2010) POPTREE2: Software for constructing population trees from allele frequency data and computing other population statistics with Windows interface. Mol Biol Evol 27: 747-752.
- Rambut A (2016) FigTreeV1.4.3. A graphical viewer of phylogenetic trees and as a program for producing publication-ready figures.
- Pritchard JK, StephensM, Donnelly P (2000) Inference of population structure using multilocus genotype data. Gene 155: 945-959.
- Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol Eco Notes 7: 574-578.
- Earl DA, Vonholdt BM (2012) structure harvester: A website and program for visualizing structure output and implementing the Evanno method. Conserv Gen Res 4: 359-361.
- Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Eco 14: 2611-2620.
- Â Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol Eco Res 15: 1179-1191.
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol Eco Res 15: 1179-1191.
- He Q, Li XW, Liang GL, Ji K, Guo QG et al. (2011) Genetic Diversity and Identity of Chinese Loquat Cultivars/Accessions (Eriobotrya japonica) Using Apple SSR Markers. Pla Mol Biol Repo 29: 197-208.
- Baraket G, Chatti K, Saddoud O, Abdelkarim AB, Mars M, et al. (2011) Comparative Assessment of SSR and AFLP Markers for Evaluation of Genetic Diversity and Conservation of Fig, Ficus carica L., Genetic Resources in Tunisia. Pla Mol Biol Repo 29: 171-184.
- Sharma SS, Negi MS, Sinha P, Kumar K, Tripathi SB (2011) Assessment of genetic diversity of biodiesel species pongamia pinnata Accessions using AFLP and three Endonuclease AFLP. Pla Mol Biol Repo 29: 12-18.
- Rampersaud SN, Perez-Brito D, Torres-Calzada C, Tapia-Tussell R, Carrington VF (2013) Genetic structure and demographic history of Colletotrichum gloeosporioides sensu lato and C. truncatum isolates from Trinidad and Mexico. Bmc Evol Biol 13: 130.
- Holsinger KE, Weir BS (2009) Genetics in geographically structured populations: Defining, estimating and interpreting F(ST). Natu rev Gen 10: 639-650.
Citation: Jifar H, Tesfaye K, Dagne K, Assefa K, Tadele Z (2020) Genetic Diversity and Population Structure of Tef [Eragrostis tef (Zucc.) Trotter] as Revealed by SSR Markers. Adv Crop Sci Tech 8: 438. DOI: 10.4172/2329-8863.1000438
Copyright: © 2020 Jifar H, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Select your language of interest to view the total content in your interested language
Share This Article
Recommended Journals
Open Access Journals
Article Tools
Article Usage
- Total views: 4256
- [From(publication date): 0-2020 - Dec 13, 2025]
- Breakdown by view type
- HTML page views: 3306
- PDF downloads: 950